Expression of the transcription factor Twist1 induces dissemination of normal mammary epithelial cells without changing the RNA levels of epithelial-specific genes such as E-cadherin.
Abstract
Dissemination of epithelial cells is a critical step in metastatic spread. Molecular models of dissemination focus on loss of E-cadherin or repression of cell adhesion through an epithelial to mesenchymal transition (EMT). We sought to define the minimum molecular events necessary to induce dissemination of cells out of primary murine mammary epithelium. Deletion of E-cadherin disrupted epithelial architecture and morphogenesis but only rarely resulted in dissemination. In contrast, expression of the EMT transcription factor Twist1 induced rapid dissemination of cytokeratin-positive epithelial cells. Twist1 induced dramatic transcriptional changes in extracellular compartment and cell–matrix adhesion genes but not in cell–cell adhesion genes. Surprisingly, we observed disseminating cells with membrane-localized E-cadherin and β-catenin, and E-cadherin knockdown strongly inhibited Twist1-induced single cell dissemination. Dissemination can therefore occur with retention of epithelial cell identity. The spread of cancer cells during metastasis could similarly involve activation of an epithelial motility program without requiring a transition from epithelial to mesenchymal character.
Introduction
Metastasis is the primary cause of death in breast cancer, and patient outcomes correlate negatively with the extent of metastatic spread at diagnosis (Bogenrieder and Herlyn, 2003; Polyak, 2010). Metastasis initiates with dissemination, the escape of epithelial cancer cells from the primary tumor into the surrounding stroma (Nguyen et al., 2009). As dissemination requires loss of epithelial cell–cell junctions, a change in the expression of intercellular adhesion genes could be the initiating event (Nelson, 2009; Polyak and Weinberg, 2009). Two related molecular models for dissemination have been proposed on this basis: genomic loss of cell adhesion genes (Hirohashi, 1998; Bogenrieder and Herlyn, 2003; Jeanes et al., 2008) and repression of cell adhesion genes through an epithelial to mesenchymal transition (EMT; Peinado et al., 2007; Yang and Weinberg, 2008).
These molecular models converge on the cell adhesion gene Epithelial cadherin (E-cad; Cdh1). E-cad is essential for early embryonic development (Larue et al., 1994); E-cad expression is frequently lost in human breast cancer (Berx et al., 1996); and E-cad loss in experimental cancer models accelerates metastatic progression (Derksen et al., 2006; Onder et al., 2008). However, a gap exists in our understanding of the relationship between the normal role of E-cad in adult tissues and its function during cancer metastasis. Analyses of E-cad’s requirement in adult epithelial tissues using Cre-mediated deletion (Boussadia et al., 2002) have revealed highly varied E-cad null phenotypes. Conditional E-cad deletion in the mammary gland results in cell death during lactation (Boussadia et al., 2002), whereas conditional E-cad deletion in the skin results in hyperproliferation of some cell types and premature degeneration of others (Tinkle et al., 2004). E-cad deletion in these developmental contexts is not associated with systemic dissemination. Nevertheless, transcriptional repression of E-cad by EMT transcription factors such as Twist1 remains a central concept in cancer metastasis (Peinado et al., 2007; Yang and Weinberg, 2008). Twist1 regulates metastasis in a mouse mammary tumor model (Yang et al., 2004), and its expression is up-regulated in both invasive lobular and invasive ductal breast cancer (Yang et al., 2004; Mironchik et al., 2005).
Collectively, previous publications have demonstrated that E-cad functions as an invasion suppressor and that induction of EMT transcription factors can accelerate malignant progression (Hirohashi, 1998; Berx and Van Roy, 2001; Bogenrieder and Herlyn, 2003; Derksen et al., 2006; Yang and Weinberg, 2008; Polyak and Weinberg, 2009). However, human breast tumors typically contain thousands of mutations in both signaling and structural genes (Wood et al., 2007; Stephens et al., 2009). These coexisting mutations obscure the contribution of individual genetic events to discrete steps in the metastatic cascade. Specifically, the presence of additional mutations in cancer cell lines has made it difficult to distinguish the individual sufficiency of E-cad deletion or Twist1 expression for dissemination. Importantly, dissemination can be induced in developmental contexts such as neural crest migration, suggesting that its molecular regulation may be distinct from other aspects of neoplasia (Barrallo-Gimeno and Nieto, 2005).
We sought to define the minimum molecular perturbations necessary to induce dissemination of normal mammary epithelial cells. To accomplish these goals, we used a combination of organotypic culture, Cre-lox–based genetic models, inducible expression systems, lentiviral gene knockdown (KD), and time-lapse imaging to test the sufficiency of E-cad deletion or Twist1 expression to induce dissemination in primary mammary epithelium. Our results demonstrate that E-cad is required for simple epithelial architecture and branching morphogenesis, but its loss is not associated with significant dissemination in 3D culture or in vivo. In contrast, Twist1 expression induces rapid epithelial dissemination. Moreover, Twist1-induced dissemination occurs without loss of epithelial gene expression and requires E-cad.
Results
E-cad is required for simple mammary epithelial architecture
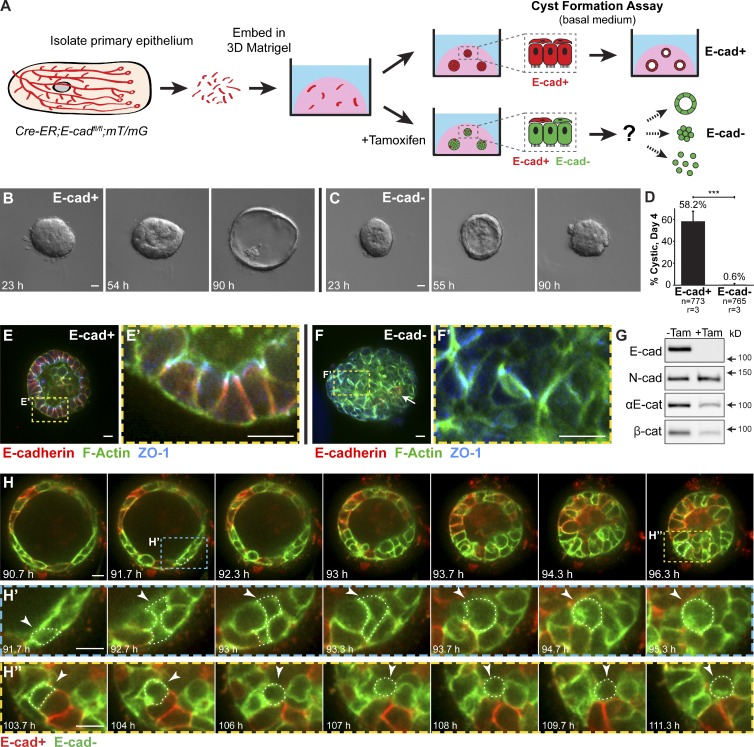
We first assayed the acute consequences of E-cad deletion in primary mammary epithelial cells using the “organoid” assay, in which mammary ducts are explanted into 3D ECM (Matrigel; Ewald et al., 2008; Fig. 1 A). In basal medium without growth factors, normal organoids form polarized cysts (Ewald et al., 2008; Fig. 1, A and B). We isolated organoids from mice carrying floxed E-cad alleles (Boussadia et al., 2002) and a ubiquitously expressed, tamoxifen-inducible Cre recombinase (Badea et al., 2003; Cre-ER;E-cadfl/fl mice). Organoids from the same mouse were divided into a control group and an experimental group in which E-cad deletion was induced with 50 nM tamoxifen.
Figure 1.
E-cad deletion induced loss of simple epithelial architecture. (A) E-cad deletion was induced in half of Cre-ER;E-cadfl/fl organoids with tamoxifen. (B) Control, E-cad+ organoids (−Tam) formed cysts. (C) E-cad− organoids (+Tam) failed to form cysts (28/42 movies across three biological replicates) or transiently established and then lost lumens (14/42 movies). (D) E-cad deletion blocked cyst formation. n, total number of organoids; r, number of biological replicates. Error bars indicate SD. ***, P = 0.0004, two-tailed Student’s t test with equal variance. (E) Control organoids formed cysts with enrichment of E-cad and ZO-1 along apicolateral membranes (E’). (F) E-cad− organoids were multilayered, lacked E-cad immunoreactivity, and displayed abnormal ZO-1 localization. Arrow indicates rare E-cad+ cells. (G) By Western blot, E-cad deletion (+Tam) resulted in complete loss of E-cad protein and significant reductions in αE-catenin and β-catenin (see also Fig. S1, A and B). Whole cell lysate samples were loaded for equal protein based on BCA analysis. (H) The Cre biosensor mT/mG was used to observe E-cad− cell behaviors by confocal microscopy (Video 1). Cre+, E-cad− cells (green) changed shape, from columnar to round, before shifting apically (H’ and H’’, arrowheads). Gamma adjustments were performed in E and F to improve image clarity. Bars: (B and C) 20 µm; (E, F, and H) 10 µm.
Control organoids efficiently formed polarized cysts, with E-cad localized to lateral points of cell–cell contact and zona occludens 1 (ZO-1) localized in apical puncta (58.2%; Fig. 1, B, D, E, and E′). In contrast, E-cad− organoids either failed to establish simple epithelial architecture (66%) or transiently established and then lost simple epithelial architecture (33%; Fig. 1, C and D). E-cad− organoids had a multilayered organization and lacked morphologically evident lumens (Fig. 1, E and F). Interestingly, a subset of lateral cell–cell contact surfaces within E-cad− organoids had continuous ZO-1 staining (Fig. 1, F and F′). Immunofluorescence (Fig. 1 F) and Western blotting (Fig. 1 G) confirmed loss of E-cad protein by day 6 in culture. E-cad loss coincided with a reduction in αE-catenin and β-catenin (Fig. 1 G) but not with a significant change in neural cadherin (N-cad; Fig. S1, A and B). By immunofluorescence, αE-catenin was absent from most internal cell–cell contact surfaces (Fig. S1, C–C″).
E-cad− cells are excluded from regions of simple epithelial organization but rarely disseminate
We next used time-lapse microscopy to observe the cell dynamics driving loss of simple organization after E-cad deletion. We monitored recombined cells using the genetically encoded Cre biosensor mT/mG (Muzumdar et al., 2007). Without Cre activity, all cells in mT/mG mice express a membrane-localized red fluorescent protein. Cre activity excises the red gene and induces heritable expression of a membrane-localized green fluorescent protein. We could thereby distinguish in real-time the behaviors of E-cad+ (red) versus E-cad− (green) cells. Loss of simple epithelial architecture in E-cad− organoids correlated with a reduction in luminal volume (Fig. 1 H and Video 1), consistent with a loss of tight junctions. This tissue-level change was accompanied by a change in E-cad− cell shape from simple columnar to round (Fig. 1, H′ and H″). Round cells shifted internally, inducing a transition from a single to multiple luminal epithelial cell layers (Fig. 1, H′ and H″, arrowheads).
We next monitored whether E-cad deletion was sufficient to induce dissemination into the ECM. Normal epithelial cysts maintain a smooth basal surface with the ECM (Fig. 2 A). Conversely, E-cad− cells accumulated on the basal surface and collectively migrated into Matrigel as single file columns (Fig. 2, B, B′, G, and G′) and as disorganized masses (Fig. 2, E, E′, F, and F′). Single file migration initiated from basally positioned E-cad− cells that rounded up but maintained cell–cell contact (Fig. 2 C). As additional cells changed shape, the initiating cell migrated further into the ECM and led a column of closely connected E-cad− cells. However, despite contact with the ECM and high motility, E-cad− cells rarely disseminated into the matrix. Each organoid consists of 300–500 cells; yet, on average, only one cell disseminated from each E-cad− organoid (n = 206 organoids imaged by time-lapse microscopy across nine biologically independent replicates). Most E-cad− cells remained adherent to other epithelial cells. The few E-cad− cells that did disseminate were rounded, migrated minimally, and had no detectable membrane protrusions.
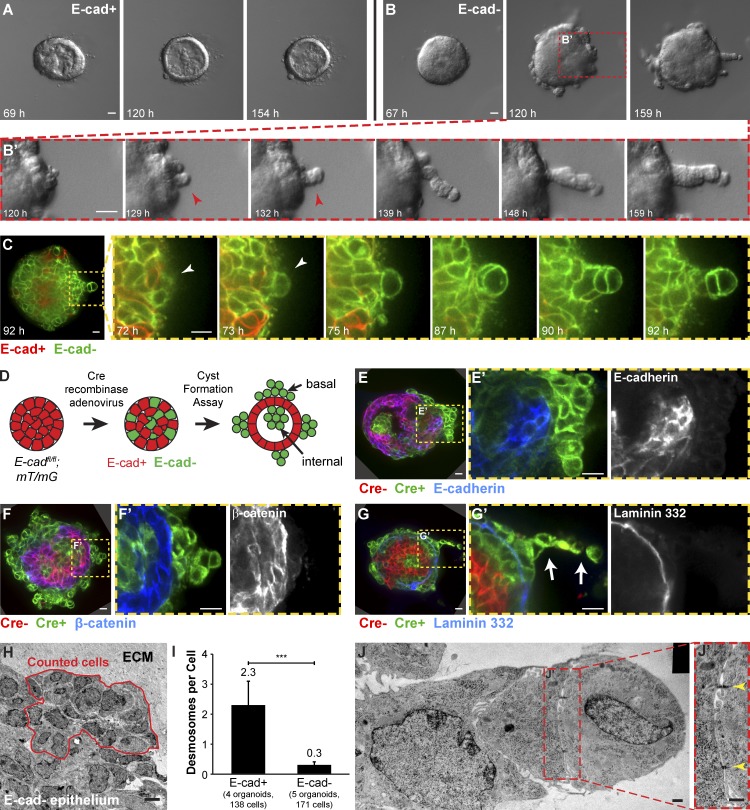
Figure 2.
E-cad− cells displayed reductions in multiple classes of intercellular junctions. Cre-ER;E-cadfl/+ and Cre-ER;E-cadfl/fl organoids were isolated, and widespread recombination was induced with tamoxifen. (A) Control organoids (E-cad+) maintained a smooth basal epithelial border. (B) E-cad− organoids collectively migrated into Matrigel as single file columns (B’, red arrowhead). (C) Basally positioned E-cad− cells (white arrowhead) were observed to round up and initiate single file cell columns. (D) Adeno-Cre was used to generate genetic mosaic organoids with a mixture of E-cad+ and E-cad− cells. (E and F) Green, Cre+ cells reliably lacked E-cad (E and E’) and β-catenin (F and F’). (G) Basally positioned E-cad− cells were located beyond the basement membrane protein laminin 332. Arrows in G′ indicate a single file column. (H) TEM was used to quantify desmosomes in Adeno-Cre–transduced E-cadfl/+ (control) and E-cadfl/fl organoids. Red outline indicates a representative region used for analysis. (I) E-cad− epithelium had significantly fewer desmosomes compared with control E-cad+ epithelium. Error bars indicate SD. ***, P = 0.0008; two-tailed Student’s t test with equal variance. (J) Small desmosomes (J’, yellow arrowheads) were detected connecting cells in single file columns. Gamma adjustments were performed in E and F to improve image clarity. Bars: (A and B) 20 µm; (C and E–G) 10 µm; (H) 5 µm; (J) 0.5 µm. The black rectangle in the top right corner of J is a region of the image mosaic where no pixels were collected.
E-cad− cells remain adherent despite reductions in multiple classes of intercellular junctions
To analyze the effects of mosaic E-cad loss, we used adenovirally delivered Cre (Adeno-Cre) to induce E-cad deletion in a subset of cells within E-cadfl/fl;mT/mG organoids (Fig. 2 D). We confirmed that green, Cre+ cells stained negative for E-cad protein (Fig. 2, E and E′) and that loss of E-cad was accompanied by loss of β-catenin from the membrane (Fig. 2, F and F′). In these genetic mosaic organoids, E-cad− cells were observed both basally at the cell–ECM interface and in interior positions apical to polarized E-cad+ cells (Fig. 2, D–F). The basally positioned E-cad− cells were frequently observed past the cell-derived basement membrane (laminin 332 immunofluorescence; Fig. 2, G and G′). E-cad+ cells within the same organoid localized β-catenin to points of cell–cell contact and were inside the basement membrane (Fig. 2, E–G).
We next sought to identify adhesion systems that could allow epithelial cells to remain adherent despite loss of E-cad and membrane-localized β-catenin. Desmosomes represent a major class of intercellular junctions in mammary epithelial cells (Bissell and Bilder, 2003). However, E-cad inhibition can induce reductions in both desmosomes and tight junctions (Gumbiner et al., 1988). We therefore used transmission electron microscopy (TEM) to quantify the effect of E-cad loss on intercellular junctions in organoids from E-cadfl/fl and E-cadfl/+ littermates. We induced recombination with Adeno-Cre and focused our analysis on basally positioned cells (Fig. 2 H). E-cad− cells typically lacked both punctate ZO-1 immunoreactivity (Fig. 1 F) and ultrastructurally identifiable tight junctions. Compared with normal E-cad+ epithelium, E-cad− epithelium also had a statistically significant, almost eightfold reduction in desmosomes (Fig. 2 I). However, we still detected small desmosomes connecting E-cad− cells, even within single file migration columns (Fig. 2, J and J′). Collectively, our data reveal that loss of E-cad results in loss of simple epithelial architecture, reductions in multiple classes of cell–cell junctions, and both apical and basal exclusion of E-cad− cells from E-cad+ simple epithelium. However, E-cad loss was not sufficient for robust single cell dissemination into the 3D ECM.
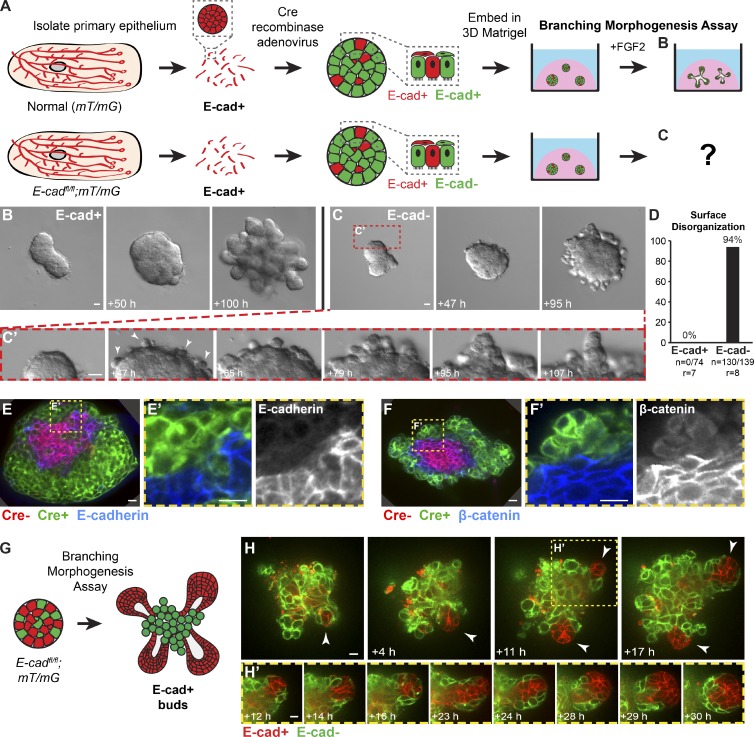
E-cad is required for branching morphogenesis in 3D culture
Normal mammary branching morphogenesis begins with a transition from simple to multilayered architecture and a concurrent reduction in apicobasal polarity and intercellular junctions (Ewald et al., 2008, 2012). Therefore, it was plausible that E-cad− cells could participate in branching morphogenesis. We thus assayed the effects of E-cad deletion under culture conditions that use FGF2 to induce branching morphogenesis (Ewald et al., 2008; Fig. 3 A).
Figure 3.
Loss of E-cad inhibited branching morphogenesis and induced epithelial disorganization in 3D culture. (A) E-cad+/+;mT/mG and E-cadfl/fl;mT/mG organoids were isolated, recombination was induced with Adeno-Cre, and branching morphogenesis was induced with FGF2. (B) E-cad+ organoids completed branching morphogenesis (Video 2). (C) E-cad− organoids developed a disorganized basal surface composed of rounded cells (C’; Video 2). (D) This disorganized surface morphology was observed in 94% of E-cad− organoids. n, number of time-lapse movies; r, number of biological replicates. (E and F) Basally positioned cells were green (Cre+), E-cad− (E), and β-catenin− (F). (G and H) In genetic mosaic organoids with a mixture of E-cad+ and E-cad− cells, E-cad+ cells (red) were observed to initiate new buds (arrowheads; H’; Video 3). Bars: (B and C) 20 µm; (E, F, and H) 10 µm.
We first induced E-cad deletion in most epithelial cells using Adeno-Cre and monitored effects on branching by time-lapse microscopy. Control E-cad+ organoids underwent normal branching morphogenesis (Fig. 3 B and Video 2). Specifically, they initiated and elongated numerous mammary buds and maintained a smooth border with the ECM. In contrast, E-cad− organoids did not undergo branching morphogenesis and instead rapidly developed a disorganized and uneven basal epithelial surface (Fig. 3, C and D; and Video 2). Cells at the basal ECM border were rounded and displayed extensive, uncoordinated motility (Fig. 3 C′). These cells were Cre biosensor+ (green) and E-cad− by immunofluorescence (Fig. 3, E and E′) and lacked β-catenin at points of cell–cell contact (Fig. 3, F and F′).
We next induced genetic mosaic E-cad deletion and observed some normal, smooth epithelial buds emerging from disorganized cell surfaces. We hypothesized that these buds were composed of E-cad+ cells that had escaped recombination (Fig. 3 G). Consistent with this model, we observed groups of red, E-cad+ cells coalesce and initiate new buds from within large disorganized groups of green, E-cad− cells (Fig. 3, H and H′; and Video 3). Our data demonstrate that E-cad− cells remain motile and adherent but fail to incorporate into epithelial buds. Furthermore, in genetic mosaic mixtures, E-cad+ cells can initiate buds from predominantly E-cad− organoids (Fig. 3, G and H).
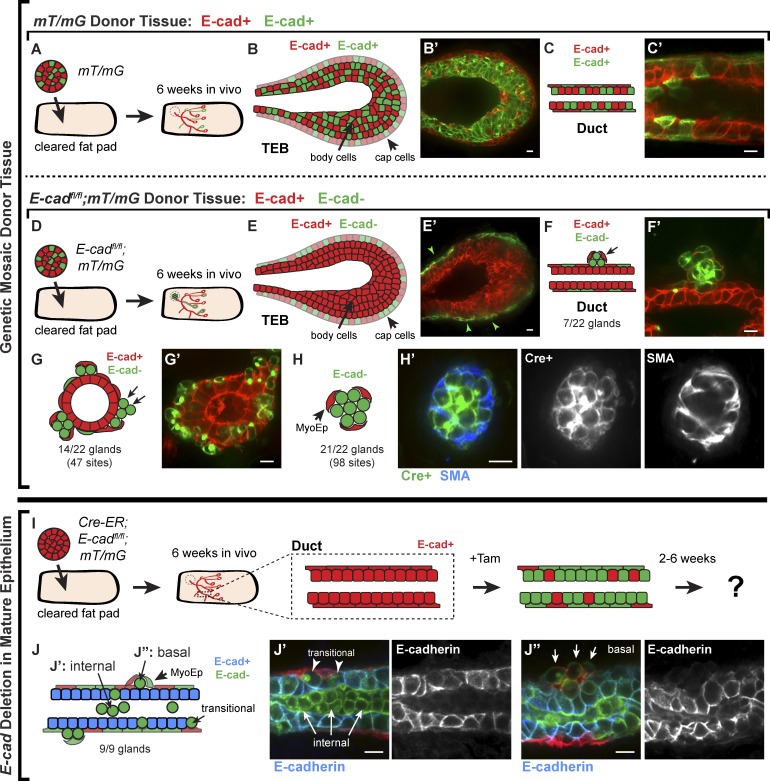
E-cad− cells are excluded from polarized ducts and the body cell compartment of the terminal end bud (TEB) in vivo
Our genetic mosaic analysis in 3D culture revealed that E-cad− cells lost simple epithelial architecture and most intercellular junctions but remained adherent to each other. However, the presence of E-cad− cells in an organoid did not prevent the initiation of E-cad+ epithelial buds. Accordingly, we predicted that, in vivo, genetic mosaic E-cad deletion would result in exclusion of E-cad− cells from regions of active branching morphogenesis and accumulation of disorganized E-cad− cell groups both apically and basally. To test this prediction, we isolated organoids from control E-cad+/+;mT/mG mice and from E-cadfl/fl;mT/mG mice, induced mosaic recombination with Adeno-Cre, and transplanted the organoids into contralateral, cleared mammary fat pads of 3-wk-old NOD/SCID mice (Fig. 4, A and D, respectively). Glands were harvested and analyzed 6 wk after transplantation.
Figure 4.
E-cad− cells were excluded from polarized ducts and the body cell compartment of the TEB in vivo. (A) Adeno-Cre–transduced mT/mG organoids (E-cad+) were transplanted into cleared mammary fat pads, and glands were harvested after 6 wk. (B and C) Both TEBs (B) and polarized ducts (C) contained a mixture of red and green cells in the luminal and myoepithelial cell layers. (D) Adeno-Cre–transduced E-cadfl/fl;mT/mG organoids were transplanted into contralateral no. 4 glands. (E) E-cad− luminal cells were markedly excluded from the body cell region of the TEB in ductal outgrowths. Arrowheads indicate green cells in the cap cell layer. (F and G) E-cad− luminal cells were observed on the basal surfaces of polarized epithelium in the gland periphery (F) and near the injection site (G). (H) E-cad− cells were also observed in disorganized clusters surrounded by myoepithelial cells (H’, SMA+). (I) Cre-ER;E-cadfl/fl;mT/mG organoids (E-cad+) were transplanted into cleared mammary fat pads and allowed to grow out for 6 wk. Tamoxifen was injected to induce E-cad deletion, and glands were harvested after 2–6 wk. (J) E-cad− cells were observed in the lumens (J’) and on the basal surfaces (J’’) of E-cad+ polarized ducts (nine glands). Bars, 10 µm.
Mammary ducts during puberty are elongated by specialized epithelial structures known as TEBs (Williams and Daniel, 1983). TEBs are composed of a single, basally positioned layer of cap cells and multiple, apically positioned body cell layers (Hinck and Silberstein, 2005). Cap cells give rise to myoepithelial lineages, whereas body cells give rise to luminal epithelial lineages. Only body cells express E-cad (Daniel et al., 1995). Ductal outgrowths from control, genetic mosaic mT/mG transplants had both red and green cells in the body and cap cell regions of the TEB (Fig. 4, B and B′) and in the luminal and myoepithelial cell layers of polarized ducts (Fig. 4, C and C′). In contrast, outgrowths from genetic mosaic E-cadfl/fl;mT/mG transplants displayed a striking exclusion of E-cad− cells from the body cell region of the TEB (Fig. 4, E and E′) and from the luminal layer of ducts (Fig. 4, F and F′). Myoepithelial cells were red and green in E-cadfl/fl;mT/mG genetic mosaic outgrowths (Fig. 4 E′, green arrowheads), but myoepithelial cells express P-cadherin instead of E-cad (Daniel et al., 1995).
Despite their exclusion from polarized ducts and body cells, E-cad− cells were observed in vivo at 6 wk after transplantation. Groups of E-cad− cells were detected on the basal surfaces of polarized E-cad+ ducts in the gland periphery (Fig. 4, F and F′) and on the basal surfaces of polarized E-cad+ epithelium near the transplantation site (Fig. 4, G and G′). We also observed small clusters of exclusively E-cad− cells, surrounded by myoepithelial cells (smooth muscle actin+ [SMA]; Fig. 4, H and H′). Consistent with our 3D culture data, green, Cre+ cells in vivo were validated by antibody staining to lack membrane-localized E-cad (Fig. S1, D–F) and β-catenin (Fig. S1G).
We next tested the in vivo consequences of E-cad deletion in polarized mammary epithelium. We isolated and transplanted organoids from Cre-ER;E-cadfl/fl;mT/mG mice into cleared mammary fat pads, waited 6 wk for mature ductal outgrowths, and induced E-cad deletion by tamoxifen injection (Fig. 4 I). Glands were harvested and analyzed 2–6 wk after tamoxifen injection. E-cad− cells (by immunofluorescence) were observed apically inside duct lumens (Fig. 4, J and J′) and basally as single cells or disorganized groups on duct surfaces (Fig. 4, J and J″). Basally positioned E-cad− cells were typically still surrounded by myoepithelial cells.
We conclude that E-cad is required in mammary epithelial cells to establish and maintain simple epithelial architecture and to initiate and elongate new buds, both in 3D culture and in vivo. E-cad− cells are viable in both contexts but extrude apically and basally from polarized epithelium and fail to contribute to mammary branching morphogenesis in the luminal cell compartment.
Twist1 induces robust cell dissemination of otherwise normal primary epithelial cells
Our data reveal that E-cad loss is not sufficient for robust cell dissemination in 3D culture or in vivo. This conclusion has implications for our conceptual understanding of EMT, as repression of E-cad is considered a core effector of EMT (Yang et al., 2004; Peinado et al., 2007; Vesuna et al., 2008; Thiery et al., 2009). The bHLH transcription factor Twist1 has emerged as a candidate regulator of EMT in cancer and is thought to act through regulation of E-cad (Yang et al., 2004; Vesuna et al., 2008; Yang and Weinberg, 2008). We sought to test the hypothesis that acute expression of Twist1 would induce epithelial dissemination.
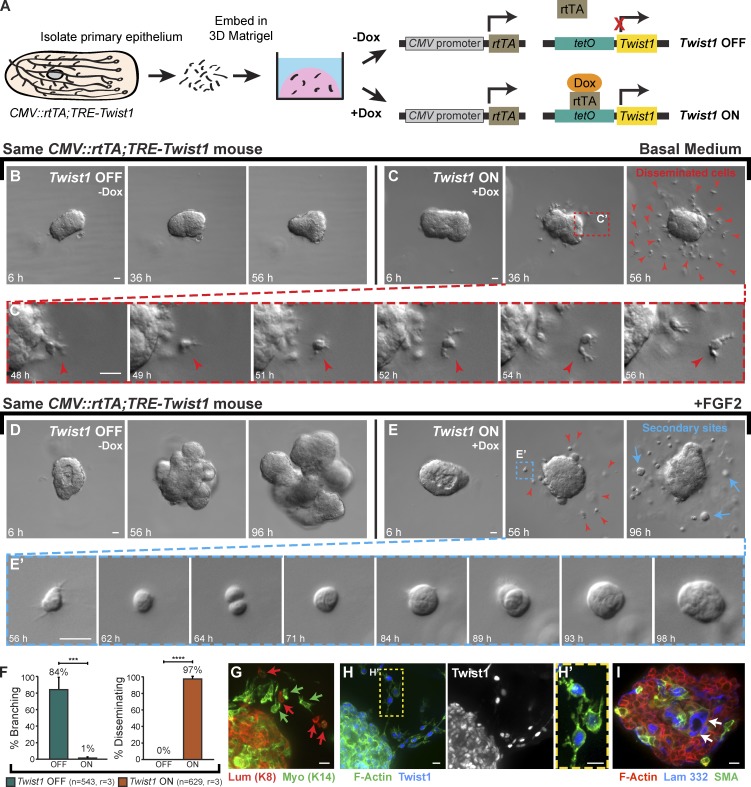
We isolated organoids from mice carrying a ubiquitously expressed reverse tetracycline transactivator (rtTA) and a Tet-responsive Twist1 allele (Fig. 5 A, CMV::rtTA;TRE-Twist1; Tran et al., 2012). In basal medium, control organoids maintained a smooth basal surface (Fig. 5 B). In contrast, Twist1 expression induced rapid dissemination of protrusive, individual cells out of the epithelium as early as 24 h after Twist1 induction (Fig. 5, C and C′; and Video 4). We next tested the relationship between branching morphogenesis and dissemination. In FGF2-containing medium, control organoids branched efficiently (Fig. 5 D). In contrast, Twist1 expression inhibited FGF2-induced branching morphogenesis in 99% of organoids and induced robust dissemination in 97% of organoids (Fig. 5, D–F; and Video 5). In the presence of FGF2, disseminated cells proliferated to form secondary epithelial sites within the ECM (Fig. 5 E′). Immunofluorescent staining for cell type–specific cytokeratins revealed that both luminal epithelial (K8+) and myoepithelial (K14+) cells disseminated in response to Twist1 induction (Fig. 5 G). Interestingly, both cells within the epithelial structure and disseminated cells dislayed nuclear Twist1 immunoreactivity (Fig. 5, H and H′). The Twist1+ epithelial group displayed abnormal internal localization of myoepithelial cells (SMA+) and basement membrane (laminin 332) and corresponding gaps in basal myoepithelial and basement membrane coverage (Fig. 5 I).
Figure 5.
Twist1 induced robust dissemination of normal epithelial cells. (A) Organoids were isolated from CMV::rtTA;TRE-Twist1 mice, and Twist1 was induced in half of the organoids with doxycycline. (B and C) In basal medium, control organoids maintained epithelial organization (B), whereas Twist1 expression induced robust dissemination (C; Video 4). Disseminating cells (red arrowheads) migrated away from the epithelium with extensive protrusions (C’). (D and E) In FGF2-containing medium, control organoids completed branching morphogenesis (D), whereas Twist1 expression blocked branching and induced robust dissemination (E; Video 5). Red arrowheads in E indicate disseminated cells. With FGF2, disseminated cells proliferated to form secondary epithelial sites (blue arrows; E’; 6/9 biological replicates). (F) Less than 1% of Twist1+ organoids branched (***, P = 0.0006, two-tailed Student’s t test with equal variance), whereas 97% disseminated (****, P = 4 × 10−7, two-tailed Student’s t test with equal variance). n, total number of organoids; r, number of biological replicates. Error bars indicate SD. (G) Both luminal (K8+; red arrows) and myoepithelial (K14+; green arrows) cells disseminated. (H) Both disseminated single cells and cells within the main epithelial group were Twist1+. (I) Myoepithelial cells (SMA+) and basement membrane (laminin 332; arrows) were inappropriately localized to the organoid interior. Gamma adjustments were performed in H, H′, and I to improve image clarity. Bars: (B–E) 20 µm; (G–I) 10 µm.
Epithelial cell behaviors are restored when Twist1 expression is turned off
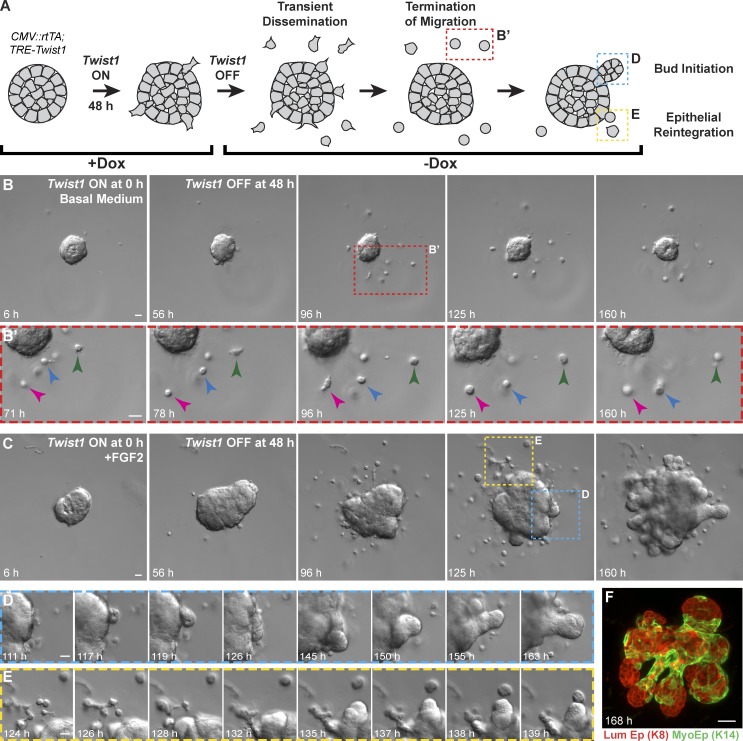
We next tested the consequences of transient expression of Twist1. We induced Twist1 for 48 h and then turned off Twist1 by removing doxycycline (Fig. 6 A). In basal medium, disseminated single cells stopped migrating within 48 h of doxycycline removal (Fig. 6, B and B′; and Video 6). In FGF2-containing medium, organoids initiated branching morphogenesis within 70 h of doxycycline removal (Fig. 6, C and D; and Video 7), and disseminated single cells were observed reintegrating with the epithelial group (Fig. 6 E). Remarkably, by day 7, the resulting branched structures had normalized epithelial organization, with internal luminal cells and basally positioned myoepithelial cells (Fig. 6 F). We conclude that epithelial cells can rapidly reestablish normal developmental programs, such as branching morphogenesis, when Twist1 expression ceases.
Figure 6.
Organoids recovered epithelial behaviors when Twist1 was turned off. (A) Twist1 was transiently activated in CMV::rtTA;TRE-Twist1 epithelium by a 48-h pulse of doxycycline. (B) In basal medium, organoids transiently disseminated, but disseminated cells stopped migrating after doxycycline removal (arrowheads; Video 6). (C–E) In the presence of FGF2, organoids initiated new buds after doxycycline removal (D), and disseminated cells reintegrated with the main organoid (E; Video 7). (F) Branched organoids displayed normal mammary epithelial organization, with inner luminal epithelial cells (K8+) and outer myoepithelial cells (K14+). Bars, 20 µm.
Twist1-induced dissemination is cell autonomous
We demonstrated that ubiquitous Twist1 activation induced rapid epithelial dissemination. We next sought to test whether a single Twist1+ cell could escape a mostly Twist1− epithelium. We reasoned that Twist1− cells could serve as a barrier to dissemination. Alternatively, Twist1+ cells could induce the dissemination of neighboring Twist1− cells. To achieve mosaic activation of Twist1, we used a Lox-Stop-Lox-rtTA (R26::LSL-rtTA [Belteki et al., 2005]) and varying titers of Adeno-Cre to modulate the fraction of cells capable of activating Twist1. We again used the mT/mG reporter (Muzumdar et al., 2007) to distinguish between Cre+rtTA+ (green) and Cre−rtTA− (red) cells. We monitored the resulting genetic mosaic tissue for dissemination in our branching morphogenesis assay (Fig. S2 A).
Without doxycycline or Twist1 induction, organoids formed branched structures with a mix of red and green cells (Fig. S2 B). Conversely, Twist1 induction resulted in dissemination of green (Cre+rtTA+Twist1+) cells across a wide range of viral titers, even in organoids in which most cells were Cre−rtTA−Twist1− (Fig. S2, C and D). Epithelium that was mostly Twist1− had a high frequency of branching morphogenesis despite the dissemination of Twist1+ cells (Fig. S2 E). As the fraction of rtTA+Twist1+ cells per organoid increased, we observed a decrease in the percentage of branching organoids (Fig. S2 E), consistent with the branching inhibition observed in epithelium with constitutive Twist1 expression (Fig. 5 F). We never observed dissemination of red, rtTA− cells, indicating that Twist1-induced dissemination is cell autonomous.
Twist1 is sufficient to induce local dissemination in vivo
Our 3D culture data predicted that Twist1 would induce epithelial dissemination in vivo. To test this prediction, we transplanted genetic mosaic organoids containing a mixture of red, Twist1− and green, Twist1+ cells into cleared mammary fat pads of 3-wk-old NOD/SCID mice (Fig. S2 F). Twist1 was induced in culture and maintained in vivo using doxycycline feed. Consistent with our 3D culture data, we observed local dissemination of green, Twist1+ cells into the surrounding stroma (Fig. S2, G–G″). We did not detect dissemination of red, Twist1− cells. Importantly, we observed groups of 10–20 green, Twist1+ epithelial cells in the stroma (Fig. S2, H and H′). We hypothesize that these groups represent secondary epithelial sites formed from disseminated Twist1+ cells. We conclude that Twist1 expression is sufficient for epithelial dissemination in 3D culture and in vivo.
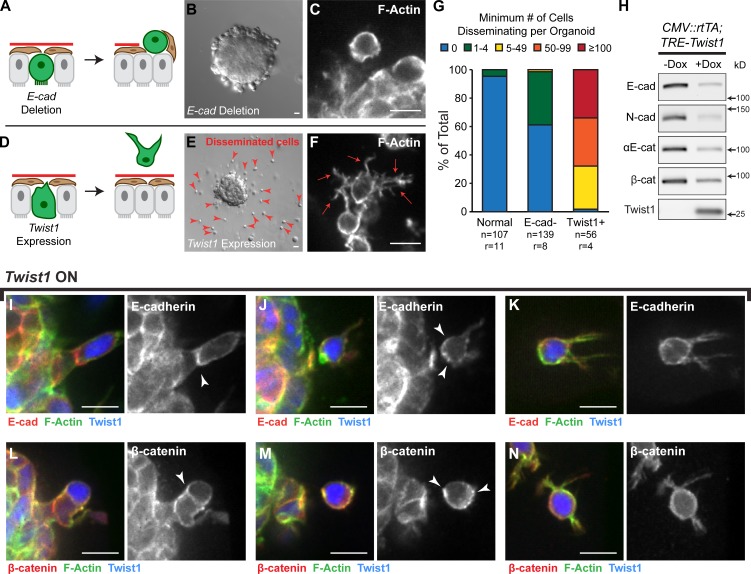
Twist1 induces dissemination without complete loss of adherens junction components
Our genetic analyses revealed that loss of E-cad and expression of Twist1 induced distinct cell behaviors and tissue-level phenotypes. E-cad deletion resulted in loss of polarized epithelial architecture, whereas Twist1 expression induced dissemination of luminal and myoepithelial cells (Fig. 7, A–G). We next used immunoblotting to compare levels of cell–cell adhesion proteins between normal (−Dox) and Twist1-expressing (+Dox) tissue. We observed significant reductions in E-cad, αE-catenin, β-catenin, and N-cad (Fig. 7 H and Fig. S3, A and B) but detected protein in all cases. This reduction in N-cad is inconsistent with a cadherin switch model in which increases in N-cad levels induce migration (Nieman et al., 1999). We next used immunofluorescence to localize E-cad and β-catenin during dissemination. Consistent with our Western blot results, E-cad staining appeared weaker in cells expressing Twist1. However, we observed E-cad and β-catenin localized at points of cell–cell contact in cells before dissemination (Fig. 7, I and L); E-cad and β-catenin localized to the rear of recently disseminated cells (Fig. 7, J and M); and E-cad and β-catenin localized to the membranes of cells migrating in the ECM (Fig. 7, K and N). We also observed E-cad− cells in the matrix, consistent with the dissemination of K14+ myoepithelial cells, which would not normally express E-cad. We conclude that Twist1 can induce dissemination of cells with membrane-localized adherens junction proteins.
Figure 7.
Twist1 induced single cell dissemination despite membrane-localized adherens junction proteins. (A and B) E-cad deletion blocks branching and induces epithelial disorganization. (C) Rare E-cad− disseminated cells maintain a rounded morphology. (D and E) Twist1 expression blocks branching and induces single cell dissemination (red arrowheads). (F) Disseminated Twist1+ cells exhibit extensive actin-rich protrusions (red arrows). (G) In time-lapse movies, E-cad− cells were only rarely observed to disseminate. In contrast, >100 cells per Twist1+ organoid were routinely observed to disseminate. n, number of time-lapse movies; r, number of biological replicates. (H) By Western blot, Twist1 expression resulted in reductions in protein levels of E-cad, N-cad, αE-catenin, and β-catenin (see also Fig. S3, A and B). Whole cell lysate samples were loaded for equal protein based on BCA analysis. (I–N) Membrane-localized E-cad and β-catenin (arrowheads) were detected in basally positioned cells protruding into the ECM (I and L), in cells that had just disseminated (J and M), and in disseminated cells migrating through the ECM (K and N). Bars: (B and E) 20 µm; (C, F, and I–N) 10 µm.
Twist1 induces transcriptional changes in cell–matrix adhesion but does not fundamentally alter epithelial identity
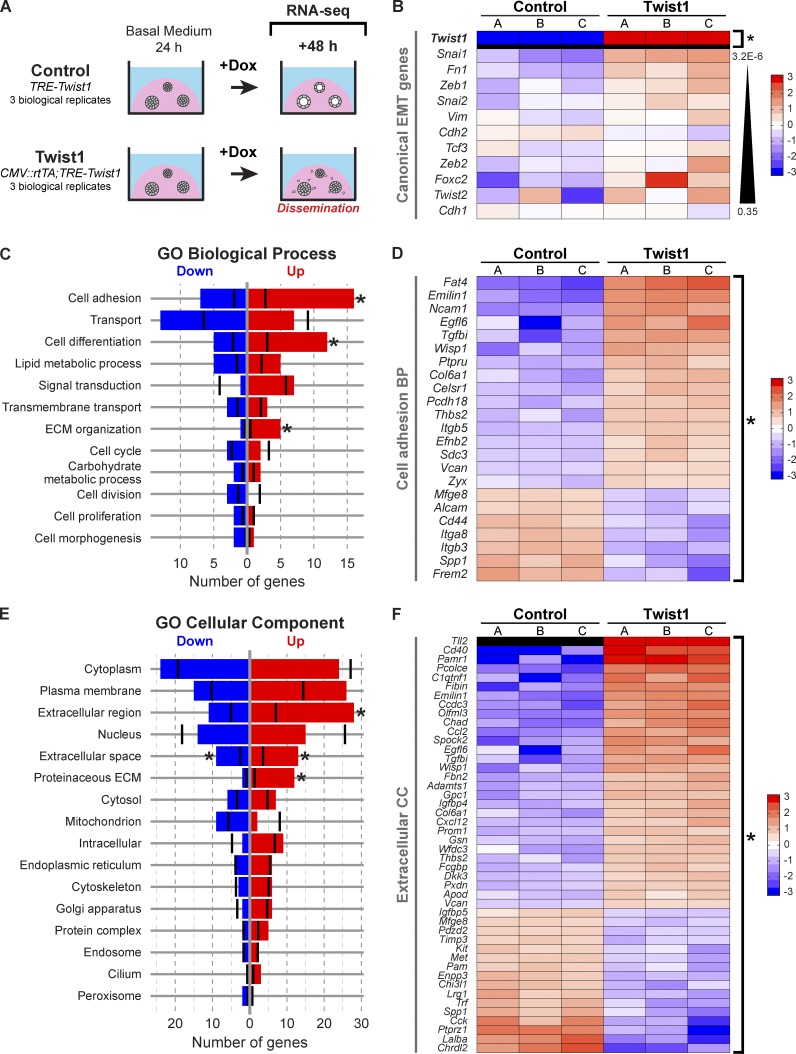
We next sought to identify the early transcriptional changes downstream of Twist1. We isolated organoids from three CMV::rtTA;TRE-Twist1 mice and three rtTA− littermate controls (TRE-Twist1), cultured these organoids for 24 h in basal medium, added doxycycline for 48 h, and then extracted RNA during active dissemination (Fig. 8 A). RNA-seq analysis identified 183 genes that were differentially expressed between control and Twist1-expressing tissue at genome-wide significance (P = 2.7 × 10−6; Table S1).
Figure 8.
Twist1 induced changes in genes regulating cell–ECM interactions and the extracellular space. (A) RNA-seq was used to compare gene expression 48 h after Twist1 induction in control versus Twist1+ organoids. (B) Heat map of canonical EMT genes. Only Twist1 was significantly differentially expressed. Genes are sorted by increasing p-value. (C–F) The 183 DE genes were mapped to direct associations with GO Slim biological process (C) and cellular component (E) terms. Black vertical bars indicate the expected number of DE genes per category. Asterisks specify significantly enriched terms. (D) DE genes associated with cell adhesion. (F) DE genes associated with extracellular space, extracellular region, and proteinaceous ECM. Genes are sorted by descending fold change in D and F. BP, biological process; CC, cellular component.
Surprisingly, none of the canonical EMT genes were significantly differentially expressed at the RNA level, including E-cad (P = 0.35). However, both Snai1 (P = 3.2 × 10−6) and Fn1 (P = 4.6 × 10−6) were close to genome-wide significance (Fig. 8 B). We next analyzed the expression of 127 genes involved in cell–cell adhesion and intercellular junctions (Table S2). Of these genes, only five were differentially expressed (Celsr1, Cldn2, Fat4, Frem2, and Pcdh18). No classical cadherins, desmosomal cadherins, catenins, or cytokeratins were significantly dysregulated at the RNA level. We conclude that Twist1 induces dissemination without loss of epithelial identity. This observation is consistent with the positive immunoreactivity for cytokeratin, E-cad, and β-catenin protein in disseminated cells.
We next analyzed gene ontology (GO) slim annotations to characterize biological process (Fig. 8 C), cellular component (Fig. 8 E), and molecular function terms (Fig. S5A) associated with our 183 differentially expressed (DE) genes (Ashburner et al., 2000). Relevant GO biological processes with several DE genes included cell adhesion (Fig. 8 D), transport (Fig. S4 A), cell differentiation (Fig. S4 B), lipid metabolic process (Fig. S4 C), and signal transduction (Fig. S4 D). Of these, the cell adhesion, cell differentiation, and ECM organization GO categories were statistically significantly enriched for DE genes relative to genes overall. Importantly, the DE cell adhesion genes were primarily associated with cell–substrate, not cell–cell, adhesion and with cell migration, cell projections, and ECM organization (Fig. 8 D). We observed significantly more DE genes than expected in GO cellular component categories for extracellular region, extracellular space, and proteinaceous ECM (Fig. 8 F). DE genes were also associated with the cytoplasm (Fig. S4 E), plasma membrane (Fig. S4 F), and nucleus.
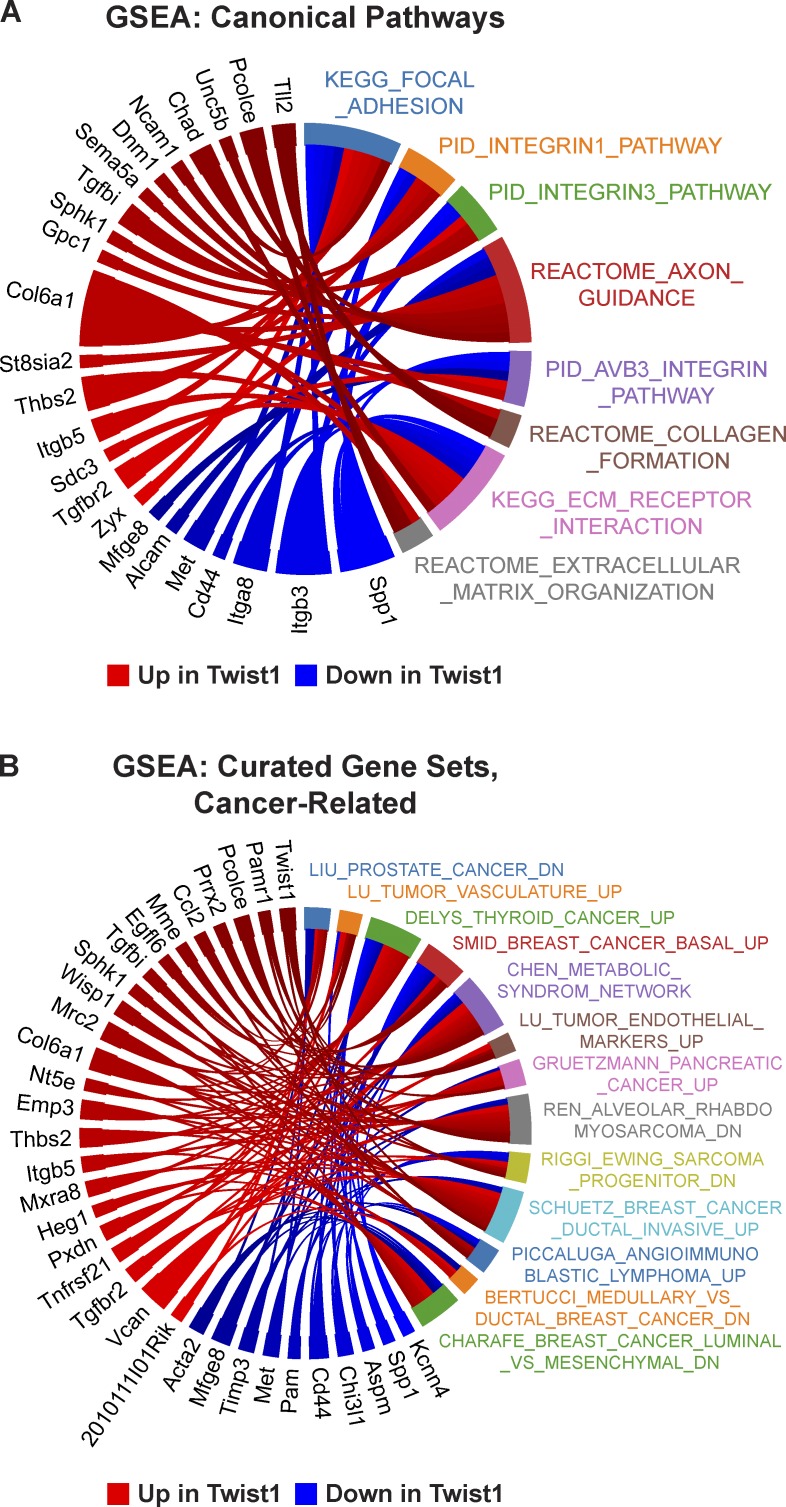
To complement the GO analysis of DE genes at genome-wide significance, we also performed a more general test of differential expression. Because we observed discordant expression changes within pathways, we used a joint test of up- and down-regulation by calculating the absolute value of the z-score for differential expression of each gene, followed by a t test for genes within versus outside each pathway. Gene sets from pathway databases (canonical pathways) were augmented to include gene lists from the literature (curated pathways). We found eight significant canonical pathways, all related to the cell interface with the extracellular space: focal adhesion, integrins, axon guidance, collagen formation, ECM–receptor interactions, and ECM organization (Fig. 9 A and Table S3). We found 51 significant curated gene sets, 13 of which we characterized as cancer related (Fig. 9 B and Table S3). Out of 183 DE genes, 33 were associated with at least three cancer-related gene sets (Fig. 9 B). We conclude that our data identify a novel set of genes regulated by Twist1 during dissemination that collectively reprogram the extracellular environment and cell interactions with the ECM. Importantly, some of the DE genes have enzymatic activity, are up-regulated in human cancers, and may represent novel targets for inhibiting dissemination (Fig. 9 B and Fig. S5 B).
Figure 9.
Significantly enriched pathways relate to cell–matrix adhesion. Gene set enrichment analysis (GSEA) identified eight significant canonical pathways (A), all related to cell–matrix adhesion and ECM organization, and 51 significant curated gene sets, 13 of which were characterized as cancer related (B). The Circos plots depict DE genes associated with each gene set. PID, Pathway Interaction Database.
E-cad loss blocks single cell dissemination of Twist1+ cells
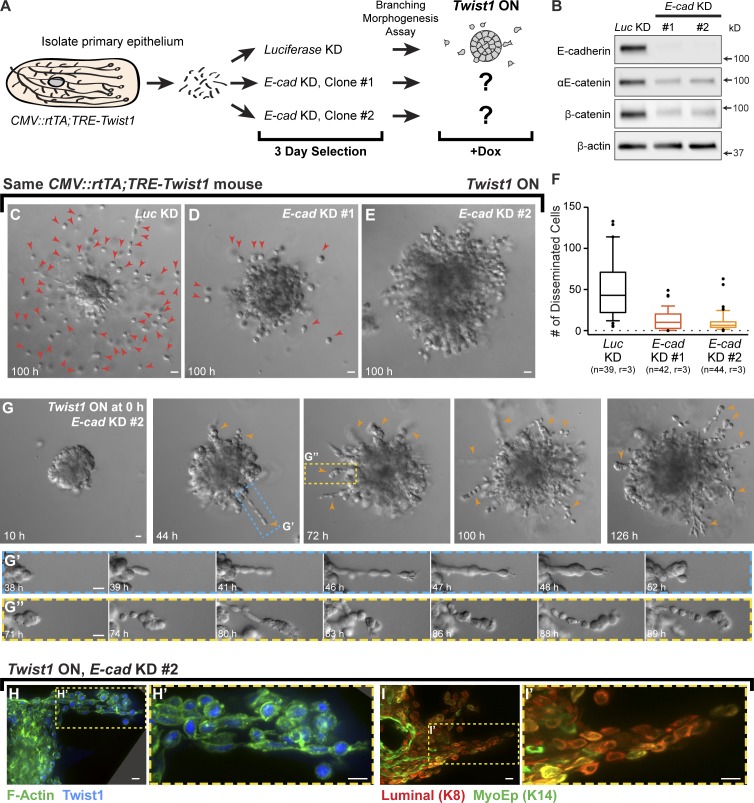
We observed that Twist1 induced dissemination of cells with membrane-localized E-cad and β-catenin and that Twist1 did not affect E-cad RNA levels. It was therefore possible that E-cad was contributing to Twist1-induced dissemination. Accordingly, we tested whether E-cad KD would inhibit single cell dissemination. We used lentiviral shRNA and puromycin selection to knock down Luciferase (Luc) or E-cad in CMV::rtTA;TRE-Twist1 organoids isolated from the same mouse (Fig. 10 A). We confirmed E-cad loss by immunoblotting (Fig. 10 B) and used Luc KD organoids as a negative control. Consistent with our E-cad deletion experiments, we observed a tandem reduction in αE-catenin and β-catenin.
Figure 10.
E-cad is required for Twist1-induced single cell dissemination. (A) Organoids from CMV::rtTA;TRE-Twist1 mice were divided into three groups for treatment with lentiviral shRNA against Luc or E-cad (two clones). Puromycin was used to select for transduced cells. Organoids were monitored for dissemination after Twist1 induction. (B) E-cad shRNA induced loss of E-cad protein and reductions in αE-catenin and β-catenin. Whole cell lysate samples were loaded for equal protein based on BCA analysis. (C–E) In FGF2-containing medium with doxycycline, E-cad KD organoids disseminated significantly fewer cells than Luc KD organoids (Video 8). Red arrowheads in C and D indicate disseminated cells. (F) Disseminated cells per organoid were quantified from movies after 100 h of Twist1 induction. E-cad KD significantly reduced single cell dissemination. Box-and-whisker plots are drawn with the box extending from the 25th to 75th percentiles and whiskers at the 10th and 90th percentiles. n, number of time-lapse movies; r, number of biological replicates. P < 0.0001 between Luc shRNA and E-cad shRNA #1 or #2; P = 0.014 between E-cad shRNA #1 and #2 (negative binomial generalized estimating equations model). (G) E-cad KD organoids extended collective chains of cells into the matrix (orange arrowheads; Video 9; G’ and G’’). (H and I) Cells within collective chains stained positive for Twist1 and luminal (K8+) and/or myoepithelial (K14+) cytokeratins. Bars: (C–E and G) 20 µm; (H and I) 10 µm.
We next used time-lapse microscopy to quantify dissemination after Twist1 induction in Luc KD and E-cad KD organoids. Surprisingly, E-cad KD resulted in a strong inhibition of single cell dissemination (Fig. 10, C–F; and Video 8). Many Twist1-induced, E-cad KD organoids had no detectable disseminated cells (Fig. 10 E). Instead, concurrent Twist1 expression and E-cad KD induced migration of long chains of cells into the ECM (Fig. 10, G–G″; and Video 9). Twist1-induced, E-cad KD cells retained nuclear Twist1+ protein (Fig. 10, H and H′), were cytokeratin+ (Fig. 10, I and I′), and were organized in collective groups (Fig. 10, H–I′). We conclude that Twist1 requires E-cad for efficient single cell dissemination.
Discussion
Our goal was to define the molecular requirements for dissemination of primary, normal epithelial cells. We used genetic techniques to contrast deletion of the cell adhesion gene E-cad with expression of the EMT transcription factor Twist1. We focused on E-cad as it is frequently mutated in human cancer (Berx et al., 1996; Hirohashi, 1998), and loss of E-cad can synergize with loss of p53 to promote metastasis (Derksen et al., 2006). In our experiments, E-cad deletion throughout the mammary epithelium resulted in loss of most intercellular junctions, loss of simple epithelial architecture, and inhibition of branching morphogenesis. However, E-cad+ cells within genetic mosaic epithelium were able to initiate and elongate normal mammary buds in 3D culture and in vivo. The associated E-cad− cells were viable and remained adherent to each other but did not contribute to polarized luminal ducts. Therefore, mammary epithelial cells must be able to maintain contact through alternate adhesion systems such as desmosomal cadherins or protocadherins. We conclude that E-cad is required for normal mammary development but that its loss alone is not sufficient for robust dissemination.
We next evaluated the consequences of Twist1 expression as Twist1 can regulate multiple aspects of metastasis and is thought to function primarily through E-cad repression (Yang et al., 2004; Peinado et al., 2007; Vesuna et al., 2009; Tran et al., 2012; Tsai et al., 2012). Our data reveal that Twist1 expression is sufficient to induce normal epithelial cells to disseminate out of an epithelium, migrate through the ECM, and establish secondary epithelial sites. Surprisingly, the disseminating cells retained cytokeratin expression, and many displayed membrane-localized E-cad and β-catenin. Transcriptional profiling revealed essentially no changes in the RNA expression of epithelial-specific cadherins, catenins, or keratins. Instead, Twist1 regulated many genes that mediate cell–matrix adhesion or modify the extracellular compartment. Because Twist1 regulates distinct targets depending on its bHLH dimer partner (Barnes and Firulli, 2009), it is likely that its regulated genes vary in different experiments. However, because our experimental induction of Twist1 was sufficient to induce dissemination, our 183 DE genes are likely particularly important in regulating the transition from adherent to motile epithelial cell phenotypes.
The conceptual framework for EMT was developed in response to classic experiments by Hay and co-workers in which definitive epithelial tissues lost polarity and disseminated as single cells into collagen I gels (Greenburg and Hay, 1982; Hay and Zuk, 1995). The appearance of these cells was most similar to that of embryonic mesenchymal cells, leading to the concept of an EMT. We are observing a similar migration of cells out of an epithelial tissue and into the surrounding ECM. However, we do not observe a loss of epithelial-specific gene expression, and the migratory cells are readily able to reestablish epithelial organization, both spontaneously and after cessation of Twist1 expression. Interestingly, we observed reductions in protein but not RNA levels of E-cad, β-catenin, and αE-catenin, suggesting posttranslational regulation of adherens junction components after Twist1 induction (Reynolds, 2007). However, we demonstrated that complete KD of E-cad dramatically inhibited Twist1-induced single cell dissemination. We speculate that the exact protein levels of E-cad may critically regulate whether Twist1 expression results in single cell dissemination or collective cell migration.
Accordingly, our data support the concept of a Twist1-dependent epithelial migratory program rather than a transition to mesenchymal cell fate or gene expression. Consistent with this framework, Twist1 regulates genes important for interactions with the stromal ECM environment. Our concept of an epithelial migratory program also finds support in breast cancer. Human breast tumors can express E-cad in both the primary tumor and in distant metastases (Kowalski et al., 2003), and primary breast tumor cells positive for the EMT transcription factor Slug can express high levels of E-cad (Côme et al., 2006).
Most cancer therapies target the increased proliferation of cancer cells relative to normal tissues and not the cell behaviors driving invasion, dissemination, and metastasis. Few of these drugs have proven clinical benefit in metastatic breast cancer patients (Carey, 2010). Twist1 is overexpressed in multiple metastatic human cancers and appears to specifically regulate metastatic cell behaviors in multiple experimental cancer models (Yang et al., 2004; Morel et al., 2012; Tran et al., 2012; Tsai et al., 2012). Twist1 therefore represents an attractive conceptual target for developing anti-metastatic therapies. However, it is very difficult to target a transcription factor with small molecule therapeutics. Our observation that Twist1 regulates many proteins in the extracellular compartment suggests that there may be essential, druggable effectors downstream of Twist1 whose repression could inhibit dissemination. Our inducible mouse model revealed that disseminated single cells rapidly cease migration after loss of Twist1 expression, suggesting that interfering with the Twist1 program could be an effective anticancer strategy. We envision our Twist1-induced dissemination assay as a rapid, reproducible, and scalable platform to build a new molecular model for effectors of Twist1 and to identify novel therapeutic compounds to antagonize cancer invasion and dissemination.
Materials and methods
Mouse strains
The R26::Cre-ER mouse line (Badea et al., 2003) was a gift from J. Nathans (Johns Hopkins University, Baltimore, MD). The CMV::rtTA transgenic line was a gift from F. Cong and H. Varmus (National Cancer Institute, Bethesda, MD). The Twist1-tetO7-luc (TRE-Twist1) transgenic line was previously described (Tran et al., 2012). E-cadfl/fl (Boussadia et al., 2002), mT/mG (Muzumdar et al., 2007), and R26::Lox-Stop-Lox-rtTA-IRES-EGFP (R26::LSL-rtTA; Belteki et al., 2005) mouse lines were acquired from The Jackson Laboratory. Mouse husbandry and procedures were all conducted under an Institutional Animal Care and Use Committee–approved animal protocol.
Isolation and 3D culture of primary mammary epithelial organoids
We used mechanical disruption, collagenase/trypsin digestion, and differential centrifugation to purify fragments of primary mammary epithelial ducts, termed organoids, as previously described (Ewald et al., 2008, 2012; Nguyen-Ngoc et al., 2012). In brief, mammary glands were harvested from mice 8–12 wk old, minced with a scalpel, and shaken for 40 min at 37°C in collagenase solution: DMEM (10565–018; Gibco) with 2 mg/ml collagenase (C2139; Sigma-Aldrich), 2 mg/ml trypsin (27250–018; Gibco), 5% FBS (F0926; Sigma-Aldrich), 5 µg/ml insulin (I9278; Sigma-Aldrich), and 50 µg/ml gentamicin (15750; Gibco). Suspensions were centrifuged at 1,250 rcf to remove a floating layer of adipocytes, and pellets were treated with 2 U/µl DNase (D4263; Sigma-Aldrich) to detach organoids from stromal cells. Enzymes and single cells were removed by four quick spins at 1,250 rcf such that the final pellet consisted mostly of organoids, each containing several hundred cells. Organoids were embedded in 3D Matrigel (354230; BD) at 2–3 organoids/µl and plated as 100-µl suspensions in 24-well coverslip-bottomed plates (662892; Greiner Bio-One) over a 37°C heating block. Gels were allowed to polymerize for 30 min at 37°C and then cultured in organoid medium: DMEM with 1% insulin-transferrin-selenium (51500–056; Gibco) and 1% penicillin-streptomycin (P4333; Sigma-Aldrich). Basal organoid medium was used to induce cyst formation, whereas addition of 2.5 nM FGF2 (F0291; Sigma-Aldrich) was used to induce branching morphogenesis. Branching was scored as organoids with three or more elongated buds. Cysts were scored as unbranched organoids with lumens detectable by light microscopy. Dissemination was scored as organoids with one or more adjacent single cells that were clearly separated from the epithelial group.
Tamoxifen-inducible Cre-mediated deletion in 3D culture
Cre activity was induced in Cre-ER;E-cadfl/fl;mT/mG epithelium by culturing organoids overnight with 50 nM tamoxifen once embedded in Matrigel. To wash out tamoxifen, samples were rinsed with PBS, incubated in organoid medium for 20 min at 37°C, and then cultured in fresh organoid medium. The tamoxifen-inducible system resulted in Cre activity in almost all cells and did not affect branching morphogenesis in control organoids (e.g., Cre-ER;E-cadfl/+;mT/mG).
Adenoviral delivery of Cre recombinase
Before embedding in Matrigel, mammary organoids were infected with Adeno-Cre (1045; Vector Laboratories) at a ratio of ∼107 PFU per 1,000 organoids. Infections were conducted in 50 µl DMEM for 1–2 h at 37°C to yield recombination in 50–75% of cells. Percentage of recombination was raised by increasing viral titer or by overnight incubation with virus.
Twist1 activation in 3D culture
Twist1 expression was induced in 3D Matrigel cultures by supplementing organoid medium with 5 µg/ml doxycycline (Shanghai RenYoung Pharmaceutical Co., Ltd). Because doxycycline is labile, medium was replaced every 48 h. To turn Twist1 expression off, doxycycline-containing medium was removed and samples were rinsed with sterile PBS. Samples were then incubated with organoid medium without doxycyline for 20 min at 37°C. This medium was then discarded, samples were rinsed again with PBS, and fresh organoid medium without doxycycline was added back.
Mammary fat pad transplantation
For transplantation of genetic mosaic E-cad or Twist1 tissue, we isolated organoids, induced recombination with Adeno-Cre, and washed organoids twice with 500 µl DMEM to remove viral particles. For all experiments, organoids were incubated at 37°C overnight in organoid medium with 2.5 nM FGF2 in HydroCell 96-well microplates (174907; Thermo Fisher Scientific). For Twist1 experiments, organoid medium was supplemented with 5 µg/ml doxycycline. The next day, organoids were resuspended in a 50% DMEM/50% Matrigel solution at a density of 20–40 organoids/µl and stored on ice. We conducted orthotopic transplantation into 3-wk-old NOD/SCID mice in a sterile hood. In brief, mice were anesthetized with 2–2.5% isoflurane and immobilized, and the surgical site was cleaned with ethanol. A 1-cm incision was made at the midline and a 0.5-cm incision was made from the midline to one hip. The skin was retracted to expose the no. 4 mammary gland. The no. 5 mammary fat pad and the region of the no. 4 mammary fat pad proximal to the lymph node were removed. The organoid suspension was loaded into a syringe (702RN [7636–01]; Hamilton; custom 1-in needles, 26 gauge), and 10–20 µl were injected into the cleared no. 4 fat pad. The skin was then locally infiltrated with 5–10 µl of 0.25% bupivacaine. The same procedure was repeated for the contralateral mammary gland. For each mouse, we transplanted control organoids (e.g., mT/mG) in one gland and experimental organoids (E-cadfl/fl;mT/mG, Cre-ER;E-cadfl/fl;mT/mG, or R26::LSL-rtTA;TRE-Twist1;mT/mG) in the other. The surface of the peritoneum was wet with PBS (without Ca2+ and Mg2+), and wounds were closed with 9-mm autoclips. Triple antibiotic ointment was applied to the incision site as needed. For Twist1 experiments, i.p. injections of 100 µg doxycycline in PBS (without Ca2+ and Mg2+) were also performed at the time of surgery. Twist1 activation in vivo was maintained with doxycycline feed (TD.01306; Harlan Laboratories). For deletion of E-cad in mature ductal networks, Cre-ER;E-cadfl/fl;mT/mG organoids were transplanted into NOD/SCID mice and allowed to grow for 6 wk. To induce E-cad deletion, we injected tamoxifen i.p. every other day for 5 d (three total injections) using a 1-ml syringe and a 30G1/2 needle (305106; BD). Each injection consisted of 100 µl of 10 mg/ml tamoxifen dissolved in sunflower seed oil. Glands were harvested 2–6 wk after injection.
Differential interference contrast (DIC) microscopy
Time-lapse imaging of mammary organoids was conducted using an LD Plan-Neofluar 20×/0.4 Korr Ph2 objective lens and a Cell Observer system with an AxioObserver Z1 and an AxioCam MRM camera (Carl Zeiss). In general, we recorded 100–200 positions in parallel for 5–7 d, with images acquired at 20-min intervals. Temperature was maintained at 37°C and CO2 at 5%. AxioVision (Carl Zeiss) was used to acquire and analyze time-lapse movies, place scale bars, and export individual TIFFs. Photoshop CS5 (Adobe) was used to adjust levels on entire images to maximize image clarity.
Confocal microscopy
Confocal imaging was performed on a spinning-disk confocal microscope (Solamere Technology Group Inc.) with an XR/MEGA-10 S30 camera (Stanford Photonics, Inc.), as previously described (Ewald et al., 2011; Ewald, 2013). A Fluar 20×/0.75 objective lens (Carl Zeiss) was used for intermediate magnification images. An LD C-Apochromat 40×/1.1 W Korr objective lens (Carl Zeiss) was used for high magnification single and time-lapse image acquisition, with water and oil used as the imaging mediums, respectively. Acquisition of both fixed and time-lapse images was performed using a combination of μManager (Edelstein et al., 2010) and Piper (Stanford Photonics, Inc.). For time-lapse imaging, images were collected at 20-min intervals for 2–4 d, and temperature was maintained at 37°C and CO2 at 5%. Imaris (Bitplane) was used to analyze time-lapse movies, place scale bars, and export individual TIFFs. Photoshop CS5 was used as needed to adjust levels and gamma for each channel on entire images to maximize image clarity.
High-pressure freezing and freeze substitution processing
We isolated epithelium from E-cadfl/fl and E-cadfl/+ littermates, induced recombination with Adeno-Cre, and cultured organoids for 5–7 d in Matrigel. Embedded organoids were then fixed in 3% glutaraldehyde to preserve for shipping to Lawrence Berkeley National Laboratory. There, samples were placed in 1-mm-wide by 200-µm-deep aluminum freezing hats and, before freezing, were surrounded with 20% BSA, used as a cryoprotectant. Samples were then cryoimmobilized using a high-pressure freezer (HPM-010; Bal-tec, Inc.) and freeze substituted in 1% osmium tetroxide and 0.1% uranyl acetate in acetone with 5% DDH2O, as previously described (McDonald and Webb, 2011). Upon completion of freeze substitution, samples were progressively infiltrated with an epon-Araldite resin (McDonald and Müller-Reichert, 2002). Polymerization in epon-Araldite resin was performed by flat embedding between two glass slides to allow for precise localization of features of interest (Müller-Reichert et al., 2003).
TEM
Samples were sectioned into 70–100-nm-thin and 500-nm-thick sections using an Ultramicrotome (UC6; Leica). Sections were then collected onto formvar-coated, rhodium-enforced copper 2-mm slot grids. The grids were post-stained with 2% uranyl acetate followed by Reynold’s lead citrate, for 5 min each. The sections were imaged using a Tecnai 12 TEM (FEI), operated between 690× and 11,000× at 120 kV under normal conditions. Images were recorded using an Orius SC1000B CCD with Digital Micrograph 3 software (Gatan Inc.). Serial electron microscopy software was used to collect wide-field montages for overview TEM images of complete organoid cross sections (Mastronarde, 2005). ImageJ software (Abramoff et al., 2004) and Photoshop CS4 were used to crop images, place scale bars, and adjust brightness and contrast on entire images, as needed.
Desmosome quantification
Desmosomes were counted among basally positioned cells in five E-cadfl/fl organoids and four E-cadfl/+ organoids imaged by TEM. For each organoid, we selected one to four regions of 20–30 cells that were no more than two cells deep from the organoid–ECM interface. Regions were free of single file columns or epithelial buds. For desmosomes located between cells in the second and third layers deep to the surface, we counted the desmosome but not the third-layer cell. Photoshop CS4 was used to track the desmosomes and regions used for quantification.
Immunofluorescence
Organoids grown in 3D Matrigel were fixed in 4% paraformaldehyde for 10 min, rinsed three times in PBS for 10 min, embedded in Optimal Cutting Temperature compound (OCT), and frozen at −80°C. OCT blocks were sectioned at 50-µm thickness by cryostat at −20°C. Sections were placed on Superfrost Plus Gold microscope slides (15-188-48; Fisherbrand) and stored at −80°C. For antibody staining, samples were thawed at room temperature, rinsed twice in PBS for 10 min to remove OCT, permeabilized with 0.5% Triton X-100 for 1 h, and rinsed twice in PBS for 10 min. Samples were blocked for 1–3 h with 10% FBS/1% BSA, incubated with primary antibodies overnight at 4°C in 1% FBS/1% BSA, and rinsed three times in 1% FBS/1% BSA for 15 min. Incubation with secondary antibodies was conducted in 1% FBS/1% BSA overnight at 4°C or for 2 h at room temperature. Slides were rinsed three times in PBS for 10 min, mounted with Fluoromount (F4680; Sigma-Aldrich), and sealed with coverslips. F-Actin was stained with Alexa Fluor Phalloidin (1:100; Invitrogen), and nuclei were stained with DAPI (1:1,000; D3571; Invitrogen). Immunofluorescence staining for each antibody was performed at least three independent times for a minimum of 10–15 organoids. Primary antibodies used were rat anti–E-cad (1:250; 13-1900; Invitrogen), rabbit anti–ZO-1 (1:500; 40-2300; Invitrogen), rabbit anti–β-catenin (1:1,000; C2206; Sigma-Aldrich), mouse anti–αE-catenin (1:100; ALX-804-101; Enzo Life Sciences), rabbit anti–laminin 332 (1:1,000; gifts of P. Marinkovich, Stanford University, Stanford, CA, and M. Aumailley, University of Cologne, Cologne, Germany), rat anti–cytokeratin-8 (1:100; TROMA-I; Developmental Studies Hybridoma Bank), rabbit anti–cytokeratin-14 (1:500; PRB-155P; Covance), mouse anti-smooth muscle α-actin (1:250; A5228; Sigma-Aldrich), and mouse anti-Twist1 (1:50; sc-81417; Santa Cruz Biotechnology Inc.). Secondary antibodies used were all Alexa Fluor–conjugated antibodies (1:200; Invitrogen).
Transplanted no. 4 mammary glands were dissected, fixed in 4% paraformaldehyde for 4 h at room temperature, rinsed three times in PBS for 15 min, and embedded in OCT. OCT blocks were sectioned at 50–100-µm thickness by cryostat with OT at −40°C and CT at −30°C. Samples on slides were stained as above but incubated in primary antibody for 48 h at 4°C and in secondary antibody for 24 h at 4°C or for 6–8 h at room temperature.
Protein extraction
Lysis buffer for protein extraction was prepared by diluting 10× RIPA buffer (20–188; EMD Millipore) in ultrapure water and chilling the mixture at 4°C for at least 2 h. Immediately before use, lysis buffer was supplemented with 0.1% SDS, 5% glycerol, 3 mM EDTA, 1 mM NaF, 1 mM PMSF, 1.5 mM NaVO4, Aprotinin (A6279; Sigma-Aldrich), and a mini protease inhibitor tablet (11836153001; Roche). Organoids embedded in 3D Matrigel were collected using freshly made PBS/EDTA buffer (5 mM, 1 mM NaVO4, 1.5 mM NaF, and 1 mM PMSF in PBS). Medium was aspirated from 3D culture wells, and all wells were rinsed once quickly with 1 ml of cold PBS. Approximately 1 ml of cold PBS/EDTA buffer was used to dissolve two 100-µl gels. Solutions were transferred to centrifuge tubes and mixed well by pipetting. Tubes were left on a 4°C shaker for 1 h to dissolve the Matrigel and then centrifuged at 400 rcf for 5 min at 4°C. Supernatants were removed and, if necessary, pellets were washed with additional PBS/EDTA to remove residual Matrigel. After another 5-min spin, pellets were resuspended in 100 µl of RIPA lysis buffer, vortexed, and left on ice for 30–40 min. Tubes were centrifuged for 10 min at 18,400 rcf at 4°C and supernatants transferred to new centrifuge tubes and stored at −80°C. For lentivirus experiments, organoids were collected after 3-d puromycin selection, washed once with cold PBS to remove trace medium, and resuspended in RIPA lysis buffer as described.
Western blotting
Whole cell protein lysates were thawed on ice for 20 min, vortexed, and centrifuged for 5 min at 18,400 rcf at 4°C. Samples in Laemmli sample buffer (161–0747; Bio-Rad Laboratories) and β-mercaptoethanol were heated at 70°C for 10 min and loaded for equal protein based on bicinchoninic acid (BCA) analysis (Thermo Fisher Scientific) in 4–15% Mini-PROTEAN TGX precast gels (456–1084; Bio-Rad Laboratories). SDS-PAGE was performed at 40 V for 30 min and 80 V for ∼90 min until the dye front ran off the gels. Gels were transferred onto nitrocellulose membranes (45-000-948; GE Healthcare) at 100 V for 1 h at 4°C. Membranes were blocked with 5% milk in TBST (03-500-537; Thermo Fisher Scientific) for 1 h at room temperature. Primary antibodies were prepared in blocking buffer and added overnight at 4°C. Primary antibodies used were rat anti–E-cad (1:1,000; 13-1900; Invitrogen), rabbit anti–β-catenin (1:2,000; C2206; Sigma-Aldrich), mouse anti–αE-catenin (1:1,000; ALX-804-101; Enzo Life Sciences), rabbit anti–N-cad (1:1,000; ab18203; Abcam), mouse anti-Twist1 (1:500; sc-81417; Santa Cruz Biotechnology, Inc.), and mouse anti–β-actin (1:1,000; A2228; Sigma-Aldrich). The N-cad antibody detected protein by Western blotting but not by immunofluorescence. Membranes were washed three times with TBST for 5 min and incubated with HRP-conjugated secondary antibodies (1:2,000; Invitrogen) in blocking buffer for 1 h at room temperature. Bands were detected with ECL reagents (34075 or 34095; Thermo Fisher Scientific), and membranes were imaged using an imager and software (Alpha-InnoTec). Band intensities acquired under autoexposure were quantified using Fiji. To probe for more than one protein, membranes were incubated with stripping buffer (21059; Thermo Fisher Scientific) for 30 min at 37°C, washed three times with TBST for 5 min, and reblocked. Primary antibodies used on the same membrane were from different hosts.
E-cad KD experiments
Approximately 1,000 CMV::rtTA;TRE-Twist1 organoids resuspended in 200 µl of basal organoid medium were added to each of four wells of a HydroCell 96-well microplate. Organoids were allowed to settle for 1 h at 37°C. Lentiviral transduction particles were thawed on ice: (1) MISSION pLKO.1-puro Luciferase shRNA (106 TU/ml; SHC007V; Sigma-Aldrich); (2) MISSION pLKO.1-puro Cdh1 shRNA #1 (106 TU/ml; TRCN0000042578; Sigma-Aldrich); (3) MISSION pLKO.1-puro Cdh1 shRNA #2 (106 TU/ml; TRCN0000042581; Sigma-Aldrich). In separate centrifuge tubes, 3 µl of ViroMag R/L nanoparticles (RL40200; OZ Biosciences) were mixed with 47 µl of lentivirus and incubated at room temperature for 30 min. In three of the organoid wells, 150 µl of medium were carefully removed, and 50 µl of the ViroMag/lentivirus mix were added. Suspensions were mixed well to disperse the organoids and prevent aggregation. The fourth organoid well served as a no-virus, puromycin control to evaluate killing efficiency. The 96-well plate was incubated on top of a magnetic plate (MF10000; OZ Biosciences) at 37°C for 1.5 h and then taken off the magnet and incubated overnight at 37°C. On day 2, ∼70 µl of medium were removed from each well with virus, 200 µl of fresh organoid medium were added, and the suspensions were mixed well to redisperse the organoids. On day 3, ∼200 µl of medium were removed from all wells, and 200 µl of organoid medium with 2.5 nM FGF2 and 4 µg/ml puromycin were added. Selection was performed for 3 d, and the surviving virus-treated organoids were collected for 3D culture and imaged by DIC time-lapse microscopy. In parallel, TRE-Twist1 (rtTA−) organoids from littermate controls were treated with lentiviruses and, after selection, used for protein extraction to evaluate E-cad KD efficiency. Time-lapse movies were used to track and count cells that had disseminated by 100 h of Twist1 induction in each of the three treatment groups. A cell was considered disseminated if it had a clear space between itself and the main organoid (visible ECM) and was observed to be migrating away persistently. Cells within collective chains that temporarily detached in only a few frames were not counted as disseminated.
RNA isolation and sequencing
Organoids were isolated from three CMV::rtTA;TRE-Twist1 mice (“Twist1”) and three TRE-Twist1 (rtTA−; “Control”) littermates (all inbred FVB/N). For each mouse, organoids were embedded in 3D Matrigel at 3–10 organoids/µl and plated as six 50-µl suspensions in a 35-mm dish. Organoids were cultured in basal organoid medium for 24 h and in basal organoid medium supplemented with 5 µg/ml doxycycline for an additional 48 h. All CMV::rtTA;TRE-Twist1 samples were disseminating at 48 h of Twist1 induction, whereas no control samples were disseminating. Total RNA was extracted using 1 ml of TRIzol per dish (15596-026; Life Technologies) and RNeasy (QIAGEN). With 10–100 ng of RNA collected per sample, we generated barcoded NuGen RNA-seq v.2 libraries and ran paired end, 75-bp, 50-cycle sequencing on a HiSeq 2000 (Johns Hopkins Medical Institutions Deep Sequencing and Microarray Core Facility).
Paired-end, non-strand–specific RNA-seq reads were mapped to the mouse reference genome (Genomic Reference Consortium build 38) using read mapper Bowtie (Langmead et al., 2009) and splice junction mapper TopHat (Trapnell et al., 2009). We achieved a mean of 51.4 million uniquely mapped reads per sample and estimated the number of reads mapped to each gene using HTSeq (Anders, 2010), with gene coordinates from the reference genome Generic Feature Format file. Raw counts were normalized and p-values were calculated for Twist1 versus control differential expression from negative binomial distributions using DESeq (Anders and Huber, 2010). Based on the number of genes tested, the genome-wide significance level for 0.05 family-wise error rate was 2.74 × 10−6, and 183 genes were significant at this level. Sequence data has been uploaded to the Sequence Read Archive (project accession no. SRP033275).
GO analysis
Significant genes were characterized using GO Slim categories for biological process, cellular component, and molecular function (Ashburner et al., 2000; GO file downloaded on 9/12/13 and mapping from 3/5/13). We first computed the overall fraction of genes that were up-regulated (fup = 107/18,260) and down-regulated (fdown = 76/18,260). For each GO term, we then calculated the number of genes n annotated to the category and also among the 18,260 genes sequenced. Categories with n < 2 were not considered further. For categories with n ≥ 2, we conducted separate two-sided tests corresponding to enrichment or depletion based on Poisson distributions for up-regulated (expected number = fupn) and down-regulated (expected number = fdownn) DE genes. For each of the three ontologies, the p-value threshold for 0.05 family-wise error rate was set to 0.05/(4 × number of categories tested). This conservative approach was selected as more computationally convenient than the corresponding Fisher’s exact tests.
Pathway-level differential expression
To assess pathway-level differential expression, we used gene sets available through MSigDB v4.0 (Subramanian et al., 2005). The 1,320 canonical gene sets within MSigDB are aggregated from major pathway databases: BioCarta, KEGG, Reactome, the Pathway Interaction Database, the Sigma-Aldrich database, the Signaling Gateway database, the Signal Transduction KE database, and the SuperArray database. These are then augmented to 4,722 curated gene sets by including gene lists from published studies, the L2L gene sets from mammalian microarray studies (Newman and Weiner, 2005), the MYC Target Gene Database (Zeller et al., 2003), and other public resources. For each gene, including genes not differentially expressed at genome-wide significance, the p-value from DESeq was converted to the equivalent z-score for a two-sided test, with z > 0 for Twist1 > Control and z < 0 for Twist1 < Control. Each pathway was tested using a one-sided Student’s t test of |z| within pathway > |z| outside pathway, corresponding to an increased number of DE genes without regard to direction (up or down in Twist1 versus Control). Significance thresholds for 0.05 family-wise error rates at the pathway level were estimated using 500 permutations of gene z-scores, recording the best p-value from each permutation, and taking the 25th-best p-value as the 0.05 FWER threshold. The resulting thresholds were 1.44 × 10−13 for canonical gene sets and 5.31 × 10−19 for curated gene sets. These thresholds are more stringent than a standard Bonferroni correction.
Online supplemental material
Fig. S1 shows that E-cad deletion results in complete loss of E-cad protein and reduced levels of other adherens junction components in 3D culture and in vivo. Fig. S2 shows that Twist1-induced dissemination is cell autonomous and occurs in vivo. Fig. S3 shows that Twist1 expression results in reduced levels of adherens junction proteins. Fig. S4 shows heat maps of genes directly associated with relevant GO Slim biological process and cellular component terms. Fig. S5 shows the mapped GO Slim molecular function terms and a heat map of Twist1-regulated genes with enzymatic activity. Video 1 shows confocal time-lapse imaging of a Cre-ER;E-cadfl/fl;mT/mG organoid transitioning from simple to multilayered organization after E-cad deletion. Video 2 shows DIC time-lapse imaging of representative E-cad+ and E-cad− organoids grown in the branching morphogenesis assay. Video 3 shows confocal time-lapse imaging of E-cad+ bud initiation in a genetic mosaic E-cadfl/fl;mT/mG organoid. Videos 4 and 5 show DIC time-lapse imaging of representative CMV::rtTA;TRE-Twist1 organoids grown in basal medium and FGF2-containing medium, respectively, with and without Twist1 induction. Videos 6 and 7 show DIC time-lapse imaging of a CMV::rtTA;TRE-Twist1 organoid grown in basal medium and FGF2-containing medium, respectively, with transient Twist1 induction. Video 8 shows DIC time-lapse imaging of representative Twist1-expressing organoids with Luc and E-cad KD. Video 9 shows DIC time-lapse imaging of collective epithelial migration in a Twist1-expressing, E-cad KD organoid. Table S1 lists all genes sequenced by RNA-seq, sorted by p-value, and the 183 DE genes along with their associated GO Slim terms. Table S2 lists RNA-seq raw counts and p-values for 127 epithelial genes involved in cell–cell adhesion and intercellular junctions. Table S3 lists the significant canonical pathways and curated gene sets identified by gene set enrichment analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306088/DC1.
Supplementary Material
Acknowledgments
We thank Jen Beck for technical support, Ryan Gray for preliminary experiments, Xian Zhou for statistical support, and the Johns Hopkins Medical Institutions Deep Sequencing and Microarray Core Facility for sequencing support. We thank Feng Cong and Harold Varmus for donating the CMV::rtTA mouse line.
This work was supported by a Research Scholar Grant (RSG-12-141-01 - CSM) from the American Cancer Society (to A.J. Ewald), by a Research Scholar Grant (RSG-12-196-01 - TBG) from the American Cancer Society (to P.T. Tran), by funds from the National Institutes of Health/National Cancer Institute (P50CA103175; to P.T. Tran and A.J. Ewald), by a grant from the Safeway Foundation for Breast Cancer Research (to A.J. Ewald), by the Avon Foundation for Women (to A.J. Ewald), by the Isaac Morris Hay and Lucille Elizabeth Hay Graduate Fellowship Award (to E.R. Shamir), by funds from the National Institutes of Health/National Institute of General Medical Sciences (P01 GM051487; to M. Auer), by the University of California, Berkeley Physical Sciences in Oncology Center (to M. Auer and D.M. Jorgens), by funds from the National Institutes of Health/National Center for Research Resources (U54RR020839; to J.S. Bader), and by the Robert J. Kleberg and Helen C. Kleberg Foundation (to J.S. Bader).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BCA
- bicinchoninic acid
- DE
- differentially expressed
- DIC
- differential interference contrast
- E-cad
- epithelial cadherin
- EMT
- epithelial to mesenchymal transition
- GO
- gene ontology
- KD
- knockdown
- Luc
- luciferase
- N-cad
- neural cadherin
- OCT
- Optimal Cutting Temperature compound
- rtTA
- reverse tetracycline transactivator
- SMA
- smooth muscle actin
- TEB
- terminal end bud
- TEM
- transmission electron microscopy
- ZO-1
- zona occludens 1
References
- Abramoff M.D., Magalhaes P.J., Ram S.J. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42 [Google Scholar]
- Anders S. 2010. HTSeq: Analysing high-throughput sequencing data with Python. Available at http://www-huber.embl.de/users/anders/HTSeq/doc/index.html (Accessed on December 26, 2012) [DOI] [PMC free article] [PubMed]
- Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea T.C., Wang Y., Nathans J. 2003. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23:2314–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes R.M., Firulli A.B. 2009. A twist of insight - the role of Twist-family bHLH factors in development. Int. J. Dev. Biol. 53:909–924 10.1387/ijdb.082747rb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrallo-Gimeno A., Nieto M.A. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 132:3151–3161 10.1242/dev.01907 [DOI] [PubMed] [Google Scholar]
- Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S.E., Nagy A. 2005. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33:e51 10.1093/nar/gni051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G., Van Roy F. 2001. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 3:289–293 10.1186/bcr309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G., Cleton-Jansen A.M., Strumane K., de Leeuw W.J., Nollet F., van Roy F., Cornelisse C. 1996. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 13:1919–1925 [PubMed] [Google Scholar]
- Bissell M.J., Bilder D. 2003. Polarity determination in breast tissue: desmosomal adhesion, myoepithelial cells, and laminin 1. Breast Cancer Res. 5:117–119 10.1186/bcr579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenrieder T., Herlyn M. 2003. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene. 22:6524–6536 10.1038/sj.onc.1206757 [DOI] [PubMed] [Google Scholar]
- Boussadia O., Kutsch S., Hierholzer A., Delmas V., Kemler R. 2002. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech. Dev. 115:53–62 10.1016/S0925-4773(02)00090-4 [DOI] [PubMed] [Google Scholar]
- Carey L.A. 2010. Through a glass darkly: advances in understanding breast cancer biology, 2000–2010. Clin. Breast Cancer. 10:188–195 10.3816/CBC.2010.n.026 [DOI] [PubMed] [Google Scholar]
- Côme C., Magnino F., Bibeau F., De Santa Barbara P., Becker K.F., Theillet C., Savagner P. 2006. Snail and slug play distinct roles during breast carcinoma progression. Clin. Cancer Res. 12:5395–5402 10.1158/1078-0432.CCR-06-0478 [DOI] [PubMed] [Google Scholar]
- Daniel C.W., Strickland P., Friedmann Y. 1995. Expression and functional role of E- and P-cadherins in mouse mammary ductal morphogenesis and growth. Dev. Biol. 169:511–519 10.1006/dbio.1995.1165 [DOI] [PubMed] [Google Scholar]
- Derksen P.W., Liu X., Saridin F., van der Gulden H., Zevenhoven J., Evers B., van Beijnum J.R., Griffioen A.W., Vink J., Krimpenfort P., et al. 2006. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 10:437–449 10.1016/j.ccr.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:Unit14.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J. 2013. Practical considerations for long-term time-lapse imaging of epithelial morphogenesis in three-dimensional organotypic cultures. Cold Spring Harb. Protoc. 2013:100–117 10.1101/pdb.top072884 [DOI] [PubMed] [Google Scholar]
- Ewald A.J., Brenot A., Duong M., Chan B.S., Werb Z. 2008. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 14:570–581 10.1016/j.devcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Werb Z., Egeblad M. 2011. Dynamic, long-term in vivo imaging of tumor–stroma interactions in mouse models of breast cancer using spinning-disk confocal microscopy. Cold Spring Harb. Protoc. 2011:pdb.top97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald A.J., Huebner R.J., Palsdottir H., Lee J.K., Perez M.J., Jorgens D.M., Tauscher A.N., Cheung K.J., Werb Z., Auer M. 2012. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J. Cell Sci. 125:2638–2654 10.1242/jcs.096875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenburg G., Hay E.D. 1982. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 95:333–339 10.1083/jcb.95.1.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B., Stevenson B., Grimaldi A. 1988. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 107:1575–1587 10.1083/jcb.107.4.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E.D., Zuk A. 1995. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am. J. Kidney Dis. 26:678–690 10.1016/0272-6386(95)90610-X [DOI] [PubMed] [Google Scholar]
- Hinck L., Silberstein G.B. 2005. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 7:245–251 10.1186/bcr1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi S. 1998. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am. J. Pathol. 153:333–339 10.1016/S0002-9440(10)65575-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes A., Gottardi C.J., Yap A.S. 2008. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 27:6920–6929 10.1038/onc.2008.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski P.J., Rubin M.A., Kleer C.G. 2003. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 5:R217–R222 10.1186/bcr651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L., Ohsugi M., Hirchenhain J., Kemler R. 1994. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA. 91:8263–8267 10.1073/pnas.91.17.8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- McDonald K., Müller-Reichert T. 2002. Cryomethods for thin section electron microscopy. Methods Enzymol. 351:96–123 10.1016/S0076-6879(02)51843-7 [DOI] [PubMed] [Google Scholar]
- McDonald K.L., Webb R.I. 2011. Freeze substitution in 3 hours or less. J. Microsc. 243:227–233 10.1111/j.1365-2818.2011.03526.x [DOI] [PubMed] [Google Scholar]
- Mironchik Y., Winnard P.T., Jr, Vesuna F., Kato Y., Wildes F., Pathak A.P., Kominsky S., Artemov D., Bhujwalla Z., Van Diest P., et al. 2005. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 65:10801–10809 10.1158/0008-5472.CAN-05-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A.P., Hinkal G.W., Thomas C., Fauvet F., Courtois-Cox S., Wierinckx A., Devouassoux-Shisheboran M., Treilleux I., Tissier A., Gras B., et al. 2012. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 8:e1002723 10.1371/journal.pgen.1002723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Reichert T., Hohenberg H., O’Toole E.T., McDonald K. 2003. Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. J. Microsc. 212:71–80 10.1046/j.1365-2818.2003.01250.x [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nelson W.J. 2009. Remodeling epithelial cell organization: transitions between front–rear and apical–basal polarity. Cold Spring Harb. Perspect. Biol. 1:a000513 10.1101/cshperspect.a000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.C., Weiner A.M. 2005. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 6:R81 10.1186/gb-2005-6-9-r81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.X., Bos P.D., Massagué J. 2009. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer. 9:274–284 10.1038/nrc2622 [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc K.V., Cheung K.J., Brenot A., Shamir E.R., Gray R.S., Hines W.C., Yaswen P., Werb Z., Ewald A.J. 2012. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl. Acad. Sci. USA. 109:E2595–E2604 10.1073/pnas.1212834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman M.T., Prudoff R.S., Johnson K.R., Wheelock M.J. 1999. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 147:631–644 10.1083/jcb.147.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Gupta P.B., Mani S.A., Yang J., Lander E.S., Weinberg R.A. 2008. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68:3645–3654 10.1158/0008-5472.CAN-07-2938 [DOI] [PubMed] [Google Scholar]
- Peinado H., Olmeda D., Cano A. 2007. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 7:415–428 10.1038/nrc2131 [DOI] [PubMed] [Google Scholar]
- Polyak K. 2010. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J. Natl. Cancer Inst. Monogr. 2010:210–213 10.1093/jncimonographs/lgq019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Weinberg R.A. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Reynolds A.B. 2007. p120-catenin: Past and present. Biochim. Biophys. Acta. 1773:2–7 10.1016/j.bbamcr.2006.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J., et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 462:1005–1010 10.1038/nature08645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tinkle C.L., Lechler T., Pasolli H.A., Fuchs E. 2004. Conditional targeting of E-cadherin in skin: insights into hyperproliferative and degenerative responses. Proc. Natl. Acad. Sci. USA. 101:552–557 10.1073/pnas.0307437100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T., Shroff E.H., Burns T.F., Thiyagarajan S., Das S.T., Zabuawala T., Chen J., Cho Y.J., Luong R., Tamayo P., et al. 2012. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 8:e1002650 10.1371/journal.pgen.1002650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. 2012. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 22:725–736 10.1016/j.ccr.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna F., van Diest P., Chen J.H., Raman V. 2008. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem. Biophys. Res. Commun. 367:235–241 10.1016/j.bbrc.2007.11.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna F., Lisok A., Kimble B., Raman V. 2009. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 11:1318–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.M., Daniel C.W. 1983. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev. Biol. 97:274–290 10.1016/0012-1606(83)90086-6 [DOI] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science. 318:1108–1113 10.1126/science.1145720 [DOI] [PubMed] [Google Scholar]
- Yang J., Weinberg R.A. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14:818–829 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Zeller K.I., Jegga A.G., Aronow B.J., O’Donnell K.A., Dang C.V. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69 10.1186/gb-2003-4-10-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.