Abstract
Transforming growth factor-beta (TGF-β), a pluripotent cytokine expressed in the colon, has a crucial but paradoxical role in colorectal cancer (CRC). TGF-β is a potent proliferation inhibitor of normal colon epithelial cells and acts as a tumor suppressor. However, TGF-β also promotes invasion and metastasis during late-stage CRC, thereby acting as an oncogene. Thus, understanding the factors behind the paradoxical roles of TGF-β and elucidating the mechanisms by which TGF-β-induced proliferation inhibition is impaired in CRC are necessary. Here, we found that the N-Myc tumor suppressor gene downstream-regulated gene NDRG2 (N-Myc downstream-regulated gene 2), which is a TGF-β-responsive gene, abrogated TGF-β-induced epithelial–mesenchymal transition (EMT) and further inhibited the invasion and migration of CRC cells. TGF-β positively induced NDRG2 expression through direct transactivation mediated by Sp1 and by abrogation of the repressive c-Myc/Miz-1 complex on NDRG2 promoter in normal epithelial cells. Aberrant hypermethylation of NDRG2, which could respond to TGF-β growth inhibition signaling, abrogated the inhibitory effect of NDRG2 in TGF-β-induced EMT in CRCs. Reduced NDRG2 expression was highly correlated with the invasion stage and metastasis of CRC. Our study establishes that NDRG2 is a new tumor suppressor gene that responds to TGF-β anti-proliferative signaling and tips the balance of oncogenic TGF-β during late-stage CRC.
Keywords: colorectal cancer, TGF-β, NDRG2, EMT, methylation
Introduction
Colorectal cancer (CRC) is a commonly diagnosed cancer in men and women, and it is the third most common fatal malignancy and second leading cause of cancer-related deaths in the western world.1 The etiology of CRC is multifactorial, and it is widely accepted that genetic factors, such as Ras/Raf, transforming growth factor (TGF)-β, β-catenin/T-cell factor and the p53 pathway, have key roles in the predisposition to cancer development and progression.2 Although genetic changes have been identified at multiple levels throughout the multistage carcinogenesis process, the initial factors corresponding to these changes need to be further elucidated.
TGF-β is a secreted multifunctional cytokine that regulates a variety of cellular processes. The TGF-β receptor complex activates the Smad signaling pathway, which includes Smad2, Smad3 and Smad4, and non-Smad signaling cascades (for example, PI3K/AKT, p38MAPK, MAPK-ERK and JNK) to produce the full spectrum of TGF-β responses.3, 4, 5 The role of TGF-β signaling in carcinogenesis is complicated. Originally named for its transforming activity in in vitro assays, TGF-β now unequivocally demonstrates both tumor suppressor and oncogenic activities. The tumor suppressor activities dominate in normal tissue and mainly occur through the direct regulation of cell-cycle inhibitors, such as p21Cip1 and p15INK4B,6, 7 and cell-cycle activator c-Myc via transcriptional and post-transcriptional mechanisms.8 However, during tumorigenesis, changes in TGF-β expression and cellular responses tip the balance in favor of oncogenic activities by inducing the epithelial–mesenchymal transition (EMT), which is mediated by Fibronectin, Twist, Snail and so on, and finally accelerating tumor invasion and metastasis.9, 10, 11 There is considerable genetic evidence that the loss of sensitivity to growth inhibition by TGF-β is an important event in colorectal carcinogenesis. Much of the evidence is derived from studies in human CRCs demonstrating inactivating mutations in genes encoding proteins involved in TGF-β signal transduction, including SMAD4,12 SMAD2,13 and TGFBR2.14 However, it has also been reported that restoration of an impaired TGF-β pathway cannot restore the anti-proliferative response to TGF-β in CRC cells.15, 16 Therefore, to fully understand the paradoxical effect of TGF-β in carcinogenesis, other factors and mechanisms need to be uncovered and elucidated.
In recent years, a new tumor suppressor gene, NDRG2 (N-Myc downstream-regulated gene 2), belonging to the NDRG family that consists of four identified members (NDRG1–4), has been implicated in the regulation of cell differentiation and proliferation. The role of NDRG2 in cancer was supported by the initial observation that NDRG2 inhibited the proliferation of glioblastoma cells17 and promoted the differentiation of various cell types, including dendritic cells,18 PC12 cells19 and CRC cells.20 Consistent with the potential function of NDRG2 as a tumor suppressor, downregulation of NDRG2 expression was found in multiple human cancer cell lines and tumors, particularly colorectal carcinomas and high-risk adenoma precursor lesions.21 Most notably, our recent data showed that a decrease in NDRG2 expression in primary human CRC is a powerful and independent predictor for poor prognosis of CRC, particularly those with advanced tumor node metastasis (TNM) stage and lymph-node metastasis.22 Thus, the relevance of NDRG2 to the paradoxical roles of TGF-β in CRC should be investigated.
In addition, accumulating evidence indicates that in addition to genetic changes, epigenetic alterations have a major role in the initiation and progression of CRC.23, 24 More importantly, tumor suppressor genes inactivated by mutations and promoter CpG island methylation are more frequently affected by hypermethylation than mutations in CRC.25 At the single-cell level, mutation/copy number changes and hypermethylation are most frequently mutually exclusive.25 In addition, at the genome level, there is an inverse relationship between frequent epigenetic changes, the CpG island methylator phenotype, large-scale genomic gains or losses and chromosomal instability,26, 27 which has been characterized in ∼85% of CRCs with increased chromosomal losses and heterozygosity.28, 29, 30
Our study demonstrates for the first time that NDRG2 is a TGF-β responsive gene that abrogates TGF-β-induced EMT in CRC cells. TGF-β upregulated NDRG2 via transcriptional activation mediated by the transcription factor Sp1 and by abrogation of the repressive c-Myc/Miz-1 complex in epithelial cells. Furthermore, aberrant NDRG2 hypermethylation, which could respond to TGF-β growth inhibition signaling, abrogated the inhibitory effect of NDRG2 in TGF-β-induced EMT in CRCs. Most importantly, the reduced NDRG2 expression was highly correlated with the invasion stage and metastasis of CRC. Together, our data show that NDRG2, as a TGF-β responsive tumor suppressor gene, tips the balance of the TGF-β role toward oncogenesis in late-stage CRCs.
Results
Positive correlation between NDRG2 expression and the Smad2/3 phosphorylation level in CRC samples
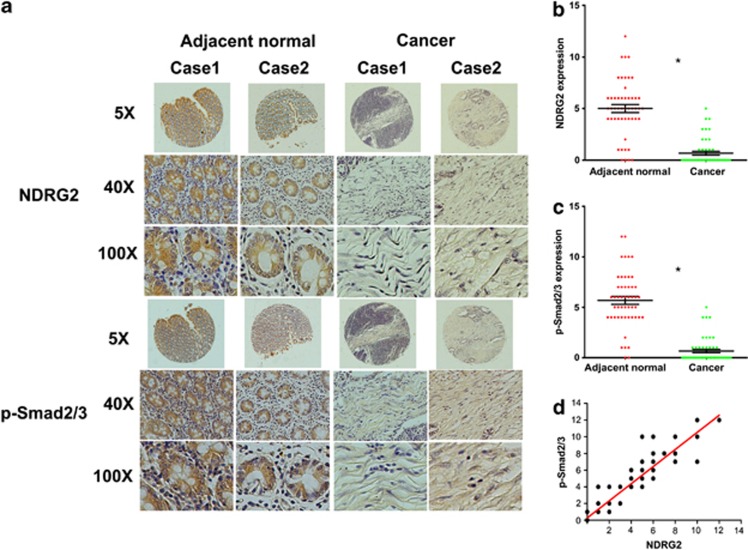
Our group recently demonstrated that NDRG2 is an important tumor suppressor that correlates with metastasis-free and overall survival in CRC patients.22 Because accumulated data suggest that the phosphorylation of Smad2/3 at its C-terminal region correlates with TGF-β-induced cell-cycle arrest and growth inhibition,31, 32 we analyzed phosphorylated Smad2/3 levels to evaluate the tumor suppressive activity of TGF-β. Immunohistochemical (IHC) analysis of 50 pairs of human colorectal tumors and adjacent normal colon samples revealed that NDRG2 and phosphorylated Smad2/3 were highly expressed in the adjacent normal colon tissues compared with the colorectal tumor samples and were inversely correlated with tumor stage (Figure 1a). Moreover, phosphorylated Smad2/3 was predominantly located in nuclei, suggesting a role of Smad complex in transcriptional regulation. Indeed, in normal versus cancer tissue, p-Smad2/3 levels were 5.11 versus 0.65 (Figure 1b), respectively, and NDRG2 levels were 5.79 versus 0.65 (Figure 1c), respectively. Additionally, even less staining was observed in the more aggressive CRCs (Tables 1 and 2). Furthermore, a correlation study determined that NDRG2 expression was positively correlated with phosphorylated Smad2/3 expression in these tissue samples (P<0.001, ANOVA test) (Figure 1d), which suggests that NDRG2 might be involved in TGF-β downstream signaling.
Figure 1.
Positive correlation between NDRG2 and p-Smad2/3 in colorectal cancer tissues. (a) IHC staining of NDRG2 and p-Smad2/3 in human adjacent normal and cancerous colon samples from patients at different stages. Case 1 is from a stage III patient, and Case 2 is from a stage II patient. Original magnification: × 5, × 40 or × 100. (b, c) Relative expression level of p-Smad2/3 and NDRG2 in human tissue samples. Student's t-test was applied for statistical analyses. *P<0.05, which was considered significantly different. (d) Correlation between NDRG2 and p-Smad2/3 expression with linear regression and Pearson's correlation significance (P<0.001, ANOVA test).
Table 1. Statistical results of the immunohistology.
| n |
C-terminal p-Smad2/3 |
P-value | ||
|---|---|---|---|---|
| |
|
− |
+ to +++ |
|
| Total | 50 | 34 | 16 | |
| Sex | 0.609a | |||
| Male | 26 | 18 | 8 | |
| Female | 24 | 16 | 8 | |
| Age at diagnosis, years | 0.805a | |||
| <60 | 25 | 16 | 9 | |
| >60 | 25 | 18 | 7 | |
| Differentiation status | <0.001b | |||
| Poor | 9 | 7 | 2 | |
| Moderate | 22 | 17 | 5 | |
| Well | 19 | 10 | 9 | |
| TNM stage | <0.001b | |||
| I–II | 8 | 1 | 7 | |
| III | 16 | 12 | 4 | |
| IV | 26 | 21 | 5 | |
Abbreviation: TNM, tumor node metastasis.
P-value when expression levels were compared using Fisher's exact test.
P-value when expression levels were compared using Kruskal Wallis.
Table 2. Statistical results of immunohistology.
| n |
NDRG2 |
P-value | ||
|---|---|---|---|---|
| |
|
− |
+ to +++ |
|
| Total | 50 | 38 | 12 | |
| Sex | 0.147a | |||
| Male | 26 | 20 | 6 | |
| Female | 24 | 16 | 8 | |
| Age at diagnosis, years | 0.173a | |||
| <60 | 25 | 18 | 7 | |
| >60 | 25 | 18 | 7 | |
| Differentiation status | <0.001b | |||
| Poor | 9 | 8 | 1 | |
| Moderate | 22 | 18 | 4 | |
| Well | 19 | 10 | 9 | |
| TNM stage | <0.001b | |||
| I–II | 8 | 1 | 7 | |
| III | 16 | 11 | 5 | |
| IV | 26 | 24 | 2 | |
Abbreviations: NDRG2, N-Myc downstream-regulated gene 2; TNM, tumor node metastasis.
P-value when expression levels were compared using Fisher's exact test.
P-value when expression levels were compared using Kruskal Wallis.
NDRG2 is a new direct responsive target of TGF-β
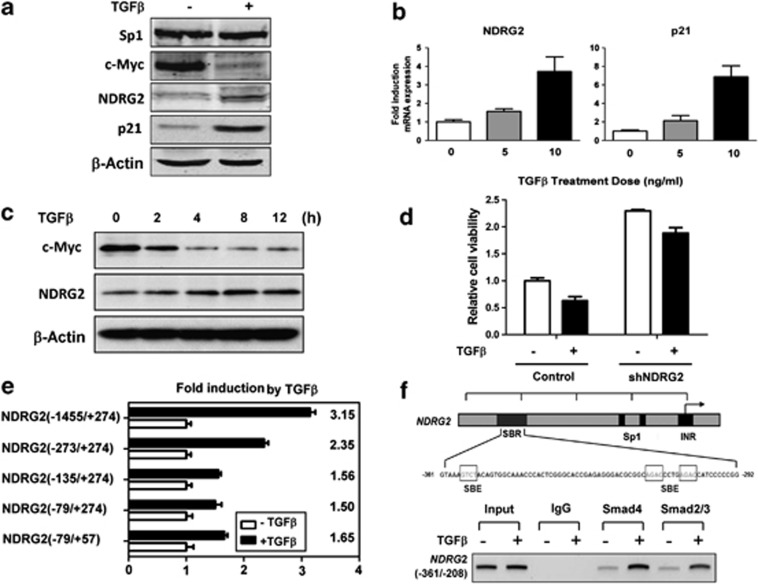
To further determine the regulation of NDRG2 by TGF-β, we first confirmed the inhibition of cellular proliferation and cell-cycle arrest due to TGF-β in the human keratinocyte cell line HaCaT, which has an intact TGF-β-Smad signaling pathway (Supplementary Figures S1A and B). Intriguingly, the NDRG2 protein and messenger RNA (mRNA) expression levels were increased in the presence of TGF-β (Figures 2a and b), which indicates the transcriptional regulation of NDRG2 by TGF-β. Since it has been shown that c-Myc has a key role in NDRG2 inhibition20 and TGF-β-induced cell-cycle arrest,33 we observed a decrease in the expression of c-Myc and increased NDRG2 expression in the presence of TGF-β (Figures 2a and c). In addition, TGF-β-induced growth inhibition was plotted against NDRG2 and the control shRNA (Figure 2d). A key step in defining direct transcriptional regulation is determining the core region within the NDRG2 promoter responsible for TGF-β induction. Thus, a series of previously created processive NDRG2 promoter truncation constructs20 were used in the present study. The presence of TGF-β increased the full-length NDRG2 promoter construct (−1455/+274) activity >3-fold, and even a 1.6-fold induction was observed when using a minimal core promoter construct (−79/+57) (Figure 2e). Finally, a chromatin immunoprecipitation (ChIP) assay was performed to validate the direct binding of the TGF-β downstream targets Smad2/3 and Smad4 with the NDRG2 promoter, which was enhanced by TGF-β (Figure 2f). In contrast, mutation of the Smad binding site showed little effect on NDRG2 promoter activity (data not shown), indicating that other transcription factors might be involved in TGF-β-mediated NDRG2 induction. Thus, the NDRG2 tumor suppressor gene was determined to be a new downstream target gene in the TGF-β pathway in epithelial cells that responds to TGF-β growth inhibitory effects.
Figure 2.
TGF-β directly transactivates NDRG2 at the transcriptional level. (a, b) HaCaT cells were treated with TGF-β for 24 h, and the protein and mRNA expression levels of NDRG2 and p21Cip1 were detected. β-Actin served as a control to ensure equal loading. (c) HaCaT cells were treated with TGF-β for the indicated time, and the c-Myc and NDRG2 protein levels were detected. β-Actin served as a control to ensure equal loading. (d) HaCaT cells were treated with 10 ng/ml TGF-β for 48 h. Relative cell viability was detected with an MTT assay (n=3). (e) A panel of NDRG2 promoter constructs were transfected into HaCaT cells. At 24 h post transfection, the cells were treated with 10 ng/ml TGF-β for 20 h. The data shown are the means±s.d. from triplicate analysis. (f) Top: Representation of the human NDRG2 promoter regions depicting the sequence of the Smad-binding region (SBR). Bottom: HaCaT cells after TGF-β treatment were extracted and subjected to a chromatin immunoprecipitation assay. The presence of the NDRG2 promoter was detected with RT–PCR and visualized with ethidium bromide staining.
Sp1 is necessary for the transcriptional activation of NDRG2 by TGF-β
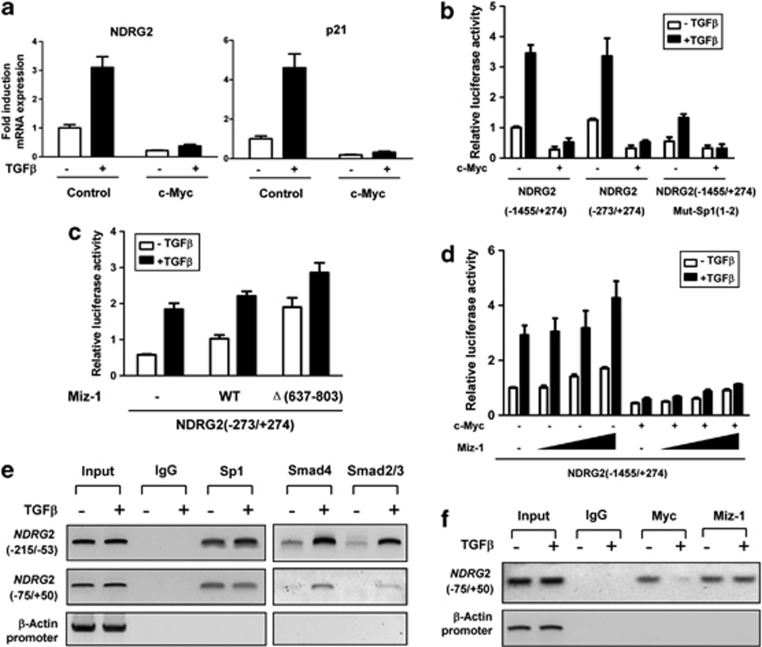
To fully understand the regulatory mechanism of NDRG2 by TGF-β, we analyzed the structure of the NDRG2 promoter, which was considered necessary and sufficient for induction by TGF-β. Three potential Sp1 binding sites were identified that are GC-rich motif in NDRG2 (−250/+274) (Supplementary Figure S2). Intriguingly, Sp1 could upregulate NDRG2 at the protein and mRNA levels (Figure 3a). NDRG2 promoter activity was also induced by Sp1 overexpression (Figures 3a and b). However, Sp1 induction was dramatically reduced on NDRG2 promoter mutants that had the first two Sp1 consensus sites deleted or mutated (Figure 3b), suggesting that these two sites are critical for the Sp1 regulation of NDRG2 promoter activity. This binding activity was also confirmed by EMSA (Supplementary Figure S3) and ChIP assays, which showed the interaction between Sp1 and the NDRG2 core promoter region (−215 to −53 and −75 to +50) (Figure 3c). Moreover, the presence of TGF-β and overexpression of Sp1 synergistically induced a greater than 10-fold increase in the promoter activity of wild-type NDRG2 (−1455/+274 and −273/+274), and there was only a slight influence on the mutant NDRG2 (−1455/+274) promoter (Figure 3d). Accordingly, an endogenous Sp1 knockdown via shRNA (psilencer-Sp1) or functional blocking using a dominant-negative Sp1 (pEBG-DN-Sp1) decreased NDRG2 expression (Supplementary Figures S4A–C) and abolished TGF-β responsiveness either on wild-type NDRG2 or on Sp1 consensus sites mutant constructs in advance (Figure 3e). In addition, Mithramycin A, an inhibitor that inhibited Sp1 binding with DNA by modifying GC-rich sites, dose dependently reduced NDRG2 expression and promoter activity (Supplementary Figures S4D and E) and blocked TGF-β-induced NDRG2 activity (Figure 3f). Therefore, Sp1 binding to consensus sites in the NDRG2 promoter was required for the TGF-β-mediated transcriptional activation of NDRG2.
Figure 3.
TGF-β-induced NDRG2 induction requires the Sp1 transcription factor. (a) Cells were transfected with pcDNA3 or pcDNA3-HA-Sp1. At 12 or 24 h after transfection, protein was extracted for western blotting (top), quantitative real-time PCR (middle) and a reporter assay using NDRG2 (−1455/+274) (bottom). (b) HaCaT cells were transfected with the indicated NDRG2 reporter constructs with or without a Sp1 expression vector. Luciferase activities were detected 24 h after transfection. (c) Human intestinal epithelial cells (HIEC) transfected with pcDNA3 or pcDNA3-HA-Sp1 were immunoprecipitated with the indicated antibodies. The presence of the NDRG2 promoter containing Sp1 binding sites was detected by RT–PCR and visualized with ethidium bromide staining. (d, e) HaCaT cells were transfected with the indicated NDRG2 reporter constructs with or without Sp1 expression or shRNA vector. The cells were incubated in the presence or absence of 10 ng/ml TGF-β for 20 h before lysis and then analyzed for luciferase activity. (f) Quantitative real-time PCR (left) and luciferase reporter assay (right) analysis for the involvement of mithramycin in TGF-β-mediated NDRG2 expression. HaCaT cells were pretreated with or without mithramycin for 1 h at the indicated concentrations and then stimulated with 10 ng/ml TGF-β for 20 h. (a–f) The data are presented as the means±s.d. from triplicate analyses.
TGF-β abrogates the c-Myc/Miz-1 repressive complex to activate NDRG2
On the basis of the above data, we observed that the removal of Sp1 consensus sites could not completely abolish TGF-β activity on the NDRG2 promoter (Figures 3d and e), suggesting that other factors contributed to the induction of NDRG2 by TGF-β. Our previous data revealed that c-Myc could transcriptionally repress NDRG2 by interacting with Miz-1 on the NDRG2 promoter.20 We next investigated the importance of the c-Myc/Miz-1 complex in response of NDRG2 to TGF-β. Because c-Myc is a key transcription node during EMT34 that has an important role in the TGF-β-induced expression of cyclin-dependent kinase inhibitors, we observed that in addition to the induction of NDRG2, TGF-β caused a rapid reduction in c-Myc (Figures 2a and c). Moreover, c-Myc overexpression dramatically abolished the NDRG2 and p21Cip1 expression induced by TGF-β (Figure 4a) and completely abolished TGF-β-mediated induction of the NDRG2 promoter, which contains mutant Sp1 binding sites (Figure 4b). In addition, there was TGF-β-induced NDRG2 promoter activity in cells overexpressing Miz-1, and this effect was further enhanced with overexpression of Miz-1Δ(637–803), which lacks the C-terminal residues (637–803) responsible for interaction with c-Myc35 (Figure 4c). However, c-Myc restoration counteracted the Miz-1 activating effects of the NDRG2 promoter (Figure 4d) but failed to block Miz-1Δ(637–803) activity (Supplementary Figure S5). Furthermore, treatment of cells with TGF-β did not dramatically alter the DNA binding activity of Sp1 on either of the Sp1 consensus sites (Figure 4e), whereas Smad4 and Smad2/3 binding to the Sp1 consensus sites was increased, indicating that Sp1 could mediate Smad complex binding on the NDRG2 promoter. In addition, TGF-β stimulation significantly reduced the binding of c-Myc at the NDRG2 promoter but did not alter Miz-1 occupancy (Figure 4f). Collectively, our results demonstrated that TGF-β upregulated NDRG2, and that is occurred partially through the abrogation of the interaction between c-Myc and Miz-1.
Figure 4.
TGF-β abrogates the c-Myc/Miz-1 complex and upregulates NDRG2 promoter activity. (a–d) HaCaT cells were incubated for 20 h in the presence or absence of 10 ng/ml TGF-β, and the mRNA levels and luciferase activity were determined. The data are presented as the means±s.d. from triplicate analyses. (a) Cells were transfected with either a control or a c-Myc expression vector. Quantitative real-time PCR was performed. (b) Cells were transfected with the indicated NDRG2 reporter constructs with or without the c-Myc expression vector. (c) Cells were transfected with the NDRG2 reporter construct together with wild-type Miz-1 and its mutant construct, as indicated. (d) Cells were transfected with the NDRG2 reporter construct with or without a c-Myc expression plasmid. Increasing amounts (0.01, 0.05 and 0.1 μg) of Miz-1 construct were co-transfected as indicated. (e, f) HaCaT cells treated with TGF-β were extracted and subjected to chromatin immunoprecipitation. The presence of the NDRG2 promoter was detected by RT–PCR and visualization by ethidium bromide staining.
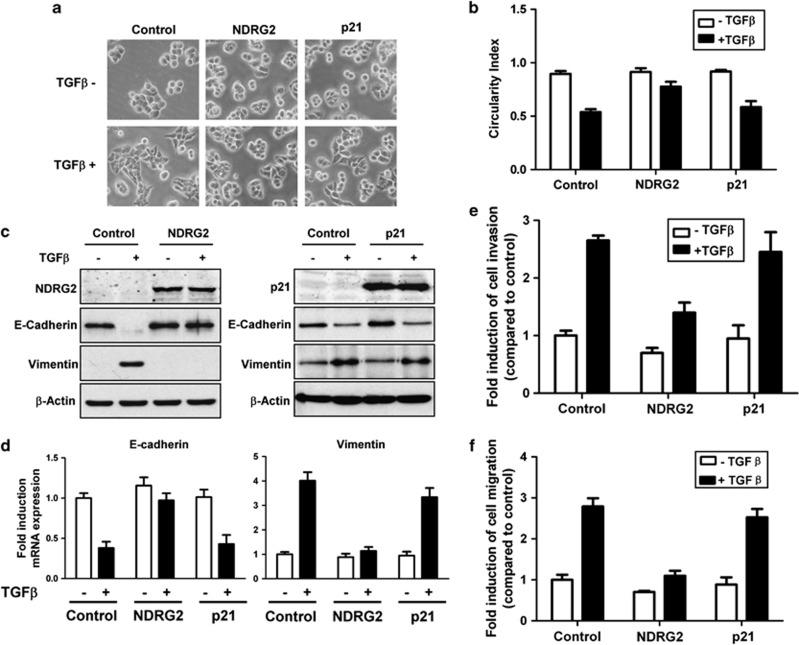
NDRG2 overexpression attenuates TGF-β-induced EMT
In agreement with previous reports,36, 37 we observed that CRC cell lines with multiple TGF-β/Smad pathway mutations lose TGF-β-induced growth inhibition characteristics (Supplementary Figure S6). To further determine whether NDRG2 is involved in the TGF-β-mediated induction of EMT in CRC cells, we established stable HT29 cell lines overexpressing NDRG2 or p21Cip1. There were no obvious morphological differences in HT29 cells after NDRG2 overexpression, which is likely because the parental cells already exhibit an epithelial phenotype. After TGF-β treatment, HT29 cells with NDRG2 overexpression showed an epithelial morphology (that is, the cells clustered together in groups, compared with the fibroblast-like shape alteration in the control group) (Figure 5a). As determined by quantification, the reduction in the circularity indices was significantly reversed by NDRG2 overexpression in the presence of TGF-β (Figure 5b). In addition, a reduction in E-cadherin and an increase in vimentin were observed in TGF-β-treated control cells. In contrast, NDRG2 overexpression inhibited the E-cadherin and vimentin alterations induced by TGF-β (Figures 5c and d). Moreover, NDRG2 overexpression decreased the migratory and invasive potential of HT29 cells and further reduced TGF-β-stimulated migration and invasion (Figures 5e and f). Notably, in HT29 cells overexpressing p21Cip1, which is one of the classical TGF-β responsive targets, EMT inhibition was not significant (Figures 5a–f). These studies indicated that NDRG2 could abrogate the migration, invasion and tumor progression induced by TGF-β in late-stage CRC via EMT inhibition.
Figure 5.
NDRG2 overexpression attenuates TGF-β-induced EMT. (a) Phase-contrast photomicrographs of control cells and cells treated with 10 ng/ml TGF-β for 48 h. (b) Circularity index of untreated and TGF-β-treated cells. (c) Western blot analysis of E-cadherin and vimentin expression from total lysates of untreated cells and cells treated with 10 ng/ml TGF-β for 48 h. (d) Relative mRNA levels of E-cadherin and vimentin in control cells and cells treated with 10 ng/ml TGF-β for 48 h. The control values were normalized to 1, and the data are expressed as fold-change in treated cells. (e, f) The migratory and invasive behavior of untreated cells and cells treated with 10 ng/ml TGF-β for 48 h. The control values were normalized to 1, and the data are expressed as fold-change in treated cells. (d–f) The data are presented as the means±s.d. from triplicate analyses.
NDRG2 knockdown promotes EMT by upregulating the key factors involved in TGF-β-induced EMT
To further confirm the EMT inhibitory ability of NDRG2, we established stable HT29 cell lines with either NDRG2 or p21Cip1 knockdown and analyzed cell motility and invasion. The cell morphology of HT29 cells with the NDRG2 knockdown converted to a more elongated, fibroblast-like shape (Figures 6a and b), and TGF-β stimulation enhanced this morphological change. In addition, NDRG2 knockdown significantly induced EMT marker alterations (that is, vimentin increase and E-cadherin loss), which became more pronounced after the addition of TGF-β (Figure 6c). As expected, NDRG2 knockdown also significantly increased the migration and invasion of HT29 cells, and the potential for these processes was enhanced after incubation with TGF-β (Figures 6d and e). Similar results were observed in p21Cip1 knockdown cells (Figures 6a–e). Moreover, by screening for key factors involved in TGF-β-induced EMT, we observed that NDRG2 knockdown dramatically upregulated the fibronectin, Twist, Snail and N-cadherin expression levels (Figure 6f). Together, these studies demonstrate that NDRG2 knockdown could promote EMT alterations in HT29 cells via the upregulation of multiple factors essential for TGF-β-induced EMT.
Figure 6.
NDRG2 knockdown promotes TGF-β-induced EMT. (a) Phase-contrast photomicrographs of control cells and cells treated with TGF-β demonstrated that NDRG2 and p21Cip1 knockdown mimics the EMT phenotype in HT29 cells. (b) Circularity index of untreated and TGF-β1-treated cells. (c) Western blot analysis of E-cadherin and vimentin expression in total lysates from untreated cells and cells treated with 10 ng/ml TGF-β for 48 h. (d, e) Migratory and invasive behavior of untreated cells and cells treated with 10 ng/ml TGF-β for 48 h. The control values were normalized to 1, and the data are expressed as the fold-change in treated cells. (f) The expression level of mRNAs encoding E-cadherin, N-cadherin, fibronectin, Twist, Snail and vimentin in control cells and cells treated with 10 ng/ml TGF-β for 48 h. (d–f) The data are presented as the means±s.d. from triplicate analysis.
Reduced NDRG2 expression caused by promoter hypermethylation fostered the progression and metastasis of CRC
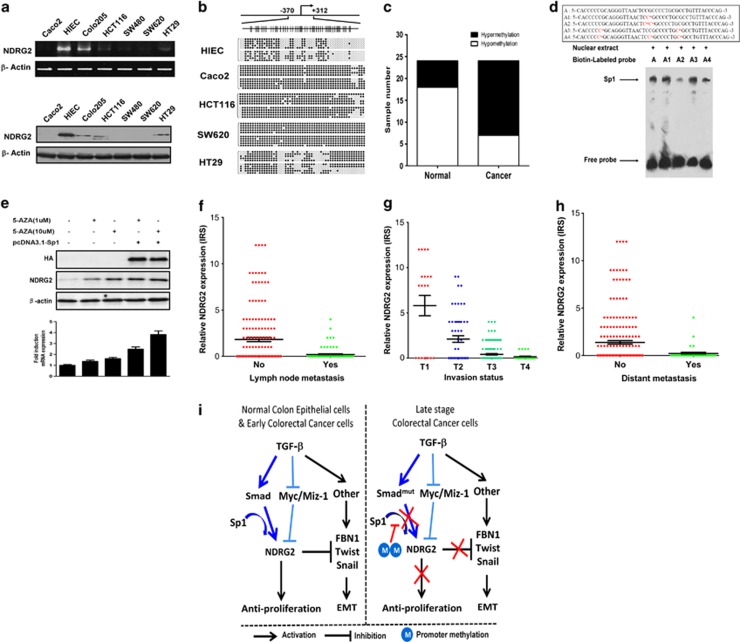
NDRG2 exhibited dramatically reduced expression at the mRNA and protein levels in the majority of CRC cells relative to normal human intestinal epithelial cells (HIEC) (Figure 7a). To determine the mechanism for the resistance to growth inhibition and inability of TGF-β to induce NDRG2 in CRC, we performed DNA methylation analysis of the NDRG2 promoter in CRC cells and patient samples. Hypermethylation of the NDRG2 promoter was detected in most CRC cell lines (Figure 7b). Moreover, ∼75% (18/24) of the clinical colorectal tumors showed hypermethylation at the NDRG2 promoter, and a low percentage of hypermethylation (6/24) was observed in the normal counterparts (Figure 7c).
Figure 7.
Promoter hypermethylation reduces NDRG2 expression and hinders the progression and metastasis of colorectal cancer. (a) The RNA and protein levels of NDRG2 in multiple colorectal cancer cell lines and the normal human intestinal epithelial cells (HIEC) were detected by comparative RT–PCR and western blotting. (b) Bisulfite sequencing analysis of the NDRG2 promoter in a panel of cell lines. (Top) Schematic of the NDRG2 promoter. The positions of the CpG dinucleotides are shown to scale by vertical lines. (Bottom) Each circle represents a CpG dinucleotide: open (white) circles denote unmethylated CpG sites and filled (black) circles indicate methylated CpG sites. Each row represents a single clone. (c) The methylation status of the NDRG2 promoter was analyzed by methylation-specific PCR in each of the 24 normal colon and colorectal cancer tissues. The white and black colors represent hypomethylation and hypermethylation, respectively. Each experiment was independently repeated three times. (d) The binding activity of Spl to methylated and unmethylated recognition sites in vitro. (Top) Double-stranded oligonucleotides were derived from the Spl recognition sites in the NDRG2 promoter. The methylated C (Cm) is shown in red. A, no methylation; A1, single methylation on recognition site; A2, double methylation on recognition site; A3, methylation on adjacent region; A4, methylation of both CpG dinucleotides within its binding site and adjacent CpG sites. (Bottom) Electrophoretic mobility shift analysis of HT29 cell nuclear extracts. The Sp1 complex and free probe are indicated by the arrow. (e) HCT116 cells were transfected with or without a Sp1 expression vector. Twenty-four hours post transfection, the cells were treated with 5-aza-dC at the indicated concentrations for 48 h before harvesting for western blotting and quantitative real-time PCR analysis. The data are the means±s.d. from triplicate analyses. (f) The immunoreactivity score (IRS) distribution of NDRG2 IHC staining among tumors of different invasion statuses. (g) The IRS distribution of NDRG2 IHC staining between tumors of different lymph-node metastasis statuses. (h) The IRS distribution of NDRG2 IHC staining between tumors of different distant metastasis statuses. (i) Model of NDRG2 tipping the balance of oncogenic TGF-β via EMT inhibition in colorectal cancer.
To determine whether promoter hypermethylation in CRC leads to the reduced sensitivity of NDRG2 to multiple growth inhibition signals, including TGF-β, we analyzed the NDRG2 expression induced by Sp1 in NDRG2 hypermethylated CRC cells. Intriguingly, NDRG2 induction by Sp1 was completely abolished in NDRG2 hypermethylated CRC cells (Supplementary Figure S7). Furthermore, Sp1 binding activity was blocked by hypermethylation of the CpG dinucleotide within its binding site (A2) or adjacent CpG sites (A4) (Figure 7d). As expected, the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) restored NDRG2 expression, promoted the induction of NDRG2 by Sp1 (Figure 7e), and further inhibited CRC proliferation (Supplementary Figure S8), thus indicating that epigenetic alteration is a critical step during tumor progression. However, the inhibition of EMT by 5-aza-dC was not dramatic (data not shown), which might be attributed to the global effects of 5-aza-dC on multiple target genes. Collectively, NDRG2 promoter methylation abrogated the Sp1-meditated induction of NDRG2, which may be responsible for the reduction in the NDRG2 response to TGF-β in CRC.
On the basis of the above data, the NDRG2 tumor suppressor has an important role in impairing the tumor progression induced by TGF-β in CRC cells through inhibition of EMT. To further understand the role of NDRG2-mediated EMT inhibition in patients, we analyzed NDRG2 expression status and tumor invasion and metastasis characteristics in 260 CRC patients (Table 3). Consistent with the in vitro data, CRC patients with reduced NDRG2 expression showed a relatively higher invasion stage than those with a normal NDRG2 expression level (Figure 7f). Most importantly, reduced NDRG2 was significantly correlated with lymph-node involvement and distant metastasis (Figures 7g and h), confirming that NDRG2 is a potent tumor suppressor that acts not only through anti-proliferation effects but also via the inhibition of invasion and metastasis.
Table 3. Statistical results of IHC assay for NDRG2 in cohort of 260 CRC.
| n |
NDRG2 |
P-value | ||
|---|---|---|---|---|
| |
|
Negative |
Positive |
|
| Total | 260 | 184 | 76 | |
| Gender | 0.863a | |||
| Male | 139 | 99 | 40 | |
| Female | 121 | 85 | 36 | |
| Age at diagnosis, years | 0.493a | |||
| <60 | 142 | 103 | 39 | |
| >60 | 118 | 81 | 37 | |
| Depth of invasion | 0.002b | |||
| T1 | 26 | 14 | 12 | |
| T2 | 62 | 36 | 26 | |
| T3 | 138 | 104 | 34 | |
| T4 | 34 | 30 | 4 | |
| Lymph-node metastasis | <0.001a | |||
| Absent (N0) | 163 | 98 | 65 | |
| Present (N1–3) | 97 | 86 | 11 | |
| Distant metastasis | 0.006a | |||
| Absent (M0) | 222 | 150 | 72 | |
| Present (M1) | 38 | 34 | 4 | |
Abbreviations: CRC, colorectal cancer; NDRG2, N-Myc downstream-regulated gene 2; IHC, immunohistochemical.
P-value when expression levels were compared using Fisher's exact test.
P-value when expression levels were compared using Kruskal Wallis.
As described in the working model (Figure 7i), our study identified NDRG2 as a new tumor suppressor gene that is a TGF-β-responsive target that tips the balance of oncogenic TGF-β via EMT inhibition in late-stage CRCs.
Discussion
TGF-β serves as a tumor suppressor primarily by inhibiting cell proliferation during early tumor stages, and it behaves as a tumor promoter by stimulating both invasion and metastasis during late tumor stages. There is growing evidence that restoration of the impaired TGF-β pathway cannot rescue the anti-proliferative response to TGF-β in CRC cells.9, 16 Therefore, other mechanisms must be elucidated to determine how selective alterations in the TGF-β signaling pathway contribute to the development, progression and metastasis of CRC.
In this study, we first demonstrated a clinical correlation between the protein accumulation of NDRG2 and C-terminal phosphorylated Smad2/3, which responds to TGF-β anti-proliferative signaling, by the IHC staining of colon cancers; these two proteins are also negatively correlated with colon cancer progression. Our analysis of 50 pairs of colon cancer and adjacent normal colon samples demonstrated that TGF-β-mediated activation likely serves as an important guardian for the expression of NDRG2 in vivo. It should be noted that, as is often observed among clinical samples, there was not a 1:1 correlation between the expression of phosphorylated Smad2/3 and NDRG2 in tissues. Thus, other factors may also be involved in TGF-β induced tumor suppressor gene expression and colon cancer progression, and some of these factors could be considered epigenetic alterations.
Moreover, NDRG2 was found to be one of the intact TGF-β/Smad signaling target genes induced in TGF-β-treated keratinocytes. A cluster of sites located in the core region of the NDRG2 promoter was further confirmed to contribute to inducibility by TGF-β in reporter assays. Although Sp1 was considered necessary for the basal transcription of multiple genes, we found that Sp1 binding at consensus sites in the core region of the NDRG2 promoter was required for TGF-β-induced NDRG2 upregulation. Our data were similar to the observation of increased p15Ink4B transcription in response to TGF-β, which is mediated through the interaction between Sp1 and the Smad2/3/4 complex.38 In addition, we also found that TGF-β induced a rapid reduction in c-Myc levels, thus abrogating the interaction between c-Myc and Miz-1, which was responsible for the transcriptional repression of NDRG2. Interference of the c-Myc/Miz-1 interaction with a dominant-negative construct, Miz-1Δ(637–803), enhanced the TGF-β activating effect on the NDRG2 promoter. This finding further supports the hypothesis that TGF-β relieves the repression of c-Myc and increases Miz-1 transcriptional activity in NDRG2 transcription. On the one hand, TGF-β/Smad signaling directly transactivated NDRG2, which is dependent on the recruitment of Sp1 to the NDRG2 promoter. On the other hand, as a second input for NDRG2 activation, TGF-β reduced c-Myc expression and abrogated the repression of the c-Myc/Miz-1 complex to allow for NDRG2 transcription (Figure 7I; Supplementary Figure S9). A related mechanism was reported for p21Cip1 as a target of TGF-β and c-Myc.39
It is well established that most CRCs are resistant to the tumor suppressor effects of TGF-β and prone to TGF-β-induced EMT,37 which is associated with features of advanced disease including metastasis, resistance to chemotherapy and the generation of cancer cells with stem cell-like characteristics.40 Specifically, the loss or abnormal intracellular localization of E-cadherin in colon cancer has been associated with advanced histological grade, metastasis and a reduction in disease-free and overall survival.41 Herein, we found that the induction of NDRG2 by TGF-β was completely abolished in CRC and that this induction accounted for the loss in anti-proliferative response. Furthermore, NDRG2 overexpression remarkably attenuated TGF-β-induced EMT and decreased the migratory and invasive potential of HT29 cells in response to TGF-β. The reduction was stronger than that caused by p21Cip1, one of the classic TGF-β responsive targets. Moreover, NDRG2 knockdown dramatically promoted EMT in HT29 cells mainly through the upregulation of key factors involved in TGF-β-induced EMT, including fibronectin, Twist, Snail and N-cadherin. It will be interesting to investigate how NDRG2 regulates these key EMT factors. Recently obtained evidence has demonstrated that NDRG2 significantly suppressed tumor metastasis by attenuating active autocrine TGF-β production in breast cancers, which suggests the need for an NDRG2-mediated anti-metastatic effects study.42 There are data showing that NDRG2 was negatively regulated by TGF-β during the progression of hepatocellular carcinomas.43 This observation may be due to impairment in the TGF-β/Smad signaling pathway or the activation of non-Smad signaling cascades (PI3K/AKT, p38MAPK and so on) in these cell lines in response to TGF-β. In addition, related evidence has shown that the enhancement of GSK-3β activity by NDRG2 overexpression causes proteasomal degradation of the Snail transcription factor and subsequent transcriptional regulation of EMT-related genes,44 which is consistent with this study. Recent observations suggest that crosstalk occurs between the cell-cycle regulation and EMT pathways,45 and p21Cip1 has been shown to attenuate EMT in Ras- and c-Myc-dependent breast cancer.46 We observed that p21 loss is necessary for EMT promotion, whereas p21 overexpression has little effect on EMT, indicating that p21 is necessary for maintaining the epithelial phenotype but that it is not the sole reason. Despite this finding, the underlying mechanism requires further investigation. Nonetheless, our current data suggest that the NDRG2 tumor suppressor could inhibit TGF-β-induced EMT as well as cell invasion and migration in late-stage CRCs.
To further understand whether NDRG2 controls the switch from TGF-β-mediated anti-proliferative gene responses to oncogenic potential in CRCs, we used clinical tumor samples to analyze the methylation status of the NDRG2 promoter, which is an important epigenetic regulation mechanism. Most colorectal tumors displayed aberrant hypermethylation in the NDRG2 promoter compared with the hypomethylation in normal counterparts. Furthermore, in addition to being prone to carcinogenesis, NDRG2 methylation also tended to occur in late-stage subjects, which was suggestive of the abrogation of TGF-β-mediated growth inhibition and increasing instances of oncogenic TGF-β properties in late-stage CRC. Thus, we hypothesized that repression of NDRG2 resulted from promoter methylation, which occurs in late-stage colon cancer patients, and that this repression tipped the balance of the TGF-β role toward oncogenesis in CRC.
To support this conclusion, the blocking intensity of promoter methylation was detected in NDRG2-hypermethylated colon cancer cells that overexpressed Sp1. Strikingly, the induction of NDRG2 by Sp1 was abolished in these cells primarily due to failed binding of Sp1 to the hypermethylated NDRG2 promoter, whereas treatment with DNA methyltransferase inhibitor significantly restored this regulation. Therefore, the expression of TGF-β-responsive tumor suppressor genes and their promoter methylation statuses should be valuable for the prediction of the role of TGF-β-mediated oncogenesis in CRCs. Additional studies on a much larger scale are needed to obtain more definitive conclusions.
In summary, our data provide the first evidence that the NDRG2 tumor suppressor gene is a new TGF-β responsive target and inhibits TGF-β-induced EMT in CRCs. The loss of NDRG2 expression, which is attributed to promoter methylation, tipped the balance of the TGF-β role toward oncogenesis in late-stage CRC. Our observations elucidate how selective alterations in the TGF-β signaling pathway contribute to CRC development and progression. These data will likely permit the identification of an additional subset of inherited CRC cases and provide novel opportunities for therapeutic intervention.
Materials and methods
Tissue samples and study cohort
This study was approved by the ethics committee of the Fourth Military Medical University. All 50 patients who provided CRC and adjacent normal colon mucosa colectomy specimens and the 260 subjects in the CRC sample study cohort provided full consent for the study. The recruited patients were consecutively diagnosed with CRC in the Department of Gastrointestinal Surgery, Xijing Hospital, Fourth Military Medical University. Patients who met any of the following criteria were excluded: (1) received treatment before surgery that included neoadjuvant chemotherapy; (2) had insufficient specimens for tissue microarray preparation; (3) were diagnosed with gastrointestinal stromal tumor or lymphoma; (4) were diagnosed with additional cancers; or (5) refused to provide authorized consent. All of these clinical cancerous specimens were collected through surgery or endoscopy. All of the specimens were histologically diagnosed by the Department of Pathology, Xijing Hospital, Fourth Military Medical University. The clinicopathologic information for the remaining patients was entered into a database. For the study cohorts, study physicians who reviewed the CRC records and recorded the data into a database were blinded to the data.
IHC detection
IHC was performed on all CRC samples using the avidin-biotin-peroxidase method. All sections were deparaffinized in xylene and dehydrated using a concentration gradient of alcohol before endogenous peroxidase activity was blocked using 0.5% H2O2 in methanol for 10 min. After non-specific binding was blocked, the slides were incubated with a NDRG2 polyclonal antibody (1:200; ABNOVA, Jhongli, Taiwan) or p-Smad2/3 (1:100, sc-11769; Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) in phosphate-buffered saline at 4 °C overnight in a humidified container. Biotinylated goat anti-rabbit IgG (1:400; Sigma, St. Louis, MO, USA) was incubated with the sections for 1 h at room temperature and detected with a streptavidin-peroxidase complex. The brown color indicative of peroxidase activity was developed by incubating with 0.1% 3,3-diaminobenzidine (Sigma) in phosphate-buffered saline with 0.05% H2O2 for 5 min at room temperature. The appropriate positive and negative controls were included in each IHC run.
Evaluation of IHC staining
Multiple tissue arrays were independently scored by two pathologists blinded to the clinicopathologic information and outcome of the patients. Staining was evaluated by scanning the entire tissue specimen under low magnification ( × 40) and then confirmed at higher magnification ( × 200 and × 400). An immunoreactivity score system based on the proportion and intensity of positively stained cancer cells was used. The extensional standard used (1) the number of cells with positive staining (⩽5%: 0; 6–25%: 1; 26–50%: 2; 51–75%: 3; and >75%: 4) and (2) the staining intensity (colorless: 0; pallide-flavens: 1; yellow: 2; brown: 3). We multiplied the numbers scored in 1 and 2 from the extensional standard, and the staining grade was stratified as absent (0 score), weak (1–4 score), moderate (5–8 score) or strong (9–12 score). All slides were interpreted by another pathologist unaware of the data. Specimens were rescored if the difference in score from the two pathologists was >3. Tumors with weak, moderate or strong immunostaining intensity were classified as staining positive (+), whereas tumors with no immunostaining were classified as staining negative (−).
Cell culture and reagents
The HaCaT human keratinocyte cell line, the Caco2 human colon cancer cell line and the HEK293 human embryo kidney cell line (ATCC) were maintained at 37 °C under 5% CO2 in Dulbecco's minimal essential medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The human colon cancer cell lines HT29 and HCT116 (ATCC) were maintained in McCoy's 5A medium supplemented with 10% fetal calf serum. TGF-β1 (100–21) was purchased from Pepro Tech (Rocky Hill, NJ, USA). Cells were treated with 10 ng/ml TGF-β1 for 20, 24 or 48 h, washed with phosphate-buffered saline, and harvested for downstream assays.
MTT assay
For cell growth and viability assays, cells were seeded into each well of a 96-well plate with five replicates for each group at each time point. After the stated incubation time, 20 μl of MTT solution (5 mg/ml) was added to each well, and after 4 h of incubation, the medium was aspirated as much as possible without disturbing the formazan crystals. Then, 150 μl of DMSO was added to each well, and the plates were placed on a plate shaker for 10 min. The OD values were determined at 570 nm using a Sunrise microplate reader (Tecan, Groedig, Austria).
Real-time PCR
For RT–PCR, cDNA was synthesized from RNA using the PrimeScript RT–PCR Kit (TAKARA, Dalian, China). We then subjected the cDNA (1–100 ng) to PCR amplification. Quantitative real-time PCRs were performed using specific primers (Supplementary Table 1). The mean Ct values for the target genes were normalized to the mean Ct value for the endogenous control GAPDH. The ratio of mRNA expression of target gene versus GAPDH was defined as 2 (–ΔCt).
Plasmids
Human NDRG2 promoter truncated regions were previously cloned into the pGL3-basic vector (Promega, Madison, WI, USA) to generate NDRG2/luciferase reporter plasmids.20 The NDRG2 promoter mutants were generated using the mutation primers listed in Supplementary Table 1. pcDNA3.1 (+)-c-Myc was generously provided by Dr Michael Cole (Lewis Thomas Laboratory, Princeton University, Princeton, NJ, USA). pcDNA3.1 (+)-Miz-1 was kindly provided by Dr Ben-Zion Levi (Department of Biotechnology and Food Engineering, Technion, Haifa, Israel).
Western blotting
Cells were collected and lysed in lysis buffer. Protein concentrations were measured using the BCA protein assay kit from Thermo Scientific (Waltham, MA, USA). Forty micrograms of protein were separated by SDS-PAGE and transferred onto Hybond ECL nitrocellulose membranes. Western blot analyses were performed using the following primary antibodies: anti-NDRG2 mouse monoclonal antibody (H00057447-M03), which was purchased from ABNOVA; anti-Sp1 rabbit polyclonal antibody (sc-59), anti-E-cadherin polyclonal antibody (sc-7870) and anti-vimentin mouse monoclonal antibody (sc-373717), which were purchased from Santa Cruz Biotechnology, Inc.; anti-p21Cip1 rabbit polyclonal antibody (#2947), which was purchased from Cell Signaling Technology (Beverly, MA, USA); and mouse monoclonal anti-human β-actin (1:5000 dilution), which was purchased from Sigma. Secondary antibodies were chosen according to the species of origin of the primary antibodies and detected using enhanced chemiluminescence (Pierce, Rockford, IL, USA) or the Odyssey Imaging System (Li-Cor Biosciences, Lincoln, NE, USA).
Bisulfite-PCR and sequencing and methylation-specific PCR
Total genomic DNA was isolated from multiple cell lines and the tumor and normal colon epithelium samples using a DNA extraction kit according to the manufacturer's instructions (Tiangen, Beijing, China). Two micrograms of genomic DNA was subjected to a denaturation/incubation cycle in sodium bisulfite solution, as previously described.47 For bisulfite sequencing analysis, 10 clones were sequenced for each cell line. For methylation-specific PCR, the intensity of the methylated and loading control bands was compared with the base pairs of the amplicons, and the cycles of individual PCRs were considered. When the relative ratio of methylated DNA to the total amount of DNA was greater than 50%, the methylation status was considered to be high, whereas it was considered low when the ratio was less than 50%. Methylation was considered absent in samples with no obvious bands (see Supplementary Table 1 for primer sequences).
Luciferase reporter assays
Cells were transfected at ∼70–80% confluence using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were treated with 10 ng/ml TGF-β for 20 h. Luciferase assays were performed using the dual luciferase reporter assay system (Promega). The values are expressed as the means±s.d. of at least three independent experiments.
Chromatin immunoprecipitation
ChIP analysis was performed using the ChIP Assay kit (Upstate Biotechnology, Charlottesville, VA, USA). The cells were crosslinked using 1% formaldehyde for 10 min at 37 °C and then washed, lysed and sonicated to generate 200–500 bp DNA fragments. The samples were precleared using 60 μl of salmon sperm DNA-protein A-agarose and subsequently incubated at 4 °C overnight with 2 μg of HA or Myc antibody; rabbit IgG was used as a control. Immunocomplexes were recovered, washed thoroughly and eluted with ChIP elution buffer. Following the reversal of crosslinks at 65 °C for 4 h, the DNA from each sample was extracted using phenol/chloroform, precipitated with ethanol and then used as a template for PCR amplification. PCR primers were used to detect the NDRG2 promoter (Supplementary Table 1).
In vitro migration and invasion assays
The migration and invasion of CRC cells were examined using polycarbonate transwell filters containing 8 μm pores (Becton Dickinson Labware, Franklin Lakes, NJ, USA). After treatment, the cells were seeded in serum-free media on the upper side of a transwell chamber that was either uncoated, for the migration assay, or coated with Matrigel (BD Biosciences, Bedford, MA, USA), for the invasion assay. The cells were allowed to migrate toward media containing 10% fetal bovine serum for 24 h. After the incubation period, the cells on the lower side of the membrane were fixed, stained with crystal violet, and counted. The migration and invasion indices were calculated as the mean number of cells in 10 random fields at × 20 magnification.
Circularity index analysis
Circularity index analysis of control cells and cells treated with 5 ng/ml of TGF-β1 for 48 h was performed. For each condition, the circularity of >100 cells in at least two separate areas was determined, and their average±s.d. was determined. *P<0.05 compared with control cells.
Statistical analyses
The data are expressed as the means±s.d. Statistical analyses using Student's T-test for independent groups was performed using the SPSS 16.0 software package (SPSS Inc, Chicago, IL, USA) for Windows. Associations between NDRG2 expression and categorical variables were analyzed by the Mann–Whitney U-test or the Kruskal–Wallis test, as appropriate. *P<0.05 was considered as statistically significant.
Acknowledgments
We thank the human study participants and all members of the Department of Biochemistry and Molecular Biology of the Fourth Military Medical University. This study was supported by National Program on Key Basic Research Project (2010CB529705 and 2009CB521704) and National Natural Science Foundation of China (No. 30830054, 81230043, 81172292 and 30900635).
Glossary
- TGF-β
transforming growth factor β
- NDRG2
N-Myc downstream-regulated gene 2
- EMT
epithelial–mesenchymal transition
- 5-aza-dC
5-aza-2′-deoxycytidine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Engel ME, Datta PK, Moses HL. Signal transduction by transforming growth factor-beta: a cooperative paradigm with extensive negative regulation. J Cell Biochem Suppl. 1998;30-31:111–122. [PubMed] [Google Scholar]
- Shin I, Bakin AV, Rodeck U, Brunet A, Arteaga CL. Transforming growth factor beta enhances epithelial cell survival via Akt-dependent regulation of FKHRL1. Mol Biol Cell. 2001;12:3328–3339. doi: 10.1091/mbc.12.11.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- Pietenpol JA, Holt JT, Stein RW, Moses HL. Transforming growth factor beta 1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc Natl Acad Sci USA. 1990;87:3758–3762. doi: 10.1073/pnas.87.10.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23:2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, et al. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- Schwarte-Waldhoff I, Klein S, Blass-Kampmann S, Hintelmann A, Eilert C, Dreschers S, et al. DPC4/SMAD4 mediated tumor suppression of colon carcinoma cells is associated with reduced urokinase expression. Oncogene. 1999;18:3152–3158. doi: 10.1038/sj.onc.1202641. [DOI] [PubMed] [Google Scholar]
- Calonge MJ, Massague J. Smad4/DPC4 silencing and hyperactive Ras jointly disrupt transforming growth factor-beta antiproliferative responses in colon cancer cells. J Biol Chem. 1999;274:33637–33643. doi: 10.1074/jbc.274.47.33637. [DOI] [PubMed] [Google Scholar]
- Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–347. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- Choi SC, Kim KD, Kim JT, Kim JW, Yoon DY, Choe YK, et al. Expression and regulation of NDRG2 (N-myc downstream regulated gene 2) during the differentiation of dendritic cells. FEBS Lett. 2003;553:413–418. doi: 10.1016/s0014-5793(03)01030-5. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamada M, Ohata H, Honda K. Ndrg2 promotes neurite outgrowth of NGF-differentiated PC12 cells. Neurosci Lett. 2005;388:157–162. doi: 10.1016/j.neulet.2005.06.055. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li F, Liu X, Shen L, Liu J, Su J, et al. The repression of human differentiation-related gene NDRG2 expression by Myc via Miz-1-dependent interaction with the NDRG2 core promoter. J Biol Chem. 2006;281:39159–39168. doi: 10.1074/jbc.M605820200. [DOI] [PubMed] [Google Scholar]
- Lorentzen A, Vogel LK, Lewinsky RH, Saebo M, Skjelbred CF, Godiksen S, et al. Expression of NDRG2 is down-regulated in high-risk adenomas and colorectal carcinoma. BMC Cancer. 2007;7:192. doi: 10.1186/1471-2407-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Zhang Z, Li Y, Wu L, Zhang J, Wang W. Prediction of colorectal cancer relapse and prognosis by tissue mRNA levels of NDRG2. Mol Cancer Ther. 2011;10:47–56. doi: 10.1158/1535-7163.MCT-10-0614. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140–148. doi: 10.1136/gut.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Derks S, Postma C, Carvalho B, van den Bosch SM, Moerkerk PT, Herman JG, et al. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29:434–439. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- Martin ES, Tonon G, Sinha R, Xiao Y, Feng B, Kimmelman AC, et al. Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Res. 2007;67:10736–10743. doi: 10.1158/0008-5472.CAN-07-2742. [DOI] [PubMed] [Google Scholar]
- Wrana JL. Crossing Smads. Sci STKE. 2000;2000:re1. doi: 10.1126/stke.2000.23.re1. [DOI] [PubMed] [Google Scholar]
- Wrighton KH, Lin X, Feng XH. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009;19:8–20. doi: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B. Integrating Myc and TGF-beta signalling in cell-cycle control. Nat Cell Biol. 2001;3:E112–E113. doi: 10.1038/35074634. [DOI] [PubMed] [Google Scholar]
- Thomson S, Petti F, Sujka-Kwok I, Mercado P, Bean J, Monaghan M, et al. A systems view of epithelial-mesenchymal transition signaling states. Clin Exp Metastasis. 2011;28:137–155. doi: 10.1007/s10585-010-9367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Li CY, Suardet L, Little JB. Potential role of WAF1/Cip1/p21 as a mediator of TGF-beta cytoinhibitory effect. J Biol Chem. 1995;270:4971–4974. doi: 10.1074/jbc.270.10.4971. [DOI] [PubMed] [Google Scholar]
- Pino MS, Kikuchi H, Zeng M, Herraiz MT, Sperduti I, Berger D, et al. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138:1406–1417. doi: 10.1053/j.gastro.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M, Taniguchi T, Makino M, Kaibara N. Reduced E-cadherin expression and enlargement of cancer nuclei strongly correlate with hematogenic metastasis in colorectal adenocarcinoma. Scand J Gastroenterol. 2000;35:839–846. doi: 10.1080/003655200750023219. [DOI] [PubMed] [Google Scholar]
- Oh SS, Kim D, Kim DH, Chang HH, Sohn KC, Kim KH, et al. NDRG2 correlated with favorable recurrence-free survival inhibits metastasis of mouse breast cancer cells via attenuation of active TGF-β production. Carcinogenesis. 2012;33:1882–1888. doi: 10.1093/carcin/bgs211. [DOI] [PubMed] [Google Scholar]
- Lee DC, Kang YK, Kim WH, Jang YJ, Kim DJ, Park IY, et al. Functional and clinical evidence for NDRG2 as a candidate suppressor of liver cancer metastasis. Cancer Res. 2008;68:4210–4220. doi: 10.1158/0008-5472.CAN-07-5040. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Kang HB, Yim HS, Kim JH, Kim JW. NDRG2 positively regulates E-cadherin expression and prolongs overall survival in colon cancer patients. Oncol Rep. 2013;30:1890–1898. doi: 10.3892/or.2013.2642. [DOI] [PubMed] [Google Scholar]
- Smit MA, Peeper DS. Epithelial-mesenchymal transition and senescence: two cancer-related processes are crossing paths. Aging (Albany, NY) 2010;2:735–741. doi: 10.18632/aging.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.