HsSAS-6 homodimers are present in the cytoplasm and assemble into ninefold symmetrical arrays at centrosomes, thus initiating procentriole formation.
Abstract
SAS-6 proteins are thought to impart the ninefold symmetry of centrioles, but the mechanisms by which their assembly occurs within cells remain elusive. In this paper, we provide evidence that the N-terminal, coiled-coil, and C-terminal domains of HsSAS-6 are each required for procentriole formation in human cells. Moreover, the coiled coil is necessary and sufficient to mediate HsSAS-6 centrosomal targeting. High-resolution imaging reveals that GFP-tagged HsSAS-6 variants localize in a torus around the base of the parental centriole before S phase, perhaps indicative of an initial loading platform. Moreover, fluorescence recovery after photobleaching analysis demonstrates that HsSAS-6 is immobilized progressively at centrosomes during cell cycle progression. Using fluorescence correlation spectroscopy and three-dimensional stochastic optical reconstruction microscopy, we uncover that HsSAS-6 is present in the cytoplasm primarily as a homodimer and that its oligomerization into a ninefold symmetrical ring occurs at centrioles. Together, our findings lead us to propose a mechanism whereby HsSAS-6 homodimers are targeted to centrosomes where the local environment and high concentration of HsSAS-6 promote oligomerization, thus initiating procentriole formation.
Introduction
The centrosome is the main microtubule-organizing center of animal cells and contributes to correct segregation of the genetic material by directing bipolar spindle assembly. The centrosome comprises two microtubule-based cylindrical centrioles embedded in pericentriolar material (PCM; for review see Azimzadeh and Bornens, 2007; Marshall, 2009; Azimzadeh and Marshall, 2010; Carvalho-Santos et al., 2010; Gönczy, 2012). A defining feature of centrioles is a ninefold radial symmetric arrangement of microtubules that is also imparted onto cilia and flagella that originate from them. Not surprisingly given their fundamental roles in multiple cellular processes, structural and functional aberrations of centrioles can cause human diseases, including ciliopathies, microcephaly, male sterility, and cancer (Nigg and Raff, 2009; Bettencourt-Dias et al., 2011).
In proliferating cells, a new procentriole assembles next to the proximal end of each parental centriole, approximately at the G1/S transition (Kuriyama and Borisy, 1981; Chrétien et al., 1997). In most species, procentrioles form around the cartwheel, a structure ∼120 nm in diameter and ∼100 nm in height that is best visualized by EM in unicellular organisms (Dippell, 1968; Cavalier-Smith, 1974). The cartwheel consists of a central hub ∼20–25 nm in diameter from which emanate nine spokes that exhibit a ∼40° angle between them and extend toward peripheral microtubules. Given its ninefold radial symmetry and appearance at the onset of procentriole formation, the cartwheel has been proposed to impart the defining arrangement of the entire centriole (reviewed in Strnad and Gönczy, 2008). Analysis of mutants in cartwheel components, including members of the SAS-6 protein family, has validated such a role. For instance, the Chlamydomonas reinhardtii SAS-6 homologue Bld12p is required for cartwheel assembly and efficient centriole formation (Nakazawa et al., 2007). The few remaining basal bodies in Bld12p-null mutants exhibit radial symmetries that differ from the canonical ninefold (Nakazawa et al., 2007), thus highlighting the crucial role of Bld12p in establishing the ninefold symmetry of the cartwheel and ensuring robust procentriole assembly.
Proteins of the SAS-6 family harbor a globular N-terminal head, followed by a coiled-coil domain and a C-terminal tail predicted to be unstructured and whose function is poorly understood (Leidel et al., 2005; Kitagawa et al., 2011; van Breugel et al., 2011). Biophysical analyses of fragments of Caenorhabditis elegans SAS-6 and C. reinhardtii Bld12p established that the coiled-coil domain drives homodimerization of SAS-6 proteins, with a Kd of ∼1 µM (Kitagawa et al., 2011). Moreover, rotary metal shadowing EM of SAS-6 and Bld12p fragments demonstrated that homodimers of SAS-6 proteins harbor globular heads followed by elongated rods corresponding to the coiled-coil moiety (Kitagawa et al., 2011). Finally, the crystal structure of the head domain of SAS-6 proteins from C. elegans, C. reinhardtii, and Danio rerio revealed the importance of a residue (corresponding to F131 in the human homologue HsSAS-6) in mediating interaction between two head domains from adjacent homodimers (Kitagawa et al., 2011; van Breugel et al., 2011), with a Kd of ∼50 µM for HsSAS-6 (van Breugel et al., 2011). This interaction drives the formation of higher order oligomers of homodimers, hereafter referred to as oligomers. In the case of Bld12p, these oligomers are seen by rotary metal shadowing EM as V-shaped tetramers as well as ringlike structures from which the coiled-coil domains emanate (Kitagawa et al., 2011). These rings exhibit a diameter of ∼22 nm, consistent with that of the cartwheel hub observed in situ, whereas the angle between two coiled-coil rods of tetramers is ∼42°, consistent with a ninefold symmetry (Kitagawa et al., 2011). These studies suggest a mechanism whereby self-assembly of SAS-6 proteins can lead to the formation of a ninefold symmetrical structure resembling the cartwheel (Kitagawa et al., 2011; van Breugel et al., 2011). Compatible with such a mechanism, analysis of the elongated cartwheel of Trichonympha sp. by cryoelectron tomography revealed stacks of rings that could accommodate oligomers of nine SAS-6 homodimers (Guichard et al., 2012).
In cycling human cells, depletion of HsSAS-6 results in a failure of centriole formation, whereas its overexpression leads to supernumerary centrioles (Leidel et al., 2005; Strnad et al., 2007). HsSAS-6 is one of the earliest proteins recruited at the onset of procentriole assembly and is present in the proximal, cartwheel-bearing, region of the procentriole until metaphase (Kleylein-Sohn et al., 2007; Strnad et al., 2007; Sonnen et al., 2012; Lukinavičius et al., 2013). Thereafter, centrosomal HsSAS-6 is degraded in an APC (anaphase-promoting complex)- and SCF (Skp1–Cullin–F-box)-dependent manner, along with the cytoplasmic protein pool (Strnad et al., 2007; Puklowski et al., 2011). Importantly, mutation of the F131 residue abrogates the function of HsSAS-6 in procentriole formation, consistent with oligomerization being critical in human cells as well (Kitagawa et al., 2011).
Despite these advances, many important questions remain regarding the mechanisms of action of HsSAS-6 in human cells. In particular, the domains of HsSAS-6 needed for targeting the protein to centrosomes as well as for sustaining centriole formation in vivo are not known. Moreover, the oligomerization state of HsSAS-6 within the cytoplasm and whether the protein assembles into a ringlike structure at the centrosome has not been determined.
Results
Identification of HsSAS-6 domains important for centrosomal targeting and function
We set out to determine the domains of HsSAS-6 mediating centrosomal targeting and those required for centriole formation. We generated U2OS cells expressing doxycycline-inducible EGFP-tagged (hereafter referred to as GFP) HsSAS-6 truncation variants to assess the contribution of the N-terminal (N), coiled-coil (C-C), and C-terminal (C) domains (Fig. 1). We also depleted endogenous HsSAS-6 using siRNAs directed against the 3′UTR, which is absent from the GFP fusion constructs (Fig. S1).
Figure 1.
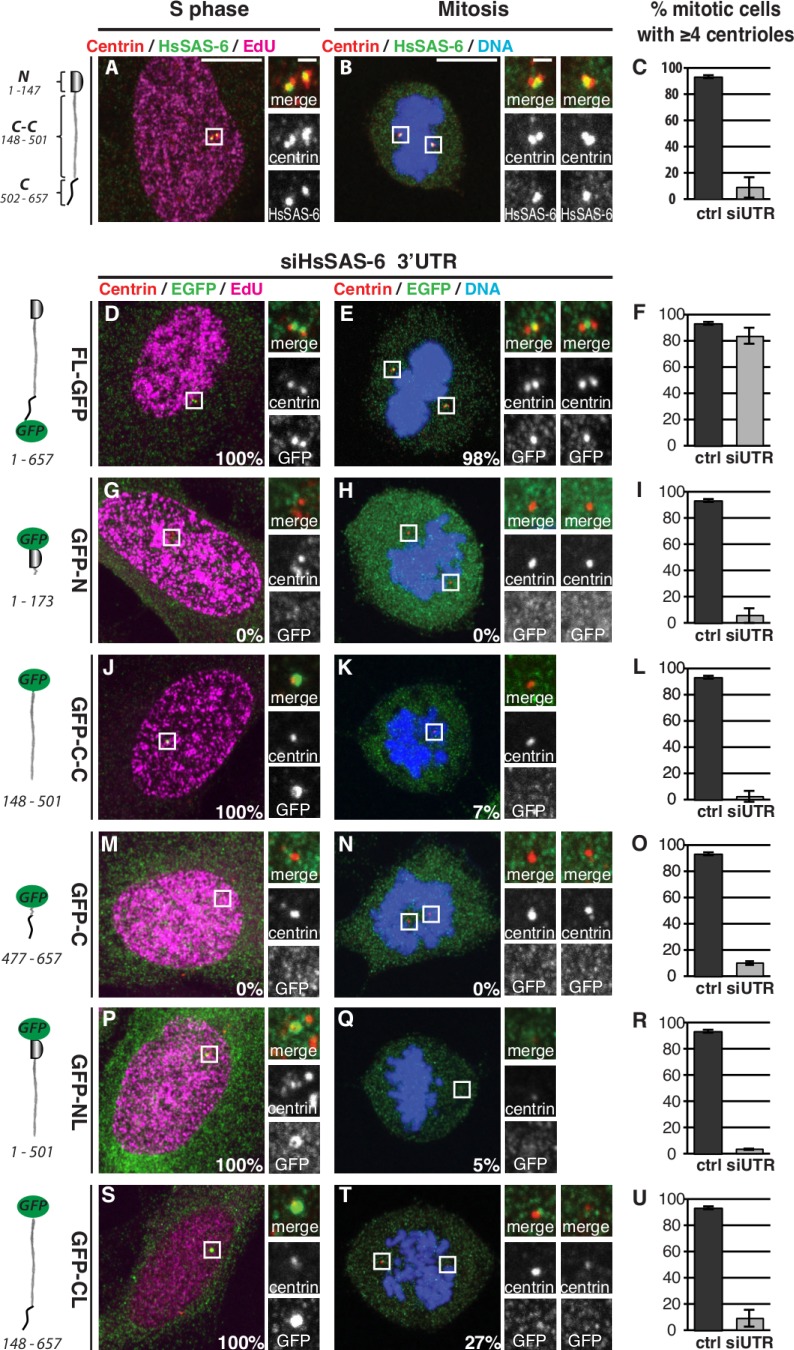
Domains of HsSAS-6 mediating centrosomal targeting and duplication. (A–U) U2OS (A–C) and iU2OS (D–U) cells in S phase (left) or mitosis (prometaphase/metaphase; right) expressing HsSAS-6 variants and stained for HsSAS-6 (A–C) or GFP (D–U) and centrin-3 as well as EdU to mark S-phase cells (left column). Schematics indicate the domains and amino acid boundaries of the variants. Doxycycline induction for 48 h was concomitant with siHsSAS-6 3′UTR (siUTR) treatment. Percentage of HsSAS-6–positive centrosomes (indicated at bottom right of images) was assessed both in EdU-positive and mitotic cells. Histograms report mean frequency of mitotic cells with at least four centrioles in control (ctrl) U2OS cells and siRNA-treated cells (siUTR). Data are from at least two experiments; error bars show standard deviations; number of cells analyzed are given in Table S1. Note that the C-terminal domain on top of the coiled coil (CL) augments the fraction of EdU-negative cells exhibiting centriolar signal. Insets are magnified regions of the boxed areas. Bars: (main images) 10 µm; (insets) 1 µm.
We analyzed cells in S phase, recognized by 5-ethynyl-2′-deoxyuridine (EdU) incorporation, and cells in prometaphase/metaphase (hereafter collectively referred to as mitosis), which were recognized by their condensed DNA. Cells were examined by immunofluorescence with antibodies against centrin to mark centrioles and against GFP to monitor the fusion proteins. As previously reported (Kitagawa et al., 2011), a full-length (FL) HsSAS-6 with a C-terminal GFP (FL-GFP) localizes to centrosomes in S phase and in mitosis, recapitulating the distribution of endogenous HsSAS-6 (Fig. 1, A, B, D, and E). An HsSAS-6 variant with an N-terminal GFP (GFP-FL) behaves similarly, though fewer cells can be analyzed because of aggregation of this fusion protein (unpublished data). Moreover, we found that neither GFP-N nor GFP-C localizes to centrosomes (Fig. 1, G, H, M, and N), whereas GFP–C-C is present at centrosomes in 100% of S-phase cells (Fig. 1 J) but in only in 7% of mitotic cells (Fig. 1 K). Cells expressing constructs with either the N- or C-terminal domain added to the coiled coil (GFP-NL and GFP-CL, respectively) behave similarly to GFP–C-C (Fig. 1, P, Q, S, and T). Because an HsSAS-6 variant lacking ∼90% of the coiled coil exhibits little or no centrosomal localization (van Breugel et al., 2011), we conclude that the coiled coil is necessary and sufficient for recruiting HsSAS-6 to centrosomes. Our data also reveal that both N- and C-terminal domains are required for the persistence of centrosomal HsSAS-6 until mitosis.
We next addressed whether the HsSAS-6 variants are functional by determining the number of centrin foci in mitotic cells. In control conditions, ∼95% of cells have at least four centrin foci (Fig. 1 C). Depletion of endogenous HsSAS-6 results in only ∼10% of cells having at least four centrin foci, a phenotype rescued to ∼85% by FL-GFP (Fig. 1, C and F). As shown in Fig. 1, no other variant rescues the depletion of endogenous HsSAS-6. Although the requirements of the N-terminal and of the coiled-coil domains were expected (Kitagawa et al., 2011; van Breugel et al., 2011), our findings uncover that the C-terminal domain is also essential for HsSAS-6 function.
HsSAS-6 variants localize in a torus around the proximal end of the parental centriole
In performing these experiments, we noted that S-phase cells expressing GFP–C-C, GFP-NL, or GFP-CL often exhibit a broader, toruslike, centrosomal signal (Fig. 1, J, P, and S). This observation was confirmed by examining single confocal sections (Fig. 2, A–C). Because these variants do not sustain centriole formation, this distribution could reflect a transient localization revealed by nonfunctional proteins being trapped during the assembly process. Indeed, we found such a torus in 17% of cells expressing the oligomerization-defective HsSAS-6–[F131E] (F131E-GFP) variant, with the remaining cells having weak or no signal at centrosomes (Fig. 2, D–E). A torus is also detectable in a smaller fraction (9%) of cells expressing FL-GFP, although most cells exhibit a focused signal, presumably corresponding to the protein being incorporated in the cartwheel (Fig. 2 E). These results establish that a torus can be observed with a functional version of HsSAS-6. However, we failed to detect a torus using antibodies against endogenous HsSAS-6, possibly because higher protein levels, as observed upon FL-GFP expression, are needed to detect what may be a very transient localization.
Figure 2.
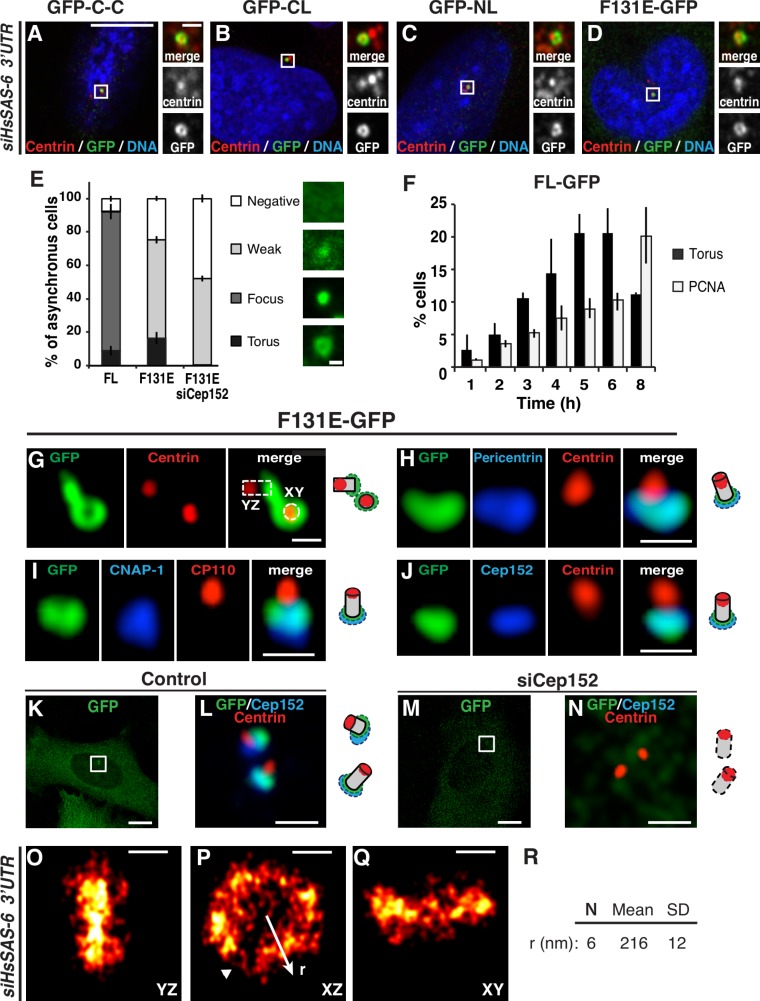
HsSAS-6 variants accumulate in a torus around the proximal end of the parental centriole. (A–D) Brightness/contrast-adjusted single planes of confocal images of GFP–C-C, GFP-CL, GFP-NL, and F131E-GFP at centrosomes showing the torus taken from iU2OS cells induced with doxycycline for 48 h concomitant with siHsSAS-6 3′UTR treatment, fixed, and stained for GFP, centrin-2, and DNA. Insets are magnified regions of the boxed areas. (E) Asynchronous population of cells expressing FL-GFP, F131E-GFP, or F131E-GFP depleted of Cep152 (siCep152) stained for GFP and centrin-2. Maximal projection images illustrate distinct GFP distributions at centrosomes (left), whereas the graph reports their occurrence (right). Error bars show standard error of the mean. Number of cells from at least two experiments: 440 for FL-GFP, 231 for F131E-GFP, and 263 for F131-GFP siCep152. (F) Time course in cells expressing FL-GFP, synchronized by mitotic shake off, and stained for GFP, centrin-2 and Pericentrin, or GFP and PCNA reveals increase in the torus-bearing fraction as cells exit mitosis and progress through G1. Error bars show standard error of the mean. Number of cells analyzed and statistical analysis are given in Table S2. (G–J) Centrosomes labeled with GFP, centrin-2 (G, H, and J), or CP110 (I) and Pericentrin (H), cNap-1 (I), or Cep152 (J) illustrate F131E-GFP torus localization at the proximal end of parental centrioles with corresponding schematics on the right. Dotted box and circle in G represent the orientation of centrioles. (K–N) Depletion of Cep152 abolishes F131E-GFP torus localization around the parental centriole. n = 253 for control and 263 for siCep152. (O–R) 2D projections along the indicated axes of 3D-STORM images of F131E-GFP–expressing cells treated with siHsSAS-6 3′UTR reveal a torus with a mean radius of 216 ± 12 nm (n = 6). Arrowhead, area with fewer molecules. Bars: (A–D [main images], K, and M) 10 µm; (A–D [insets], L, and N) 1 µm; (E and G–J) 500 nm; (O–Q) 200 nm.
We set out to determine when during the cell cycle such a torus first appears. We synchronized cells expressing FL-GFP using mitotic shake off and examined them at successive time points thereafter for the presence of a torus and the S-phase marker proliferating cell nuclear antigen (PCNA). We found that the frequency of cells with a torus increases steadily during G1, peaking 5–6 h after mitosis (Fig. 2 F). We conclude that the torus can form during the G1 phase of the cell cycle. Importantly, we found a similar profile upon depletion of endogenous HsSAS-6 (Fig. S2, A and B). Given that centriole duplication in such cells relies on the GFP-tagged protein (Fig. 1, C and F), this finding reinforces the notion that the torus may recapitulate a transient behavior of endogenous HsSAS-6. In line with its transient nature, we found that detergent extraction leads to the complete loss of the torus in cells expressing F131E-GFP, whereas the focused signal of cells expressing FL-GFP remains, as anticipated from the protein being stably incorporated in the cartwheel (Fig. S2, C–J).
To address where within the centrosome this torus resides, we conducted confocal imaging of cells expressing F131E-GFP labeled with antibodies against GFP as well as against known distal (centrin or CP110) and proximal (c-Nap1, Pericentrin, or Cep152) centriolar markers. This analysis revealed that the F131E-GFP torus is located at the proximal end of the parental centriole (Fig. 2, G–J), largely coinciding with Cep152 and Pericentrin (Lawo et al., 2012; Sonnen et al., 2012). A similar localization was found in cells expressing FL-GFP (Fig. S2, K–N). Furthermore, we found that depletion of Cep152 results in a lack of torus (Fig. 2, E and K–N), establishing that Cep152 is needed for HsSAS-6 torus formation. To characterize this torus with further precision, we turned to 3D stochastic optical reconstruction microscopy (STORM), using a depth-independent imaging protocol enabling ∼35-nm lateral and ∼50-nm axial resolutions (Olivier et al., 2013b). This allowed us to determine that the torus is 432 ± 12 nm in diameter (Fig. 2, O–R). Overall, we conclude that GFP-tagged HsSAS-6 proteins can localize around the proximal end of the parental centriole, raising the possibility that the endogenous protein also transits through that location.
HsSAS-6 centrosomal dynamics
To determine whether HsSAS-6 is immobilized after its initial recruitment to the centrosome, we performed FRAP experiments and quantified the exchange between centrosomal and cytoplasmic pools of HsSAS-6. We generated a U2OS cell line expressing tagRFP-centrin in addition to the functional FL-GFP. We conducted these experiments both in early S phase, at the onset of procentriole formation, and in late G2, when procentriole formation is completed (Fig. S3, E and F). Cells were synchronized using a double-thymidine block and imaged by dual-color confocal microscopy. Experiments performed with or without the endogenous protein were indistinguishable (Fig. S3, G and H), and the two datasets were therefore pooled for analysis.
We found that in early S phase, ∼60% of the initial fluorescence intensity of centrosomal FL-GFP recovers within 45 min, with a t1/2 of 8.5 ± 1 min (Fig. 3, A and C). Therefore, ∼40% of centrosomal HsSAS-6 does not exchange with the cytoplasmic pool within this time frame, indicating that these molecules are immobilized. In late G2, only ∼25% of the initial centrosomal fluorescence intensity recovers within 45 min, with a similar t1/2 to the one in S phase (Fig. 3, B and C), indicating that ∼75% of HsSAS-6 is now immobilized at centrosomes.
Figure 3.
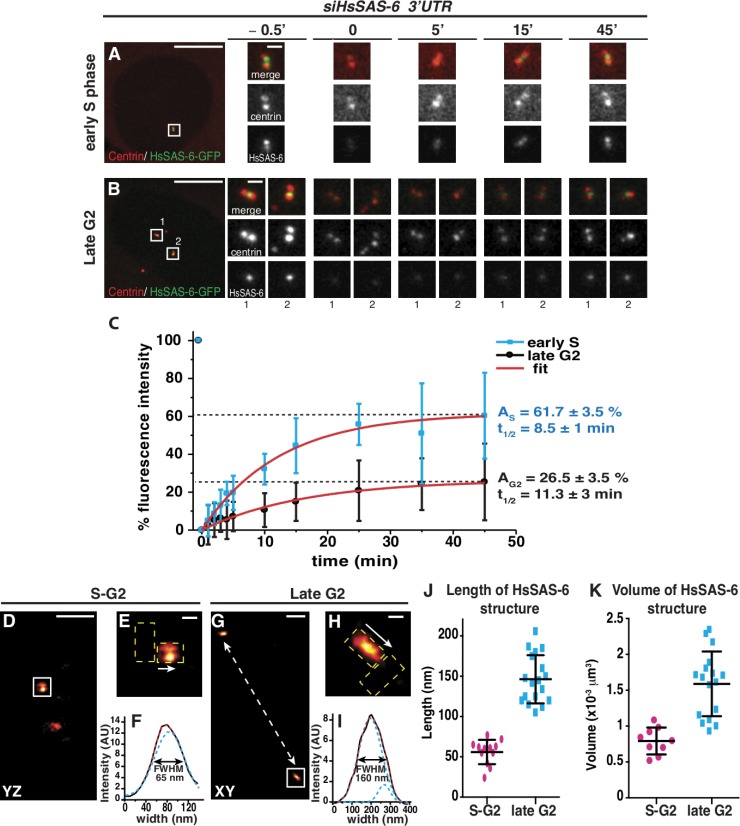
HsSAS-6 is progressively immobilized at centrosomes during the cell cycle. (A and B) Cells expressing low to medium levels of HsSAS-6–GFP were selected in early S phase (A) or late G2 (B) according to their centriolar configurations (Fig. S3, E and F), photobleached, and monitored thereafter for signal recovery. Note that given the resolution limit of confocal microscopy, no distinction can be made between pericentriolar, centriolar, or procentriolar signals. Insets are magnified regions of the boxed areas. (C) Recovery of centrosome-associated fluorescence from cells in early S phase (n = 8) or late G2 (n = 9), with 100% corresponding to prebleach intensity values, and 0% corresponding to those immediately after bleach (t0); error bars show standard deviations. Recovery curves were fitted with a single exponential function; maximum recovery amplitudes, A, and times at half-maximal recovery, t1/2, are indicated. Note that no correlation was found between levels of expression and extent of recovery among the cells analyzed. (D–H) Representative 3D-STORM images of centrosomal HsSAS-6 in S–G2 (D and E) and late G2 (G and H). Side view of HsSAS-6–containing structures shown in the indicated projections and the localization of parental and nascent centrioles (dashed yellow boxes) obtained from wide-field centrin images before STORM imaging was used to determine the length axis and quantify (F and I) the signal intensity along this axis using a line profile (arrows indicate direction; Materials and methods and J). The apparent double focus in D and E is expected from a projection of the underlying HsSAS-6 ring (Fig. 6). (J and K) Quantification of lengths (J) and volumes (K) of HsSAS-6–positive structures in 3D-STORM images reveals a significant increase from S to G2 (n = 12 for S phase, and n = 18 for G2; P < 0.0001 for both J and K). Error bars show means and standard deviations; values from individual structures are shown as circles or squares. AU, arbitrary unit; FWHM, full width at half-maximum. Bars: (A and B, main images) 10 µm; (A and B, insets) 1 µm; (D and G) 500 nm; (E and H) 100 nm.
We addressed whether this progressive immobilization is accompanied by an increase in the size of the underlying structure. 3D-STORM imaging on S–G2 and late G2 cells after detergent extraction to retain the most stably incorporated molecules revealed that the mean length of HsSAS-6–positive structures is ∼60 nm in early S phase and ∼150 nm in late G2 (Fig. 3, D–K).
Overall, these experiments establish that there is substantial recruitment of HsSAS-6 from the cytoplasm to centrosomes at the onset of S phase. More HsSAS-6 is then immobilized at centrosomes as cells progress through the cell cycle, which leads to an increase in the size of a stable HsSAS-6–containing structure.
Spatial and temporal distribution of HsSAS-6
Previous qualitative observations suggested that overall cellular levels of HsSAS-6 are low in G1 and increase gradually as cells progress through S phase and G2, whereas the signal at centrosomes becomes readily detectable in early S phase and is present until metaphase (Strnad et al., 2007). We quantified total cellular fluorescence intensity using antibodies against endogenous HsSAS-6 and found no difference between G1 and early S, a ∼2.5-fold increase as cells progress into S phase and G2, and a further approximately twofold increase as they reach late G2 (Fig. 4, A–E). In addition, we found that HsSAS-6 is clearly enriched at centrosomes starting in early S phase and increases approximately twofold as cells progress into S and G2 and a further approximately twofold by late G2 (Fig. 4, A–D and F).
Figure 4.
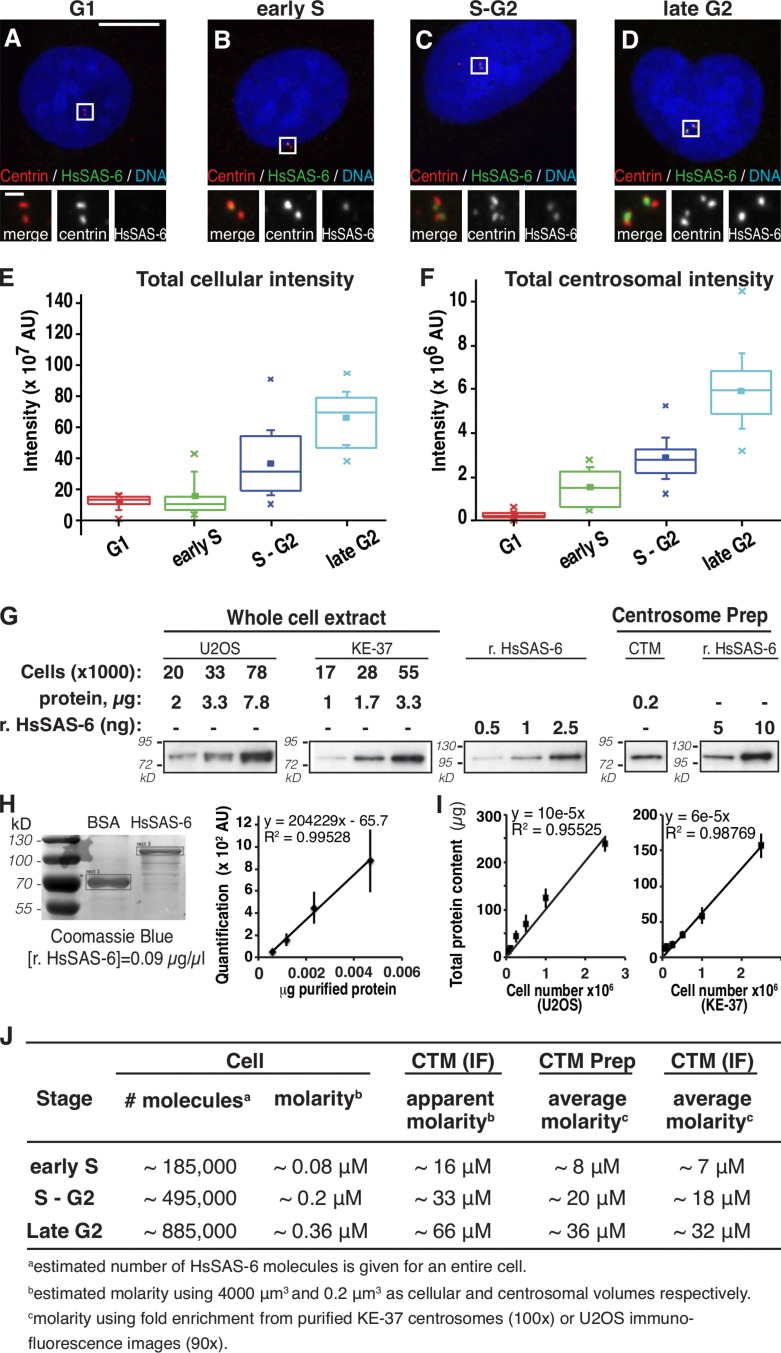
HsSAS-6 cytoplasmic and centrosomal distributions across the cell cycle. (A–D) U2OS cells synchronized by mitotic shake off, fixed, and stained at different time points thereafter for HsSAS-6, centrin-3, and DNA. Cells were binned into the indicated cell cycle stages according to centrin and HsSAS-6 configurations plus DNA condensation (Fig. S3, A–D; Materials and methods). Insets are magnified regions of the boxed areas. (E and F) Total cellular (E) and centrosomal (F) HsSAS-6 fluorescence intensity. Box plots represent lower and upper quartiles and median values as well as the means (full squares) and standard deviations, and crosses indicate minimum and maximum values. Statistical analysis: p-values and cell numbers are reported in Table S3 and Table S4. (G–J) Estimation of HsSAS-6 molarity in the cytoplasm and at centrosomes. (G) Increasing amounts of whole-cell lysates from KE-37 or U2OS cells compared with increasing amounts of recombinant HsSAS-6 (r. HsSAS-6) by Western blotting using HsSAS-6 antibodies. Proteins from purified centrosomes of asynchronous KE-37 cells (centrosome prep, CTM) were loaded in parallel to recombinant HsSAS-6. (H, left) Purified BSA and HsSAS-6 proteins were stained with Coomassie blue and quantified with GelEval software. The correlation between the indicated amounts of purified HsSAS-6 and the values obtained with GelEval is displayed on the right. Boxes define the areas taken for signal intensity measurements of BSA and HsSAS-6. (I) Increasing amounts of KE-37 and U2OS cells were lysed and the protein concentration corresponding to each lysate determined. The correlation between the cell number used and the respective total protein recovered is indicated. (H and I) Vertical lines indicate standard deviation. (J) Approximate number of HsSAS-6 molecules in the cytoplasm of early S, S–G2, and late G2 cells estimated using the mean total fluorescence intensity at these stages (E and F) and a conversion factor linking estimates by Western blotting and immunofluorescence, rounded up to the nearest 1,000, and the corresponding cellular molarity using a volume of 4,000 µm3 for U2OS cells (Beck et al., 2011). The apparent centrosomal molarity was estimated using centrosomal HsSAS-6 intensities from F and a volume of 0.2 µm3 (optical resolution limit); mean centrosomal molarities were estimated using fold enrichment values determined from KE-37 purified centrosomes (100×; CTM prep) or from U2OS immunofluorescence (images [90×; CTM (IF)]). See Materials and methods for calculation details. AU, arbitrary unit. Bars: (main images) 10 µm; (insets) 1 µm.
We set out to estimate the molarity of HsSAS-6 in the cytoplasm and at centrosomes, notably to evaluate whether the protein could readily undergo homodimerization or oligomerization in either location. To estimate the total amount of HsSAS-6 per cell, we performed Western blot analysis of lysates from an asynchronous population and compared the resulting signal intensity to that of known concentrations of recombinant protein. We determined that U2OS cells contain on average ∼0.05 pg HsSAS-6 per cell. Similar values were obtained for KE-37 cells (Materials and methods; Fig. 4, G–I). We then estimated the cytoplasmic molarity by considering the distribution of cells in each phase of the cell cycle and the fluorescence intensity for each phase (Materials and methods; Fig. 4 E), yielding concentrations of ∼0.08 µM in early S and ∼0.36 µM in late G2 (Fig. 4 J). These values are below the Kd required for homodimerization of related SAS-6 proteins in vitro (∼1 µM; Kitagawa et al., 2011; van Breugel et al., 2011) and well below that needed for oligomerization of HsSAS-6 N-terminal domains (∼50 µM; van Breugel et al., 2011).
We sought to estimate centrosomal HsSAS-6 molarity initially using the total centrosomal fluorescence intensity determined at specific stages of the cell cycle (Fig. 4 F) and considering as a volume the optical resolution of the confocal microscope (∼0.2 µm3; Fig. 4 J). This analysis yields concentrations of ∼16 µM and ∼66 µM in early S and late G2, respectively. Because centrosomal concentrations depend significantly on the volume considered, we also estimated the centrosomal molarity using two volume-independent approaches. First, we conducted Western blot analysis of centrosomes purified from asynchronously growing KE-37 cells and estimated that HsSAS-6 is ∼100-fold more concentrated at centrosomes compared with the cytoplasm (Materials and methods; Fig. 4 G). Second, we estimated by immunofluorescence the fold increase of the centrosomal signal over the cytoplasmic signal to be ∼90 in an asynchronous population of U2OS cells (n = 42; Materials and methods). These two estimates concur to indicate that the mean molarity of HsSAS-6 at centrosomes is ∼8 µM in early S and ∼36 µM in late G2 (Fig. 4 J). These values are in the same range as those estimated initially and are high enough to enable homodimerization of HsSAS-6 proteins. Moreover, given that HsSAS-6 may be more concentrated within certain centrosomal subvolumes, such as that observed in early S phase (Fig. 3, E and F), it is possible that the effective molarity in such subvolumes is high enough for oligomerization (Discussion). Overall, these experiments lead us to conclude that levels of centrosomal HsSAS-6 augment significantly between early S and late G2 and raise the possibility that concentration of HsSAS-6 within centrosomal subvolumes provides a mechanism driving oligomerization.
HsSAS-6 oligomerization state
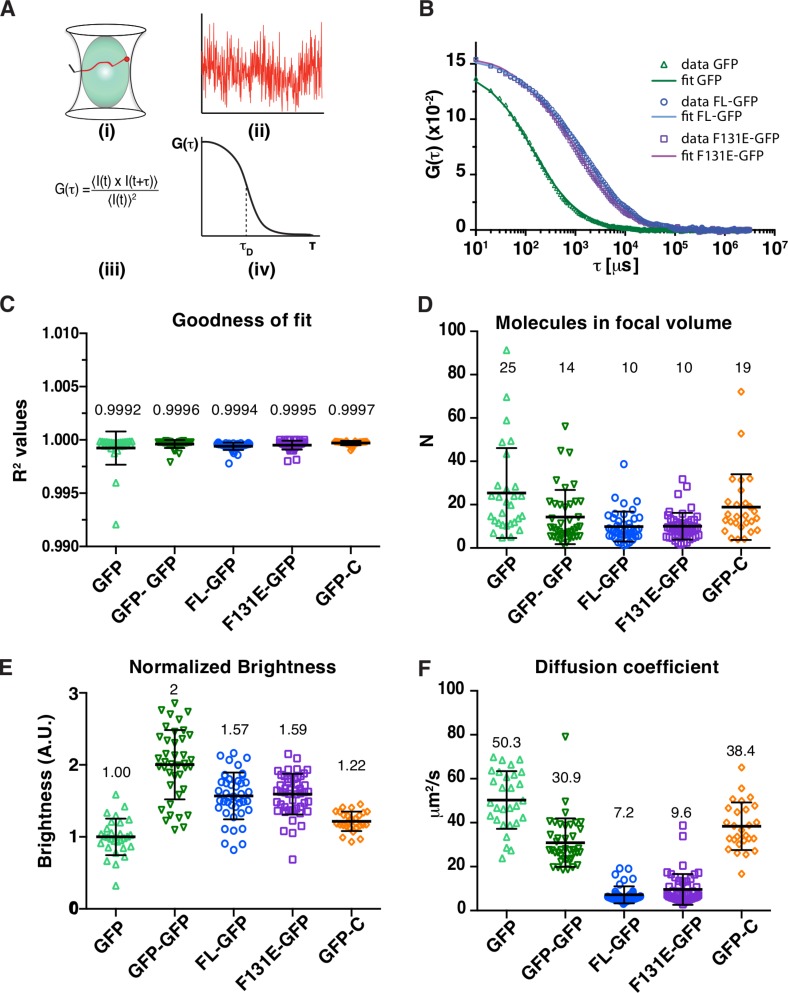
The oligomerization state of SAS-6 proteins has not been directly ascertained in vivo. We addressed this question by conducting fluorescence correlation spectroscopy (FCS) in the cytoplasm of live cells depleted of endogenous HsSAS-6 and expressing either FL-GFP, F131E-GFP, or GFP-C, which is expected to be strictly monomeric in the absence of the coiled-coil domain (Kitagawa et al., 2011; van Breugel et al., 2011). In FCS experiments (Bulseco and Wolf, 2007; Chen et al., 2008), fluctuations in fluorescence intensity are monitored as molecules pass through the observation volume (Fig. 5 A, i and ii). These fluctuations are correlated over time, yielding an autocorrelation function (ACF; Fig. 5 A, iii and iv), which is then fitted to diffusion models and yields the mean number N of molecules in the observation volume, their brightness (counts per molecule [CPM]), and their diffusion coefficients. From these parameters, the oligomerization state of molecules can be directly extracted.
Figure 5.
FCS analysis of cytoplasmic HsSAS-6. (A) Schematics of FCS measurements: intensity fluctuations of fluorescent molecules in the observation volume (i) are recorded over time (ii), analyzed using an autocorrelation function (iii), and plotted as a function of the delay times τ, thus yielding the characteristic diffusion time τD linked to the diffusion coefficient (Bulseco and Wolf, 2007; Chen et al., 2008). FCS analysis was performed with iU2OS stably expressing FL-GFP, F131E-GFP, GFP-C, or GFP–GFP as well as on U2OS cells transiently transfected with a GFP construct; endogenous HsSAS-6 was depleted concomitantly to induction of FL-GFP, F131E-GFP, and GFP-C expression for 48 h, and low- to medium-expressing cells were selected to avoid overexpression artifacts. (B) Single representative FCS curves and their fit for GFP, FL-GFP, and F131E-GFP. Note that lag times <10 µs were clipped from the graph as they represent intramolecular events rather than protein diffusion coefficients (Bulseco and Wolf, 2007). (C) R2 values from one-component anomalous diffusion fittings of the different constructs indicate that this model is a good approximation of the experimental data. Values of individual fits, calculated means (indicated above), and standard deviations are shown. (D) The mean molecule number extracted from the FCS curve fit indicates that all observation volumes contained comparable numbers of fluorescent proteins (mean values indicated above). (E and F) Normalized brightness (E) and characteristic diffusion coefficients (F) are shown (Materials and methods) as well as individual fit values, calculated means (indicated above), and standard deviations. Cell number from three experiments and statistical analysis are reported in Table S5 and Table S6. Note that no correlation was found between number of molecules detected within a focal volume (concentration) and the diffusion coefficient or brightness values, indicating that higher expression did not lead to detectable oligomeric species. A.U., arbitrary unit.
All the ACF curves could be fitted to a model for single component anomalous diffusion (Materials and methods; Fig. 5, B and C). Although the presence of low-abundant additional species cannot be excluded, this analysis suggests that all HsSAS-6 variants analyzed diffuse primarily as a single component, indicating that they each likely behave as a single macromolecular species. We also assessed the brightness detected for each protein, which is directly extracted from the fluorescent traces (Fig. 5 A, ii) and thus independent of the mathematical model used to fit the data. We used constructs carrying one or two copies of GFP to calibrate these values. As shown in Fig. 5 E, cells expressing GFP–GFP have twice the normalized brightness of those expressing single GFP and exhibit a lower diffusion coefficient, as expected from a larger protein (Fig. 5 F).
We found that GFP-C has a mean brightness similar to that of monomeric GFP (Fig. 5 E), and its diffusion coefficient is analogous to that of the similarly sized GFP–GFP (Fig. 5 F). These results establish that GFP-C exists mainly as a monomer in the cytoplasm. Furthermore, we found that FL-GFP and F131E-GFP exhibit similar brightness, ∼1.6 times that of monomeric GFP, and similar diffusion times (Fig. 5, E and F). These results demonstrate that FL-GFP and F131E-GFP exist in complexes in the cytoplasm that do not differ significantly in terms of their hydrodynamic radius. The ∼1.6-fold increased brightness of both FL-GFP and F131E-GFP as compared with GFP raises the possibility that both species are present in a mixture consisting predominantly of HsSAS-6 homodimers (brightness of two GFP) and of monomers (brightness of single GFP), thus leading to an intermediate brightness; in this case, both species should be described by a two-component diffusion model. However, we favor the alternative view whereby association of a GFP-tagged protein with residual untagged endogenous HsSAS-6 forms a dimer with the brightness of a single GFP, thus reducing the overall mean brightness. Moreover, quenching between two close GFP molecules at the C terminus of HsSAS-6 may also decrease the fluorescence exhibited by bona fide marked homodimers. Finally, we noted that the FCS measurements yield cytoplasmic concentrations of ∼0.1 µM of FL-GFP and F131E-GFP in the focal volume, which are in the range of those extracted for endogenous HsSAS-6 (Fig. 4). In conclusion, these findings reveal that wild-type and oligomerization-defective F131E mutant HsSAS-6 molecules have an indistinguishable behavior and exist predominantly as homodimers in the cytoplasm.
Centriolar HsSAS-6 is distributed in a ninefold radially symmetric manner
We wanted to address whether ring formation of HsSAS-6 occurs in the cartwheel of procentrioles in human cells. As FCS is restricted to monitoring proteins that are mobile and at low concentration, we turned to STORM microscopy to investigate HsSAS-6 distribution in situ. These experiments were performed in detergent-extracted cells so as to retain primarily the immobile centriolar fraction.
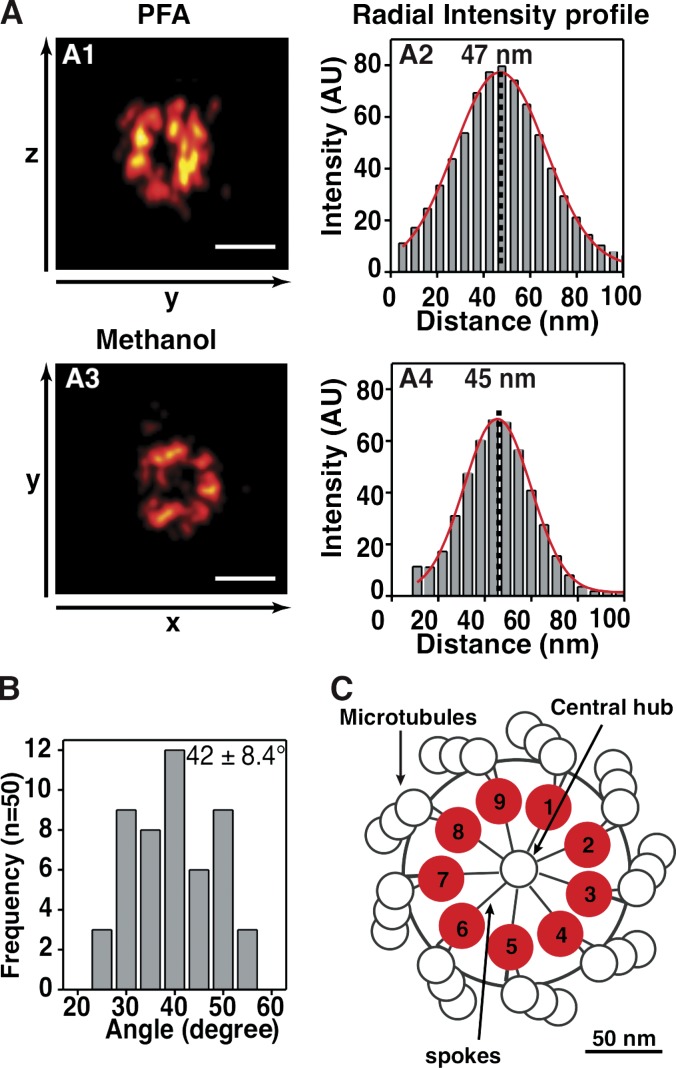
Because centrioles exhibit a variety of orientations, we took advantage of a buffer-enhanced 3D-STORM approach that enables near-isotropic lateral and axial resolutions (Olivier et al., 2013a). We analyzed cells synchronized by mitotic shake off, focusing on early S phase and thus on the early stages of procentriolar formation (Fig. S3, A–C). To best determine the organization of HsSAS-6 assemblies and their underlying symmetry, we used a monoclonal antibody directed against the C-terminal domain of HsSAS-6 (Fig. S1) labeled with Alexa Fluor 647 antibodies. Strikingly, we found that HsSAS-6 molecules are distributed in ringlike patterns with a mean radius of 45 ± 3 nm (Fig. 6 A). This allowed us to localize for the first time HsSAS-6 C termini within the centriole (Fig. 6 C). Importantly, we found that the angular distribution of the signal intensities exhibits a period of 42 ± 8° (Fig. 6 B and Fig. S4), consistent with the values published previously for in vitro assemblies (Kitagawa et al., 2011). These results strongly support the notion that N–N interaction and ring formation occurs at centrioles in human cells and that HsSAS-6 exhibits a ninefold symmetric arrangement in its cellular context (Fig. 6 C).
Figure 6.
Centriolar HsSAS-6 is distributed with a ninefold radial symmetry. (A, left) 2D projection (sum of intensities) along the indicated axes of 3D-STORM images of HsSAS-6 rings in S-phase cells, preextracted, and fixed with either PFA (A1) or methanol (A3) and stained with C-terminal–specific antibodies (Fig. S2). (right) Corresponding radial intensity histogram fitted with a Gaussian (red lines) to extract the indicated radius values. The ring radii measured in samples fixed with PFA is 45 ± 2 nm (n = 8), 45 ± 4 nm (n = 13) for MeOH-fixed samples, or 45 ± 3 nm (n = 21) for the sum of both. The black dotted lines mark the center of the Gaussian fit (red) of signal intensities, which is taken as the radius (indicated above). (B) Frequency of angular distribution on imaged HsSAS-6 rings as measured by peak-to-peak distance (n = 6 centrioles yielding 50 angular values; see also Fig. S4); means and standard deviations are indicated. (C) Model of HsSAS-6 C termini localization within the cartwheel. AU, arbitrary unit. Bars, 100 nm.
Discussion
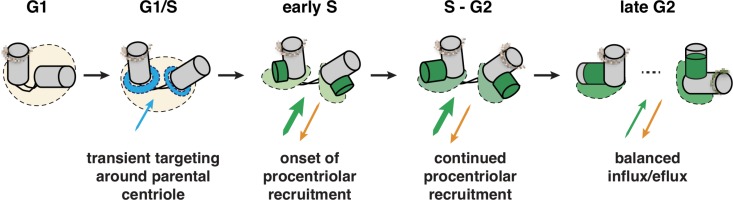
How the defining ninefold radial symmetry of centrioles is established has been a long-standing question in cell biology. The cartwheel plays a critical role in imparting this remarkable organization from algae to man. The finding that SAS-6 protein fragments can self-assemble in vitro into a structure that resembles the cartwheel has led to a model whereby this family of proteins is critical for establishing the ninefold symmetry of centrioles (reviewed in Gönczy, 2012). Many questions remained open at the onset of our work regarding the mechanisms by which this protein operates in vivo. Our analysis has uncovered a series of steps that lead from HsSAS-6 being present in the cytoplasm to its incorporation into a ninefold symmetrical structure during procentriole formation. These steps are discussed in this section and represented schematically in Fig. 7.
Figure 7.
Working model of HsSAS-6 distribution at centrosomes across the cell cycle. HsSAS-6 resides initially in the cytoplasm in a mixture of monomers and predominantly homodimers. During G1, HsSAS-6 is recruited around the parental centriole to a labile structure, the torus (blue), which coincides with Cep152 and Pericentrin. This localization might enable high enough HsSAS-6 concentrations to favor oligomerization, incorporation into a ninefold symmetrical ring structure, and initiation of cartwheel assembly. Throughout S–G2, progressive recruitment and immobilization of HsSAS-6 molecules into the assembling procentriole lead to the elongation of a stable HsSAS-6–positive structure, until a steady state of balanced exchange between centrosomal and noncentrosomal HsSAS-6 molecules is reached. See Discussion for additional details.
Cytoplasmic homodimerization: A first step in the assembly process
Considering that the coiled coil of C. elegans SAS-6 and of C. reinhardtii Bld12p homodimerizes with a Kd of ∼1 µM in vitro (Kitagawa et al., 2011) and that our estimates based on Western blotting, immunofluorescence, and FCS indicate that the concentration of HsSAS-6 in the cytoplasm is on the order of 0.08–0.36 µM, one would have expected cytoplasmic HsSAS-6 to be monomeric. However, our FCS measurements indicate that a significant fraction of cytoplasmic HsSAS-6 is homodimeric. Perhaps the Kd for the complete FL HsSAS-6 is lower than that measured for the SAS-6 and Bld12p fragments lacking the tail domain (Kitagawa et al., 2011; van Breugel et al., 2011) or else HsSAS-6 homodimerization could be aided in the cell by other proteins or an increased effective local concentration in the crowded cytoplasmic environment. Our findings using FCS establish also that HsSAS-6 oligomerization does not occur in a detectable manner in the cytoplasm, as expected given that the Kd between HsSAS-6 head domains in vitro is ∼50 µM (van Breugel et al., 2011). In contrast, DmSas-6 molecules assemble as tetramers in Drosophila melanogaster embryonic extracts (Gopalakrishnan et al., 2010), suggesting that the status of SAS-6 proteins in the cytoplasm differs between species or cell types. In some cases, multiple procentrioles can also assemble de novo in the cytoplasm of mammalian cells, as in differentiated multiciliated epithelial cells (Vladar and Stearns, 2007). In such a situation, it is possible that HsSAS-6–containing cartwheel intermediates or preassembled cartwheels exist in the cytoplasm, perhaps as a result of substantial increase in HsSAS-6 gene expression (Hoh et al., 2012).
Centrosomal recruitment: Targeting HsSAS-6 to the right place
Around the G1/S transition, HsSAS-6 becomes clearly detectable at centrosomes. Together with earlier findings (van Breugel et al., 2011), our work demonstrates that the coiled coil of HsSAS-6 is necessary and sufficient for centrosomal targeting. Given that coiled-coil domains often mediate protein–protein interactions, HsSAS-6 centrosomal targeting could be achieved through interaction with a partner protein. A plausible candidate is STIL, the human relative of C. elegans SAS-5 and Drosophila Ana2 (Stevens et al., 2010). In C. elegans, SAS-6 and SAS-5 physically interact and are mutually dependent for their centriolar localization (Leidel et al., 2005). Moreover, SAS-5 exchanges readily between the cytoplasm and centrosomes and was proposed to target SAS-6 to centrosomes (Leidel et al., 2005). STIL recapitulates several features of SAS-5, being required for centriole duplication and shuttling rapidly between cytoplasm and centrosomes; furthermore, HsSAS-6 and STIL colocalize at procentrioles and appear to be mutually dependent for their localization (Arquint et al., 2012; Vulprecht et al., 2012). However, no direct interaction has been found between STIL and HsSAS-6 (Vulprecht et al., 2012; Lukinavičius et al., 2013), indicating that the relationship between the protein pairs might differ between C. elegans and human cells. Another candidate to target HsSAS-6 centrosomal localization is Cep135. Indeed, an interaction has been detected between the C-terminal domain of Cep135 and parts of the HsSAS-6 coiled-coil and tail domains (Lin et al., 2013). However, cells depleted of Cep135 retain HsSAS-6–positive procentrioles in S phase, suggesting that Cep135 is dispensable for the initial steps of HsSAS-6 centrosomal recruitment (Lin et al., 2013).
A recruitment platform for HsSAS-6 around the parental centriole?
Further insight into the mechanisms targeting HsSAS-6 to centrosomes is suggested by our finding of a torus at the proximal end of the parental centriole in cells expressing GFP-tagged variants of the protein. We show that this localization is labile and depends on the presence of Cep152, a protein known to mediate the centriolar localization of the core centriolar proteins Plk4 and CPAP (centrosomal protein 4.1–associated protein)/SAS-4, both in human (Cizmecioglu et al., 2010; Hatch et al., 2010) and Drosophila (Dzhindzhev et al., 2010). Although the torus localization cannot be captured with endogenous HsSAS-6, perhaps because of its extremely transient nature, we propose that it reflects a recruitment platform for HsSAS-6. Interestingly, Cep152 localizes to a torus ∼400 nm in diameter around the parental centriole already in G1 (Cizmecioglu et al., 2010; Hatch et al., 2010; Lawo et al., 2012), such that it could promote HsSAS-6 recruitment to this site before S phase. The presence of a torus in cells expressing F131E-GFP is reminiscent of results in C. elegans in which oligomerization-defective SAS-6 is targeted to centrosomes but cannot be incorporated in the procentriole thereafter (Lettman et al., 2013). Intriguingly, recent evidence in human cells indicates that Plk4 also exhibits a toroid distribution early in G1, overlapping with the Cep152 torus (Kim et al., 2013). Moreover, Plk4 concentrates thereafter to a single position; because a region with less signal could be observed in the F131E-GFP torus, (Fig. 2 P, arrowhead), similar to that observed with Pericentrin (Lawo et al., 2012), it will be interesting to address whether this coincides with the Plk4 focus and perhaps marks the site of procentriole assembly.
Cartwheel assembly: HsSAS-6 oligomerization
Once recruited to centrosomes, HsSAS-6 homodimers presumably undergo oligomerization, either within the torus or the elongating cartwheel. Our estimates indicate that the apparent mean molarity of HsSAS-6 at centrosomes is ∼8 µM in early S phase, below the Kd expected to be required for oligomerization. However, higher concentrations are likely achieved within centrosomal subvolumes, thus favoring N–N interaction and oligomerization. For instance, considering the torus size of 0.04 µm3 determined by 3D-STORM, a maximum local molarity (i.e., assuming that all HsSAS-6 centrosomal proteins are in this subvolume) of ∼80 µM in early S would be achieved. Alternatively, proteins such as Cep135, which interacts with HsSAS-6 (Lin et al., 2013), or Plk4/ZYG-1, which is required for the robust presence of SAS-6 proteins at centrioles in human cells and in C. elegans (Strnad et al., 2007; Lettman et al., 2013), could help organize HsSAS-6 to favor oligomerization and incorporation within the nascent procentriole.
Superresolution methods uncovered the precise organization of several pericentriolar proteins (Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012) and the ninefold symmetrical organization of the appendage protein Cep164 (Lau et al., 2012). However, the nanoscale organization of cartwheel proteins such as HsSAS-6 has remained elusive because of the minute size of this structure. Our 3D-STORM images provide the first visualization of ninefold symmetrical HsSAS-6 rings at centrioles in the cellular context. Unexpectedly, STORM microscopy revealed a ∼2.5-fold increase in the length of stable HsSAS-6–containing structures between S and late G2 (Fig. 3, D–K), exceeding the ∼100-nm length of the cartwheel observed by EM in human cells (Guichard et al., 2010). This may reflect an unsuspected alteration in cartwheel size during procentriole formation or a fraction of stable HsSAS-6 present outside the cartwheel during late G2.
To conclude, we propose a model for HsSAS-6 assembly during procentriole formation in human cells (Fig. 7). In this model, HsSAS-6 molecules are present primarily as homodimers in the cytoplasm, before being recruited transiently around the parental centriole and then assembling into oligomers to form ninefold symmetrical rings in the cartwheel of the emerging procentriole. Elucidating the mechanisms that regulate HsSAS-6 assembly at each of these steps is expected to provide critical insight on how the evolutionarily conserved ninefold radial symmetry of centrioles is achieved.
Materials and methods
Cloning and mutagenesis
HsSAS-6 variants were cloned into a pENTR1A (Life Technologies) Gateway entry vector, modified by introducing restriction sites between the attR1-attR2 sites (5′ AgeI and XbaI 3′). HsSAS-6 inserts were generated by PCR (primers listed in Table S7), cloned into pENTR vectors using AgeI and XbaI, and sequence verified before Gateway recombination into the destination vectors pEBTet-GW-EGFP (FL and F131E) or pEBTet-EGFP-GW for all other constructs (Strnad, 2008).
The tagRFP–centrin-1 plasmid was generated by digesting pEGFP-C1–centrin-1 (gift from M. Bornens, Institut Curie, Paris, France) with BamHI and Acc65I and ligating centrin-1 into these sites in pTagRFP-C (FP141; Evrogen). The plasmid was digested with NheI and BamHI before ligation into pIRES-neomycin (gift from D. Gerlich, Institute of Molecular Biotechnology, Vienna, Austria) and transfected into U2OS cells to generate stable cell lines.
Cell lines, RNAi, and synchronization
Cell line maintenance and generation.
U2OS cells (European Collection of Cell Cultures) were maintained at 37°C and 5% CO2 in McCoy’s 5A GlutaMAX (Life Technologies) with 10% FBS or tetracycline-negative FBS (Chemie Brunschwig, AG) for the inducible episomal cell lines (iU2OS). iU2OS were generated by transfecting U2OS cells with pEBTet plasmids using Lipofectamine 2000 (Life Technologies) and selected with 1 µg/ml puromycin. Induction was performed with 1 µg/ml doxycycline for 48 h. KE-37 cells were maintained at 37°C and 5% CO2 in RPMl 1640 (Life Technologies) with 10% FBS.
RNAi.
Depletion of HsSAS-6 with Stealth siRNA (3′UTR, 5′-GAGCUGUUAAAGACUGGAUACUUUA-3′; Life Technologies) or of Cep152 with locked nucleic acid–based siRNAs (ORF, 5′-GGAUCCAACUGGAAAUCUATT-3′; Life Technologies) was performed as previously reported (Kitagawa et al., 2011; Lukinavičius et al., 2013). Stealth RNAi siRNA Lo GC was used as a negative control (Life Technologies). To assess the function of HsSAS-6 constructs and when indicated, siHsSAS-6 3′UTR treatment was concomitant to induction with doxycycline for 48 h.
Cell synchronization.
Mitotic shake off was used to synchronize cells for detection of the FL-GFP torus, quantification of HsSAS-6 levels, and STORM imaging. For FRAP experiments, U2OS cells were synchronized by a double-thymidine block (2 mM), doxycycline was added during the second thymidine block, and samples were taken at successive time points after release from the block. No synchronization was performed for cells treated with siHsSAS-6 3′UTR, but cells were binned according to their centriolar organization in that case (see section Cell cycle stage determination).
Immunofluorescence and antibodies
Fixation.
Cells were washed with PBS and fixed with methanol (Sigma-Aldrich) for 7 min at −20°C or with 2% formaldehyde (Thermo Fisher Scientific) in PBS for 20 min at room temperature. Samples were blocked in 5% BSA for ∼30 min before incubation for 2 h at room temperature with primary antibodies diluted in antibody solution (1% BSA-PBS and 0.2% Triton X-100; Sigma-Aldrich). After washing (PBS–0.2% Triton X-100), the coverslips were incubated for 45 min with secondary antibodies. After washing, coverslips were mounted in glycerol-based mounting medium or kept at 4°C in PBS with penicillin-streptomycin (Gibco) for STORM imaging.
Preextraction.
Cells were preextracted for 30–60 s in 0.5% Triton X-100 in BRB80 (80 mM K-Pipes, 1 mM MgCl2, and 1 mM EGTA; all obtained from Sigma-Aldrich), washed with PBS, and fixed with either methanol or 2% PFA.
EdU labeling.
1 µM EdU was added to the cells for 15–60 min before fixation. EdU staining was performed using EdU Alexa Fluor 647 or Alexa Fluor 488 kit (Click-iT; Life Technologies) after antibody staining.
Antibodies.
Primary antibodies for immunofluorescence were 1:4,000 mouse centrin (20H5; gift from J.L. Salisbury, Mayo Clinic Foundation, Rochester, NY), 1:2,000 rabbit anti–centrin-3 (Middendorp et al., 2000; gift from M. Bornens), 1:2,000 rabbit Cep152 (HPA039408; Sigma-Aldrich), 1:400 mouse CNAP-1 (611375; BD), 1:2,000 rabbit CP110 (12780–1-AP; ProteinTech Europe), 1:500 rabbit anti-GFP (gift from V. Simanis, Swiss Federal Institute of Technology, Lausanne, Switzerland), 1:400 goat GFP (ab6673; Abcam), 1:500 mouse anti–HsSAS-6 (sc-81431; Santa Cruz Biotechnology, Inc.), 1:1,000 mouse PCNA (sc-56; Santa Cruz Biotechnology, Inc.), and 1:1,000 goat Pericentrin (sc-28143; Santa Cruz Biotechnology, Inc.). Secondary antibodies, used at 1:1,000, were Alexa Fluor 488, 568, or 647 (Life Technologies) for confocal imaging or Alexa Fluor 647 F(ab′)2 (A-21237; Life Technologies) for STORM imaging. DNA staining was performed with ∼1 µM Hoechst in PBS.
Cell extract preparation and Western blotting
Cell extract preparation.
Extracts were prepared in lysis buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 0.5% NP-40, and 150 mM NaCl) in the presence of protease inhibitor cocktail cOmplete (Roche). Protein concentration was measured with the Bio-Rad Protein Assay kit (Bio-Rad Laboratories).
Western blotting.
SDS-PAGE was performed using 4–15% or 10% polyacrylamide gels (Criterion; Bio-Rad Laboratories) followed by transfer onto nitrocellulose membrane (GE Healthcare). The membrane was probed with primary antibodies followed by incubation with HRP-conjugated secondary antibodies, and the signal was detected with chemiluminescence. Primary antibodies were mouse HsSAS-6 (1:1,000; sc-81431), mouse tubulin (1:200,000; T5168; Sigma-Aldrich), and rabbit GFP (1:1,000; gift from V. Simanis). Rabbit or mouse HRP-conjugated secondary antibodies (65–6120 and 62–6520, respectively; Promega) were used at 1:5,000.
Microscopy
Confocal imaging.
Imaging was performed on an upright confocal microscope (LSM 710; Carl Zeiss) and using a 63× Plan Apochromat oil objective (differential interference contrast M27) with a 1.4 NA; the built-in filters main dichroic beam splitter 488/561/633 nm and main dichroic beam splitter 405 nm were used in the Frame Fast mode. 16-bit, 1,024 × 1,024–pixel (px) images were taken with a dwell time of 0.64 µs/px using the ZEN software (Carl Zeiss). A px size of 55 nm, an optical section of 0.7 µm, and z steps of 200 or 300 nm were taken, unless specified otherwise. For high-resolution confocal imaging of the HsSAS-6 torus seen in Fig. 2 (A–D), a px size of 43 nm and a z step of 120 nm were taken with a 1.27-µs/px dwell time (speed 8) and a line average of 2; moreover, a Gaussian blur of σ = 1 px was applied in Fiji, and single z planes were selected for representation. For Fig. 2 (G–J) and Fig. S2 (K–P), a px size of 170 nm and a z step of 100 nm were used with a 1.58-µs/px dwell time (speed 9) and a line average of 2.
FRAP.
FRAP was performed using the ZEN software on an inverted confocal microscope (LSM 700; Carl Zeiss) and using a 63× Plan Apochromat oil objective (differential interference contrast M27) with an NA of 1.4. px sizes of 50 nm in xy and a z step of 500 nm were used for imaging. The variable dichroic was set at 530 nm and long pass 560-nm and short pass 555-nm filters were used for the two photomultiplier tubes. Imaging was performed in an equilibrated chamber at 37°C, 5% CO2, and the cell medium was changed before to phenol red–free Leibovitz’s medium (Life Technologies) containing doxycycline and supplemented with 10% FBS. A prebleach stack was acquired before photobleaching a 2.5-µm2 region comprising the centrosome using the 488-nm laser at 100% laser power for ∼250 ms. Subsequently, z stacks covering the cell were taken at a 1-min interval for 5 min, at a 5-min interval for 10 min, and at a 10-min interval for 30 min up to ≤45 min after photobleaching. Imaging was performed at 1% 488-nm laser power and initially 1% 568-nm laser power (the 568-nm laser power was adjusted thereafter to correct for photobleaching of tagRFP–centrin-1).
Image processing and quantification
Cell cycle stage determination.
For both FRAP and HsSAS-6 levels analysis, cells were binned according to their centriolar organization, using centrin and HsSAS-6 distribution. The cell cycle stages were determined as follows: G1 (two centrin foci; no HsSAS-6), early S (two centrin foci; one or two HsSAS-6 foci), S–G2 (three to four close centrin foci; two HsSAS-6 foci), and late G2 (four well-defined centrin foci; two HsSAS-6 foci, with separated diplosomes). For each cell, the signal was measured in Fiji, and the values of mean intensity (mean), total integrated intensity (sum), and area (area, in px) were recorded and used for quantification as described and represented in Fig. S3 D.
Quantifying HsSAS-6 levels over the cell cycle.
Cells were fixed with methanol, and stained for centrin-3 and HsSAS-6. Cells were binned in different cell cycle stages according to centriolar configurations and DNA condensation. The areas and quantification procedure used are illustrated in Fig. S3 D. Total HsSAS-6 cellular intensity was quantified by drawing a region (cell) over the entire cell, including the centrosome, and measuring the mean intensity (MeanCell) and integrated intensity (SumCell) values. To remove microscope noise, the background was measured on areas outside the cells, and the mean value from 24 different such areas (MeanBG) from different coverslips was normalized to the cytoplasmic area and then subtracted from the total cytoplasmic intensity. Corrected cellular intensity (CorrSumCell) refers to the intensity after background subtraction as shown on Fig. 4 E. For each cell, one to two centrosomal areas were drawn, and the centrin signal was used to assess whether the entire centrosome was contained in the stacks. Centrosomal (PCM + centriole) intensity was measured within a circular region of 20-px diameter (∼1.1 µm) on single centrosomes. Centrosomal intensity (IntCTM) was calculated by subtracting the mean cellular intensity (not background corrected) normalized to centrosomal area from the centrosomal intensity (Fig. S3 D). As the centrosomal signal intensity represents 1% of the total cellular intensity, including the former in the latter does not significantly affect the overall cellular intensity.
FRAP image processing and signal recovery quantification.
To increase signal-to-noise ratio before quantification, the image was scaled by 0.5 (final px size of 100 nm) and a Gaussian blur of σ = 2 px was applied. Time points at which centrosomes were out of focus (judged by tagRFP–centrin-1) were excluded from the analysis. Centrosomes were identified using the centrin channel, and a circle of 22 px was used to measure centrosomal signal. Quantification of signal recovery after photobleaching was performed in Fiji as represented in Fig. S3 D and described in the section Quantifying HsSAS-6 levels over the cell cycle, but the centrosomal intensity value (IntCTM) was further divided by the mean cytoplasmic intensity (unbleached) to account for photobleaching, yielding the normalized intensity (NormIntCTL). Recovery points were fitted with a single exponential function: y = A(1 − exp(−t/τ)), yielding the maximum recovery amplitudes A and the time at half-maximal recovery t1/2 = ln(2) × τ.
Quantifying HsSAS-6 molecule numbers and molarity
Whole-cell extract (WCE) preparation.
Exponentially growing U2OS or KE-37 cells were counted with a cell counter (ADAM; Digital Bio). WCEs from increasing cell numbers (50,000–2.5 million cells) were prepared in lysis buffer (section Cell extract preparation). The correlation between the number of cells used for the WCE and the protein concentration of the respective extracts was determined. Increasing amounts of cells and of recombinant HsSAS-6 were loaded on a 10% polyacrylamide gel, and SDS-PAGE was performed.
Purified centrosome extracts.
Centrosome preparations from KE-37 cells were performed. Centrosomes were isolated from the lymphoblastic cell line KE-37 as described previously (Bornens et al., 1987). In brief, exponentially growing cells were treated with 200 nM nocodazole and 1 µg/ml cytochalasin B and lysed 1 h thereafter in a low ionic strength buffer in the presence of detergent. The lysate was cleared by centrifugation to eliminate cell debris and chromatin before isolation of centrosomes through centrifugation in a discontinuous sucrose gradient (40, 50, and 70%). Fractions were collected, and the number of centrosomes in each fraction was estimated by immunofluorescence with centrosomal markers.
Expression and purification of GST–HsSAS-6.
FL HsSAS-6 was expressed in Escherichia coli BL21(DE3) (Agilent Technologies) from a pGEX–HsSAS-6 vector. Bacteria were grown at 37°C to an OD600 of 0.5 and then at 30°C overnight after addition of IPTG to a final concentration of 1 mM. Two-step purification of GST–HsSAS-6 was performed using the Rapid GST Inclusion Body Solubilization and Renaturation kit (Cell Biolabs, Inc.) followed by glutathione–Sepharose 4B beads (GE Healthcare) at 4°C. Washes were performed with 50 mM TrisHCl, pH 7.5, 300 mM NaCl, and 1 mM DTT complemented with proteinase inhibitor cocktail. Elution of the recombinant protein was performed with 50 mM glutathione, pH 7.5, and 500 mM NaCl.
Quantification of total cellular protein content.
Western blotting was performed and the signal intensities of endogenous and recombinant HsSAS-6 were quantified with GelEval (FrogDance Software). Given that the signal intensities from recombinant HsSAS-6 correlated with known protein amounts, the total protein content for a given cell number could be then extrapolated. The calculation was performed as follows: from the amount of protein loaded (Fig. 4 G, protein, µg), we estimated the corresponding cell number using the formulas indicated in Fig. 4 I (y = 10 × 10−5x for U2OS, and y = 6 × 10−5x for KE-37, in which x = number of cells and y = micrograms of protein). Next, the signal intensity from the WCE blots (Fig. 4 G, WCE) was correlated to that of purified HsSAS-6 protein by the formula specified in Fig. 4 H (y = 204,229x − 65.7, in which x = quantification value [arbitrary unit] and y = micrograms of purified protein). Finally, these two values (number of cells and HsSAS-6 content) served to estimate the HsSAS-6 content per cell (expressed in picograms/cell), that being 0.044 pg in KE-37 and 0.055 pg in U2OS. To calculate the number of molecules per cell in U2OS, we converted the picograms/cell to molecules by taking into consideration HsSAS-6 molecular mass (74 kD) and the Avogadro number and rounded up to the nearest 1,000.
Estimating the fold enrichment of HsSAS-6 molecules at centrosomes.
The signal intensity from the WCE and centrosome blots (Fig. 4 G) was correlated to that of purified HsSAS-6 and corrected by the amount of protein loaded, yielding an HsSAS-6 fold enrichment of 61 ± 7–fold at centrosomes versus WCE. In addition, we took into consideration that a typical U2OS asynchronous population has 50% of the cells in the G1, and from those, ∼70% do not display any HsSAS-6 centriolar signal (Fig. S3, A and B). Thus, we corrected for this population that was HsSAS-6 centriolar negative, resulting in the indicated ∼100-fold (98×) enrichment. Estimates in U2OS were also calculated using background-subtracted 3D confocal images (same as in Fig. 4, E and F) on which a Gaussian blur of σ = 1 px was applied, comparing the maximum signal at centrosomes (1 px) to the mean cytoplasmic signal calculated from the mean intensity projection of the five innermost planes in the cell, yielding a value of 90 ± 27.
Estimating the molarity of HsSAS-6 molecules at centrosomes.
The number of HsSAS-6 molecules in different cell cycle phases was estimated as follows. The number of molecules estimated via Western blotting (M) and the total fluorescence intensity (I) are linked by a conversion factor α, whereby a given number of HsSAS-6 molecules M at a moment n corresponds to a fluorescence intensity of αI: Mn = αIn. It follows that the mean number of ∼500,000 HsSAS-6 molecules in WCE of asynchronously growing U2OS cell population represents the sum of molecules from the different cell cycle phases (M1, M2, and M3 in G1–S, S–G2, and late G2/M, respectively) pondered by the relative contribution of each phase (∼50% for G1/early S, ∼25% for S–G2, and ∼25% for late G2/M) giving the following equation: 0.5 M1 + 0.25 M2 + 0.25 M3 = 500,000. Similarly, the sum of the total HsSAS-6 fluorescence intensity (I1 in G1/early S, I2 for S–G2, and I3 for late G2/M) pondered by the fraction of the cell cycle gives the second equation: 0.5 αI1 + 0.25 αI2 + 0.25 αI3 = 500,000. Solving these two equations yields the value for the conversion factor α linking the fluorescence intensity to the number of HsSAS-6 molecules, whereby one HsSAS-6 molecule corresponds to a fluorescence intensity of I = 1/α, i.e., 1/0.00135 or ∼740 arbitrary units. The number of HsSAS-6 molecules in the cytoplasm was estimated using the measured mean total cellular fluorescence intensity at each cell cycle stages (Fig. 4 E) and the conversion factor. Similarly, numbers of molecules at centrosomes were estimated using the mean value of the measured total centrosomal intensity in Fig. 4 F, rounded up to the nearest 1,000. The corresponding cellular molarity was calculated using a mean volume of 4,000 µm3 for a U2OS cell (assuming a spherical shape and a 20-µm diameter as in Beck et al., 2011). A centrosomal volume of 0.2 µm3 was calculated from the optical resolution of the confocal images (1.4 NA and 600-nm emission wavelength). As the aforementioned volume calculations are limited by the confocal resolution of our images, these numbers have to be considered as estimates.
STORM
STORM sample preparation.
For HsSAS-6 staining, cells were plated on 18-mm size 1.5 precision coverslips (Carl Roth GmbH) cleaned overnight in 35% acetic acid/65% ethanol. Cells were synchronized by mitotic shake off, fixed at relevant times thereafter, and binned according to centrosome configurations. Cells were preextracted, washed with PBS, fixed with 2% PFA or methanol, and immunostained as described in the section Immunofluorescence and antibodies.
STORM imaging buffers preparation.
Buffers were prepared as previously reported (Olivier et al., 2013a,b). For Fig. 2 (M–O) and Fig. 3 (G and H), depth-independent imaging was performed in Vectashield (Vector Laboratories) as in Olivier et al. (2013a). For Fig. 3 (D and E) and Fig. 6, HsSAS-6 S-phase structures were imaged as in Olivier et al. (2013b), in a solution containing 10% (wt/vol) glucose in 10 mM Tris, pH 7.5, oxygen scavengers (0.2 mg/ml glucose oxidase [Sigma-Aldrich] and 57 µg/ml catalase [Sigma-Aldrich]), and 10 mM mercaptoethylamine combined with 50 mM β-mercaptoethanol and 2 mM cyclooctatetraene at pH ∼7–8.
STORM microscope and image analysis.
3D-STORM imaging and data analysis was performed as previously reported (Olivier et al., 2013a). In brief, imaging was performed on an inverted microscope (SR-200; Vutara), using a 60×/1.42 NA oil objective (UIS2 Plan Apochromat; Olympus). Extra magnification was used to achieve a px size of 101 nm on an electron multiplying charge-coupled device camera (Evolve 512; Photometrics). A 647-nm laser (Coherent) was used for excitation, with a power of ∼4.5 kW/cm2, and a 405-nm laser (CUBE; Coherent) for reactivation (few milliwatts/centimeter squared). Data were recorded at 25 frames per second, and typical acquisitions consisted of 10,000–30,000 raw images.
Determination of HsSAS-6 structure length and volume.
Images were projected on all three axes, and the appropriate orientation was chosen for measurements, using also centrin images acquired in wide field to help determine this. Fluorescence intensities were sampled using a 200-nm-wide line profile, and values were plotted and then fitted with a single- or double-Gaussian function in OriginPro 8.6 software (OriginLab). The full width at half-maximum of a single-Gaussian fit or the sum of full width at half-maximum for double-Gaussian fits were taken as length of the structure. Early stage structures were fit better with a single-Gaussian function, whereas for late G2 structures, a double-Gaussian function was more appropriate. Volumes occupied by HsSAS-6 molecules were calculated by projecting the localized peaks on a 2D 10-nm grid along all three dimensions, smoothing the images using an σ = 10-nm Gaussian blur, and then thresholding the images as binary images and integrating the area covered by nonzero px. The three areas were then multiplied, and the square root of the result was used as the volume.
Determination of HsSAS-6 ring radii and angular distribution.
To determine the radius of each HsSAS-6 ring and each F131E-EGFP torus, the localized peaks were rendered into a 3D stack of images, with each point represented by a Gaussian function (σ = 4 nm; Peakselector; courtesy of H. Hess, Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA). The stack was then projected along the axis of the ring to yield a 2D image. The center of mass of each image was determined using ImageJ (National Institutes of Health), and the plugin Radial Profile was used to determine the intensity distribution as a function of distance from the center of mass, which was then fitted using a Gaussian function in OriginPro 8.6. Center positions values of each fitted Gaussian were taken as the ring radius. For Fig. 6, the angular distribution and fold symmetry of HsSAS-6 rings was assessed using the angular intensity profiles given by the plugin Oval Profile. The intensity profiles obtained were then filtered using a long pass Fourier filter in OriginPro 8.6, and peak-to-peak distances were calculated using local maxima (Fig. S4).
FCS
Sample preparation.
We used iU2OS cell lines expressing GFP–GFP (55 kD), GFP-C (48 kD), FL-GFP, or F131E-GFP (both 103 kD) or U2OS transiently transfected with a pEBTet-GFP-GW construct, allowing the expression of a single GFP (as a result of a stop codon early in the Gateway recombined sequence; 27 kD). Cells were plated in 35-mm glass-bottomed dishes (MatTek Corporation), and expression was induced 48 h before imaging. For FL-GFP, F131E-GFP, or GFP-C, endogenous HsSAS-6 was depleted for 48 h concomitantly to the induction. Cells were kept at 37°C until imaging when the medium was changed to phenol red–free CO2-independent Leibovitz’s medium supplemented with 20% FCS, 2 mM glutamine, and 100 mg/ml penicillin-streptomycin. Regular U2OS cells were used to measure background fluorescence and control for lack of autocorrelation of the signal.
FCS measurements.
FCS measurements were performed with a microscope (LSM 780 ConfoCor 3; Carl Zeiss). GFP and Alexa Fluor 488 fluorescent proteins (FPs) were excited with 488 nm from the built-in Ar ion laser. An acousto-optical tunable filter was used to adjust the incident irradiance after the microscope objective (40× C-Apochromat, NA 1.2, water immersion; Carl Zeiss) below 2 kW/cm2 for the Ar ion laser (∼0.4% laser power) to reduce photobleaching and photophysical effects. Systematic determination of the lateral beam waist (ω0) of the focused laser and the structural parameter S was performed by measuring the translational diffusion time constants τD of 50 nM of soluble Alexa Fluor 488 (Life Technologies) with known diffusion coefficients D in aqueous solution (435 µm2/s) according to ω0 = (4D × τD)1/2. We obtained ω0 = 204 nm and S = 5.5, yielding effective volumes of 0.3 fl. The values of S and ω0 were kept constant for the fitting of the different samples. Thereafter, fluorescence intensity time traces and correlation curves of iU2OS cells, expressing either the different constructs, were recorded for 45 s. Cells with too high expression levels or presenting substantial fluorescence heterogeneity were not considered further.
FCS curve fitting and analysis.
Autofluorescence was assessed on five untransfected U2OS cells and subtracted from all measurements before fitting. Autocorrelation curves were generated and fitted as previously described (Schmidt et al., 2009) using a model for one- or two-component anomalous diffusion with proteinlike blinking (for GFP-containing proteins) or one-component anomalous diffusion with tripletlike blinking for Alexa Fluor 488 measurements. Initial fittings with a two-component model yielded a fraction of >97% of one component and <3% of a second much slower component (potentially reflecting cell movement) for all tested constructs (including GFP, known to diffuse as a single species). This analysis indicates that the one-component model is the most appropriate, which was thus used in all subsequent fittings. The goodness of the fit was estimated by the R2 value of each fit. The following values were extracted from each autocorrelation curve and fit: mean number N of molecules in the observation volume determined from the ACF amplitude, their brightness (CPM), and their diffusion coefficients as given by the decay time of the fitted ACF curve, τD. The relative brightness (CPM) of each construct was first normalized to the mean value obtained for GFP. The mean relative brightness (counts per second and CPM) normalized to the mean value obtained for GFP gave a value of 1.84 for GFP–GFP. This value indicates that the probability of each FP molecule to be in a fluorescent state is ∼0.92, most likely reflecting chromophore blinking and/or differences in maturation (Dickson et al., 1997; Haupts et al., 1998; Wachsmuth et al., 2000; Maeder et al., 2007). This affects the mean brightness of molecules and complexes, which carry more than one FP, whereas single FPs are either on or off. We thus corrected the brightness values for all constructs (except for the single GFP construct, as only on molecules are recorded) and reported them in Fig. 5 E. The diffusion coefficient was calculated from τD using the equation D = ω02/4 τD. For each construct, these parameters were analyzed and plotted using Prism 6 (GraphPad Software).
Statistical analyses
All statistical analyses were performed using GraphPad Prism6, and the results and p-values of multiple comparisons performed are given in Tables S2–S6. For Fig. S2 (A and B), two-way analysis of variance (ANOVA) between control and siRNA-treated cells was performed including Sidak’s multiple comparisons (α = 0.05) at different time points and between treatments. For the experiments in Fig. 4 and Fig. 5, normality of the population was assessed using D’Agostino and Pearson omnibus normality test (α = 0.05). For the comparison of HsSAS-6 structure lengths and volumes between S–G2 and late G2 (Fig. 3, J and K), an unpaired two-tailed t test with Welch’s correction for unequal variance was performed with α = 0.05, yielding a P < 0.0001 for both. For multiple comparisons, conditions that exhibited a normal distribution (Fig. 4 F) were analyzed using an ordinary one-way ANOVA test and α = 0.05, and the p-values were corrected for multiplicity (Tukey’s test). For samples with nonnormal distribution or if one of the conditions did not exhibit a normal distribution (Fig. 4 E and Fig. 5, C and D), the nonparametric Kruskal–Wallis multiple comparison was performed with α = 0.05, and the p-values were corrected for multiple comparisons (Dunn’s test).
Online supplemental material
Fig. S1 shows that HsSAS-6 variants are siRNA resistant and C-terminal specificity of the HsSAS-6 antibody. Fig. S2 shows the prevalence and localization of the torus for F131E-GFP and FL-GFP. Fig. S3 contains complementary data on quantification procedures and controls pertaining to Fig. 3 and Fig. 4. Fig. S4 describes the determination of the ninefold symmetrical organization of HsSAS-6 rings from 3D-STORM images. Table S1 contains cell numbers for Fig. 1. Table S2 shows cell numbers after shake off and results of two-way ANOVA (Fig. S2). Table S3 shows results of Kruskal–Wallis test (α = 0.05; Fig. 4 E). Table S4 shows results of ANOVA (α = 0.05; Fig. 4 F). Table S5 shows results of Kruskal–Wallis test (α = 0.05; Fig. 5 E). Table S6 shows results of Kruskal–Wallis test (α = 0.05; Fig. 5 F). Table S7 shows primer names and sequences to generate specific entry vectors. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307049/DC1.
Supplementary Material
Acknowledgments
We thank H. Vogel (Swiss Federal Institute of Technology) for help with initial FCS experiments and the Bioimaging and Optics Platform at the Swiss Federal Institute of Technology with image acquisition and processing as well as F.R. Balestra and N. Brown for critical reading of the manuscript.
D. Keller received funding from the Swiss Cancer League (grants 02024-2007 and 02584-02-2010 to P. Gönczy), M. Orpinell received funding from Federation of European Biochemical Societies, N. Olivier and S. Manley received grants from the European Research Council (ERC; 243016-PALMassembly to S. Manley), R. Wyss received funds from the National Centre of Competence in Research Chemical Biology and the Swiss National Foundation (grant 31003A-118148 to Horst Vogel), M. Wachsmuth received funding from the European Molecular Biology Laboratory, R. Mahen received funding from European Molecular Biology Organization (ALTF-416-2012) and the Wellcome Trust (Sir Henry Wellcome Fellowship), and V. Hachet received funding from the ERC (Advanced Grant 233335 to P. Gönczy). J. Ellenberg acknowledges that the research leading to these results has received funding from the European Community’s Seventh Framework Program (FP7/2007–2013; under grant agreements 241548 Mitosys and 258068 SystemsMicroscopy).
The authors declare no competing financial interests.
Author contributions: D. Keller designed and performed the research, analyzed data, and wrote the manuscript; M. Orpinell and N. Olivier performed experiments and analyzed data. M. Wachsmuth and R. Mahen provided advice and analyzed data; R. Wyss provided advice with preliminary experiments; V. Hachet produced reagents; S. Manley as well as J. Ellenberg provided advice and access to instruments and software; P. Gönczy designed and supervised the research, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used in this paper:
- ACF
- autocorrelation function
- ANOVA
- analysis of variance
- CPM
- counts per molecule
- EdU
- 5-ethynyl-2′-deoxyuridine
- FCS
- fluorescence correlation spectroscopy
- FL
- full length
- FP
- fluorescent protein
- PCM
- pericentriolar material
- PCNA
- proliferating cell nuclear antigen
- px
- pixel
- STORM
- stochastic optical reconstruction microscopy
- WCE
- whole-cell extract
References
- Arquint C., Sonnen K.F., Stierhof Y.-D., Nigg E.A. 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125:1342–1352 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M. 2007. Structure and duplication of the centrosome. J. Cell Sci. 120:2139–2142 10.1242/jcs.005231 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Marshall W.F. 2010. Building the centriole. Curr. Biol. 20:R816–R825 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. 2011. The quantitative proteome of a human cell line. Mol. Syst. Biol. 7:549 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., Godinho S.A. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M., Paintrand M., Berges J., Marty M.C., Karsenti E. 1987. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 8:238–249 10.1002/cm.970080305 [DOI] [PubMed] [Google Scholar]
- Bulseco D.A., Wolf D.E. 2007. Fluorescence correlation spectroscopy: molecular complexing in solution and in living cells. Methods Cell Biol. 81:525–559 10.1016/S0091-679X(06)81025-3 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J.B., Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123:1414–1426 10.1242/jcs.064931 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.T. 1974. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardtii. J. Cell Sci. 16:529–556 [DOI] [PubMed] [Google Scholar]
- Chen H., Farkas E.R., Webb W.W. 2008. Chapter 1: In vivo applications of fluorescence correlation spectroscopy. Methods Cell Biol. 89:3–35 10.1016/S0091-679X(08)00601-8 [DOI] [PubMed] [Google Scholar]
- Chrétien D., Buendia B., Fuller S.D., Karsenti E. 1997. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120:117–133 10.1006/jsbi.1997.3928 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., Hoffmann I. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R.M., Cubitt A.B., Tsien R.Y., Moerner W.E. 1997. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 388:355–358 10.1038/41048 [DOI] [PubMed] [Google Scholar]
- Dippell R.V. 1968. The development of basal bodies in paramecium. Proc. Natl. Acad. Sci. USA. 61:461–468 10.1073/pnas.61.2.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D.M. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Guichard P., Smith A.H., Schwarz H., Agard D.A., Marco S., Avidor-Reiss T. 2010. Self-assembling SAS-6 multimer is a core centriole building block. J. Biol. Chem. 285:8759–8770 10.1074/jbc.M109.092627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Chrétien D., Marco S., Tassin A.M. 2010. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29:1565–1572 10.1038/emboj.2010.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Desfosses A., Maheshwari A., Hachet V., Dietrich C., Brune A., Ishikawa T., Sachse C., Gönczy P. 2012. Cartwheel architecture of Trichonympha basal body. Science. 337:553 10.1126/science.1222789 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupts U., Maiti S., Schwille P., Webb W.W. 1998. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 95:13573–13578 10.1073/pnas.95.23.13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh R.A., Stowe T.R., Turk E., Stearns T. 2012. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE. 7:e52166 10.1371/journal.pone.0052166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P., Steinmetz M.O. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell. 144:364–375 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G.G. 1981. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91:814–821 10.1083/jcb.91.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L., Lee Y.L., Sahl S.J., Stearns T., Moerner W.E. 2012. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 102:2926–2935 10.1016/j.bpj.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Lettman M.M., Wong Y.L., Viscardi V., Niessen S., Chen S.H., Shiau A.K., Zhou H., Desai A., Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell. 25:284–298 10.1016/j.devcel.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Chang C.W., Hsu W.B., Tang C.J., Lin Y.N., Chou E.J., Wu C.T., Tang T.K. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32:1141–1154 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G., Lavogina D., Orpinell M., Umezawa K., Reymond L., Garin N., Gönczy P., Johnsson K. 2013. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol. 23:265–270 10.1016/j.cub.2012.12.030 [DOI] [PubMed] [Google Scholar]
- Maeder C.I., Hink M.A., Kinkhabwala A., Mayr R., Bastiaens P.I.H., Knop M. 2007. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 9:1319–1326 10.1038/ncb1652 [DOI] [PubMed] [Google Scholar]
- Marshall W.F. 2009. Centriole evolution. Curr. Opin. Cell Biol. 21:14–19 10.1016/j.ceb.2009.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., Agard D.A. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp S., Küntziger T., Abraham Y., Holmes S., Bordes N., Paintrand M., Paoletti A., Bornens M. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148:405–416 10.1083/jcb.148.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17:2169–2174 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Olivier N., Keller D., Gönczy P., Manley S. 2013a. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS ONE. 8:e69004 10.1371/journal.pone.0069004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N., Keller D., Rajan V.S., Gönczy P., Manley S. 2013b. Simple buffers for 3D STORM microscopy. Biomed. Opt. Express. 4:885–899 10.1364/BOE.4.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklowski A., Homsi Y., Keller D., May M., Chauhan S., Kossatz U., Grünwald V., Kubicka S., Pich A., Manns M.P., et al. 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13:1004–1009 10.1038/ncb2282 [DOI] [PubMed] [Google Scholar]
- Schmidt U., Im K.-B., Benzing C., Janjetovic S., Rippe K., Lichter P., Wachsmuth M. 2009. Assembly and mobility of exon-exon junction complexes in living cells. RNA. 15:862–876 10.1261/rna.1387009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., Nigg E.A. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., Raff J.W. 2010. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188:313–323 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P. 2008. The mechanisms of centriole biogenesis. PhD thesis. Faculté de biologie et médecine, Université de Lausanne, Lausanne, Switzerland: 97 pp [Google Scholar]
- Strnad P., Gönczy P. 2008. Mechanisms of procentriole formation. Trends Cell Biol. 18:389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.-O., Robinson C.V., et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science. 331:1196–1199 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Vladar E.K., Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., Krämer A. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wachsmuth M., Waldeck W., Langowski J. 2000. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298:677–689 10.1006/jmbi.2000.3692 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.