Abstract
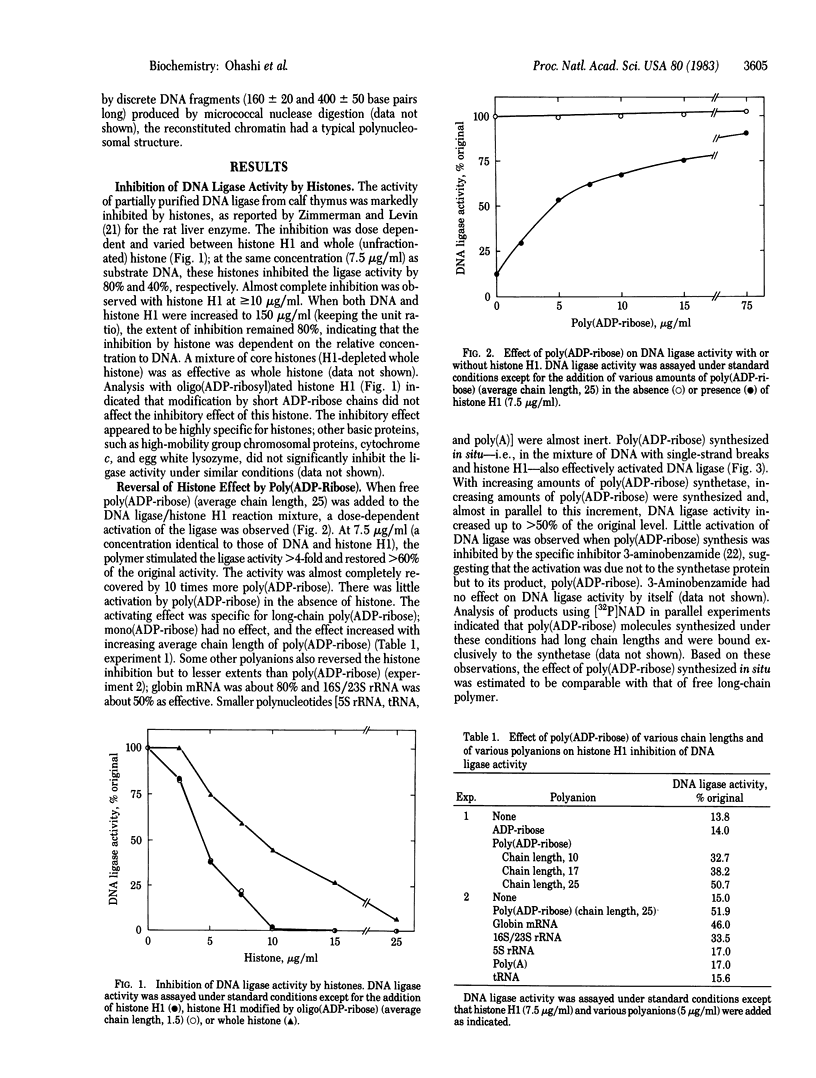
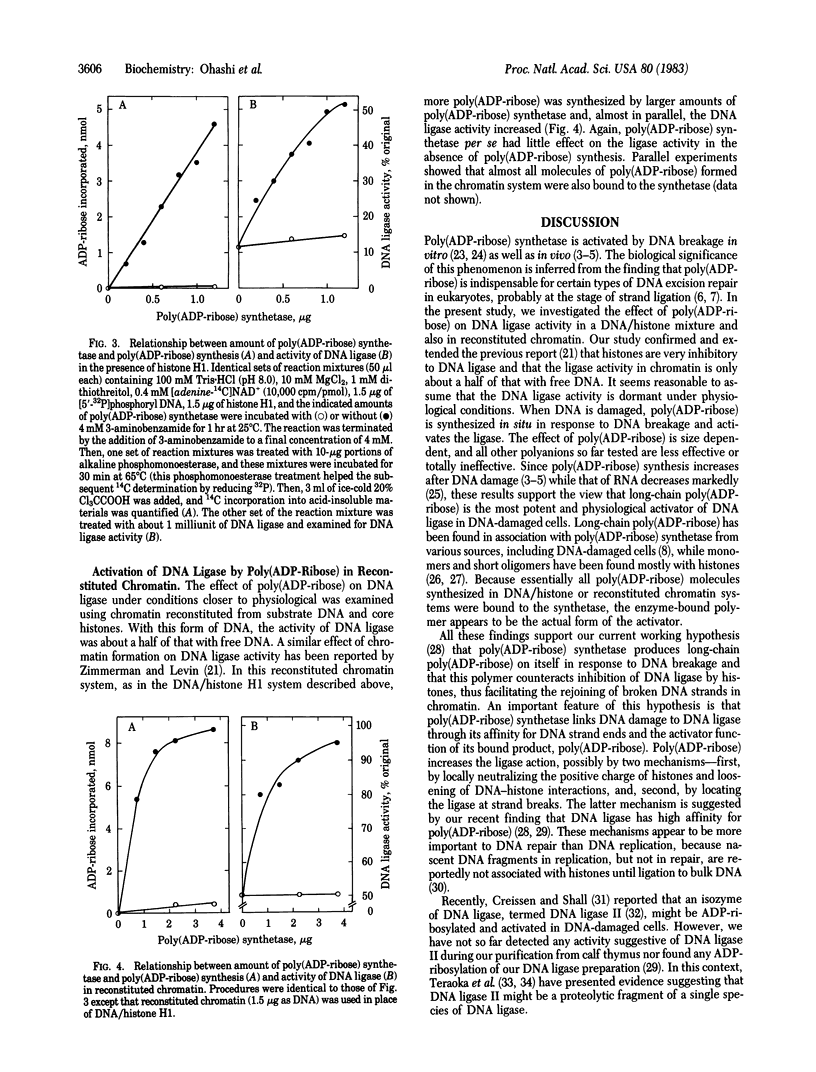
To elucidate the molecular mechanism by which poly(ADP-ribose) participates in DNA excision repair, we examined the effect of poly(ADP-ribose) on DNA ligase activity in DNA/histone and reconstituted chromatin systems. The ligase activity was markedly inhibited by histones; the inhibition varied depending on histone subfraction and DNA/histone ratio. Poly(ADP-ribose), either exogenous or synthesized in situ by poly(ADP-ribose) synthetase, reversed this inhibition by histone almost completely. This effect was specific for poly(ADP-ribose); polyanions such as mRNA, rRNAs, tRNA, and synthetic poly(A) were less effective or ineffective. The ligase activity with reconstituted chromatin as the substrate was about half of that with free DNA whereas the activities with these two substrates were almost the same in the presence of poly(ADP-ribose) synthesized in situ. The polymers synthesized under these conditions were exclusively bound to the synthetase. Together with our previous finding that the enzyme is the main acceptor of the polymer in DNA-damaged cells, these results suggest that poly(ADP-ribose) in the synthetase-bound form counteracts inhibition by histones and activates DNA ligase to rejoin DNA strands in polynucleosomal structures.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adamietz P., Hilz H. Poly(adenosine diphosphate ribose) is covalently linked to nuclear proteins by two types of bonds. Hoppe Seylers Z Physiol Chem. 1976 Apr;357(4):527–534. doi: 10.1515/bchm2.1976.357.1.527. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980 Nov 10;255(21):10502–10508. [PubMed] [Google Scholar]
- Berger N. A., Sikorski G. W., Petzold S. J., Kurohara K. K. Association of poly(adenosine diphosphoribose) synthesis with DNA damage and repair in normal human lymphocytes. J Clin Invest. 1979 Jun;63(6):1164–1171. doi: 10.1172/JCI109410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creissen D., Shall S. Regulation of DNA ligase activity by poly(ADP-ribose). Nature. 1982 Mar 18;296(5854):271–272. doi: 10.1038/296271a0. [DOI] [PubMed] [Google Scholar]
- Durkacz B. W., Omidiji O., Gray D. A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980 Feb 7;283(5747):593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Gould H. J., Maryanka D., Fey S. J., Cowling G. J., Allan J. The assay of globin gene transcription in reconstituted chromatin. Methods Cell Biol. 1978;19:387–422. doi: 10.1016/s0091-679x(08)60038-2. [DOI] [PubMed] [Google Scholar]
- Herman T. M., DePamphilis M. L., Wassarman P. M. Structure of chromatin at deoxyribonucleic acid replication forks: location of the first nucleosomes on newly synthesized simian virus 40 deoxyribonucleic acid. Biochemistry. 1981 Feb 3;20(3):621–630. doi: 10.1021/bi00506a027. [DOI] [PubMed] [Google Scholar]
- Ito S., Shizuta Y., Hayaishi O. Purification and characterization of poly(ADP-ribose) synthetase from calf thymus. J Biol Chem. 1979 May 10;254(9):3647–3651. [PubMed] [Google Scholar]
- Johns E. W. Studies on histones. 7. Preparative methods for histone fractions from calf thymus. Biochem J. 1964 Jul;92(1):55–59. doi: 10.1042/bj0920055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez-Salinas H., Sims J. L., Jacobson M. K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979 Dec 13;282(5740):740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Nishizuka Y., Ueda K., Honjo T., Hayaishi O. Enzymic adenosine diphosphate ribosylation of histone and poly adenosine diphosphate ribose synthesis in rat liver nuclei. J Biol Chem. 1968 Jul 10;243(13):3765–3767. [PubMed] [Google Scholar]
- Ohgushi H., Yoshihara K., Kamiya T. Bovine thymus poly(adenosine diphosphate ribose) polymerase. Physical properties and binding to DNA. J Biol Chem. 1980 Jul 10;255(13):6205–6211. [PubMed] [Google Scholar]
- Oka J., Ueda K., Hayaishi O. Snake venom phosphodiesterase: simple purification with Blue Sepharose and its application to poly(ADP-ribose) study. Biochem Biophys Res Commun. 1978 Feb 28;80(4):841–848. doi: 10.1016/0006-291x(78)91321-9. [DOI] [PubMed] [Google Scholar]
- Okayama H., Ueda K., Hayaishi O. Purification of ADP-ribosylated nuclear proteins by covalent chromatography on dihydroxyboryl polyacrylamide beads and their characterization. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1111–1115. doi: 10.1073/pnas.75.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell M. R., Whish W. J. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980 Mar 1;185(3):775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. A method for the fractionation of the high-mobility-group non-histome chromosomal proteins. Biochem Biophys Res Commun. 1977 Oct 10;78(3):1034–1042. doi: 10.1016/0006-291x(77)90525-3. [DOI] [PubMed] [Google Scholar]
- Skidmore C. J., Davies M. I., Goodwin P. M., Halldorsson H., Lewis P. J., Shall S., Zia'ee A. A. The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by gamma-radiation and N-methyl-N-nitrosourea. Eur J Biochem. 1979 Nov 1;101(1):135–142. doi: 10.1111/j.1432-1033.1979.tb04225.x. [DOI] [PubMed] [Google Scholar]
- Sugimura T., Yoshimura N., Miwa M., Nagai H., Nagao M. Studies on poly(adenosine diphosphate-ribose). XI. Purification of poly(adenosine diphosphate-ribose) on a hydroxylapatite column. Arch Biochem Biophys. 1971 Dec;147(2):660–665. doi: 10.1016/0003-9861(71)90425-5. [DOI] [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Two DNA ligase activities from calf thymus. Biochem Biophys Res Commun. 1973 Aug 6;53(3):910–916. doi: 10.1016/0006-291x(73)90178-2. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Agemori M., Kameshita I., Nishikimi M., Shizuta Y. Participation of poly(ADP-ribosyl)ation in the depression of RNA synthesis caused by treatment of mouse lymphoma cells with methylnitrosourea. J Biol Chem. 1982 Apr 25;257(8):4027–4030. [PubMed] [Google Scholar]
- Teraoka H., Tamura S., Tsukada K. Evidence for a single species of DNA ligase localized in nuclei of rat liver. Biochim Biophys Acta. 1979 Jul 26;563(2):535–539. doi: 10.1016/0005-2787(79)90072-8. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Tsukada K. Eukaryotic DNA ligase. Purification and properties of the enzyme from bovine thymus, and immunochemical studies of the enzyme from animal tissues. J Biol Chem. 1982 May 10;257(9):4758–4763. [PubMed] [Google Scholar]
- Ueda K., Omachi A., Kawaichi M., Hayaishi O. Natural occurrence of poly(ADP-ribosyl) histones in rat liver. Proc Natl Acad Sci U S A. 1975 Jan;72(1):205–209. doi: 10.1073/pnas.72.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Jacquemin-Sablon A., Live T. R., Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J Biol Chem. 1968 Sep 10;243(17):4543–4555. [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]
- Zimmerman S. B., Levin C. J. DNA ligase activity in chromatin and its analogs. Rejoining of DNA strands in polylysine-DNA complexes and in reconstituted chromatins. Biochemistry. 1975 Apr 22;14(8):1671–1677. doi: 10.1021/bi00679a019. [DOI] [PubMed] [Google Scholar]


