SUMMARY
Carcinomas typically invade as a cohesive multicellular unit, a process termed collective invasion. It remains unclear how different subpopulations of cancer cells contribute to this process. We developed three-dimensional (3D) organoid assays to identify the most invasive cancer cells in primary breast tumors. Collective invasion was led by specialized cancer cells that were defined by their expression of basal epithelial genes, such as cytokeratin-14 (K14) and p63. Furthermore, K14+ cells led collective invasion in the major human breast cancer subtypes. Importantly, luminal cancer cells were observed to convert phenotypically to invasive leaders following induction of basal epithelial genes. Although only a minority of cells within luminal tumors expressed basal epithelial genes, knockdown of either K14 or p63 was sufficient to block collective invasion. Our data reveal that heterotypic interactions between epithelial subpopulations are critical to collective invasion. We suggest that targeting the basal invasive program could limit metastatic progression.
INTRODUCTION
Invasion is a fundamental step in tumor progression and a driving force for metastasis. Although invasion is commonly conceptualized as a single cell process, the majority of solid tumors display features of collective invasion, in which cells invade cohesively as a multicellular unit (Friedl et al., 2012; Leighton et al., 1960). A central problem in collective invasion is how a group of adherent epithelial cancer cells acquires motile invasive behavior (Friedl and Gilmour, 2009; Gray et al., 2010; Polyak and Weinberg, 2009).
One solution is for cancer cells to rely upon the motility of migratory stromal cells, such as fibroblasts (Gaggioli et al., 2007) or macrophages (Condeelis and Pollard, 2006; DeNardo et al., 2009). However, mammary tumors also contain multiple subpopulations of tumor cells with distinct genotypic and phenotypic characteristics. Importantly, this cellular heterogeneity is associated with differences in metastatic potential and therapeutic response (Almendro et al., 2013; Fidler, 2003). It remains unclear how these subpopulations of cancer cells contribute to collective invasion.
Clinically, the transition from in situ to invasive breast cancer correlates with a strong reduction in overall survival but the molecular basis of this transition has remained elusive (Polyak, 2010). The challenge of transitioning to a motile phenotype is particularly acute in mammary luminal epithelial cells, as these cells are normally connected by extensive intercellular junctions and display less spontaneous motility than myoepithelial cells in real-time analyses (Ewald et al., 2008). Consistent with this concept, luminal breast cancers have a more favorable average prognosis, but 10–20% of cases eventually metastasize to liver, lung, or brain (Kennecke et al., 2010). Furthermore, luminal breast cancer cell lines are weakly invasive in 2D culture compared to basal subtypes (Neve et al., 2006).
We hypothesize that breast tumors accomplish collective invasion through cell-cell interactions among functionally distinct epithelial cancer cells within the primary tumor. To test this hypothesis, we developed novel 3D organoid assays to identify the most invasive cancer cells within a primary tumor in an unbiased fashion. In the present study we applied these assays to demonstrate that the cells leading collective invasion are molecularly and behaviorally distinct from the bulk tumor cells and display a conserved, basal epithelial gene expression program.
RESULTS
An Ex-vivo 3D Culture Assay Identifies Invasive Cells Within Primary Tumors
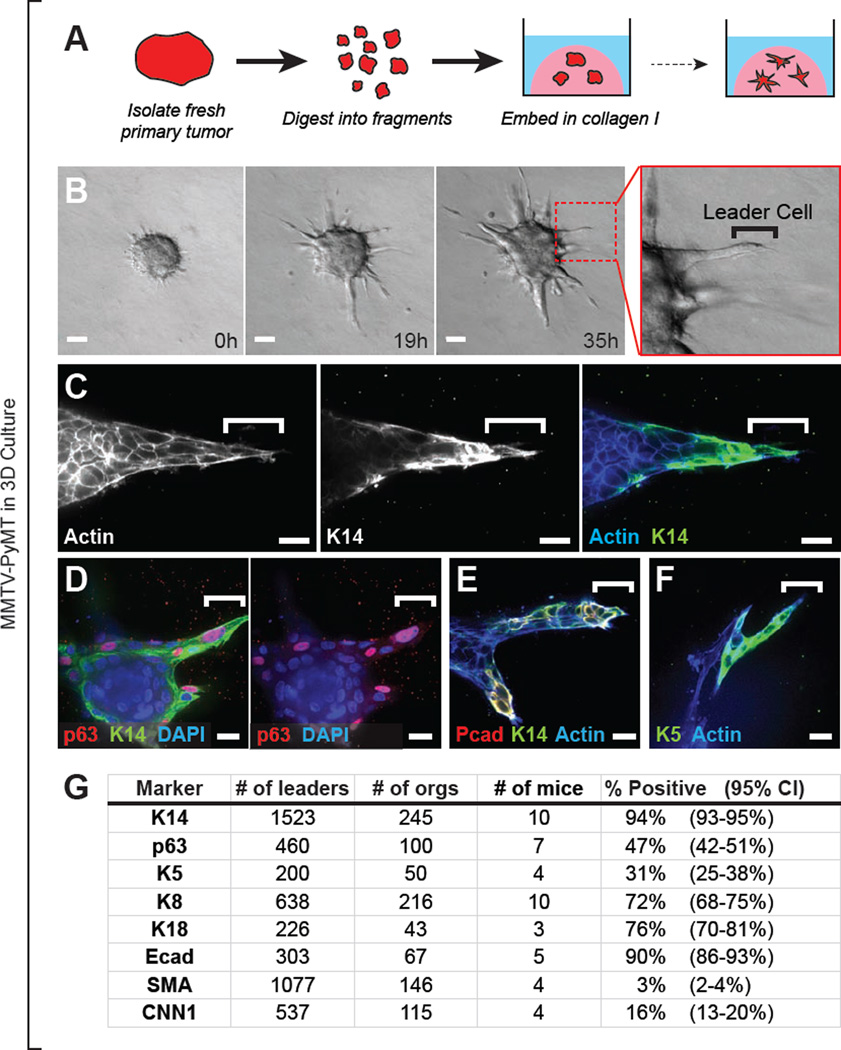
We developed a 3D primary culture model (Nguyen-Ngoc et al., 2012) that enabled us to observe cell behaviors during collective invasion and to interrogate the molecular phenotype of the most invasive cells (Figure 1A). Briefly, we isolate fresh primary tumors and use a combination of mechanical disruption and enzymatic digestion to generate "tumor organoids.” Tumor organoids are composed of 200–1000 adherent tumor cells and reflect the cellular heterogeneity present in the primary tumor. To study collective invasion, we cultured tumor organoids in 3D collagen I gels, a model for the microenvironment surrounding invasive breast cancers (Conklin et al., 2011; Nguyen-Ngoc et al., 2012; Paszek et al., 2005; Provenzano et al., 2008; Wolf et al., 2009).
Figure 1. Leaders Cells are Molecularly Distinct and Express Basal Epithelial Markers in a Luminal Mammary Carcinoma Model.
(A) Schema of leader cell assay. Primary tumor is digested to tumor organoids, each composed of 200–1000 adherent tumor cells, and embedded in 3D collagen I matrix.
(B) Time-lapse DIC microscopy of a MMTV-PyMT mouse mammary tumor organoid embedded in collagen I. Collectively migrating cells emerge from the tumor organoid. Protrusive leader cells are readily identified at the front of these invasive strands. Also see Movie S1.
(C–F) Leader cells stained with K14 and phalloidin (C), p63, K14 and DAPI (D), P-cadherin (Pcad), K14, and phalloidin (E), or K5 and phalloidin (F).
(G) Frequency of leader cells expressing K14, p63, K5, K8, K18, E-cadherin (Ecad), SMA, and CNN1 in MMTV-PyMT tumor organoids. 95% confidence intervals for each proportion denoted in parentheses.
The scale bars represent 50 µm in (B) and 20 µm in (C–F). See also Figure S1.
We first characterized invasion in organoids derived from a genetically engineered mouse model of breast cancer in which the mouse mammary tumor virus long terminal repeat drives expression of the polyoma virus middle T oncogene (MMTV-PyMT) (Guy et al., 1992a). MMTV-PyMT mice develop highly invasive mammary tumors that metastasize spontaneously to the lung (Lin et al., 2003). By gene expression profiles, MMTV-PyMT tumors cluster with the aggressive luminal B subtype of human breast cancer (Herschkowitz et al., 2007). Tumor organoids isolated from this model progressively extended multicellular strands of cancer cells into the collagen I over 48–72 hr (Figure 1B and Movie S1). Since the cells leading these invasive strands were highly protrusive and migratory, we refer to them as “invasive leader cells” (Figure 1B, right panel).
Invasive Leader Cells are Molecularly Distinct and Express Basal Epithelial Markers
We were able to determine the molecular phenotype of invasive leader cells unambiguously due to their position at the front of invasive strands and their distinct protrusive morphology. Our analysis defined four molecular features of leader cells in this luminal mouse model. First, leader cells preferentially expressed multiple basal epithelial markers including cytokeratin-14 (K14), p63, P-cadherin, and cytokeratin-5 (K5) (Figure 1C–G). Among these markers, K14 was the most frequently associated with leader cells (94% of leaders, n=1523 leaders, from 10 mice, CI [93–95%]). Second, leader cells co-expressed markers of luminal epithelium including K8, K18 and E-cadherin (Figure 1G and Figure S1A–C). However, whereas basal markers were preferentially expressed in leader cells, K8 and E-cadherin also strongly stained the noninvasive K14− core of cells within the tumor organoid (Figure S1A,C). Third, leader cells typically did not express markers of smooth muscle contractility, such as SMA, MYH11, and CNN1 (Figure 1G and Figure S1D–E), and did not contract in response to the hormone oxytocin, which induces smooth muscle contractility (Figure S1F–G). Because the smooth muscle program is a defining feature of myoepithelial cells in the normal mammary gland, we conclude that invasive leader cells are distinct from myoepithelial cells. Fourth, leader cells did not typically express Twist, Slug, or vimentin (Figure S1H), common markers of a molecular epithelial-mesenchymal transition (EMT). Finally, leader cells retained membrane localized E-cadherin at sites of cell-cell contact with follower cells (Figure S1C, blue arrows). Taken together, our observations reveal that invasive leader cells in this luminal breast cancer model are molecularly distinct from bulk tumor cells and preferentially express K14 and other basal epithelial markers.
K14+ Cells are Enriched at the Invasive Border in Primary Tumor and in Lung Metastases
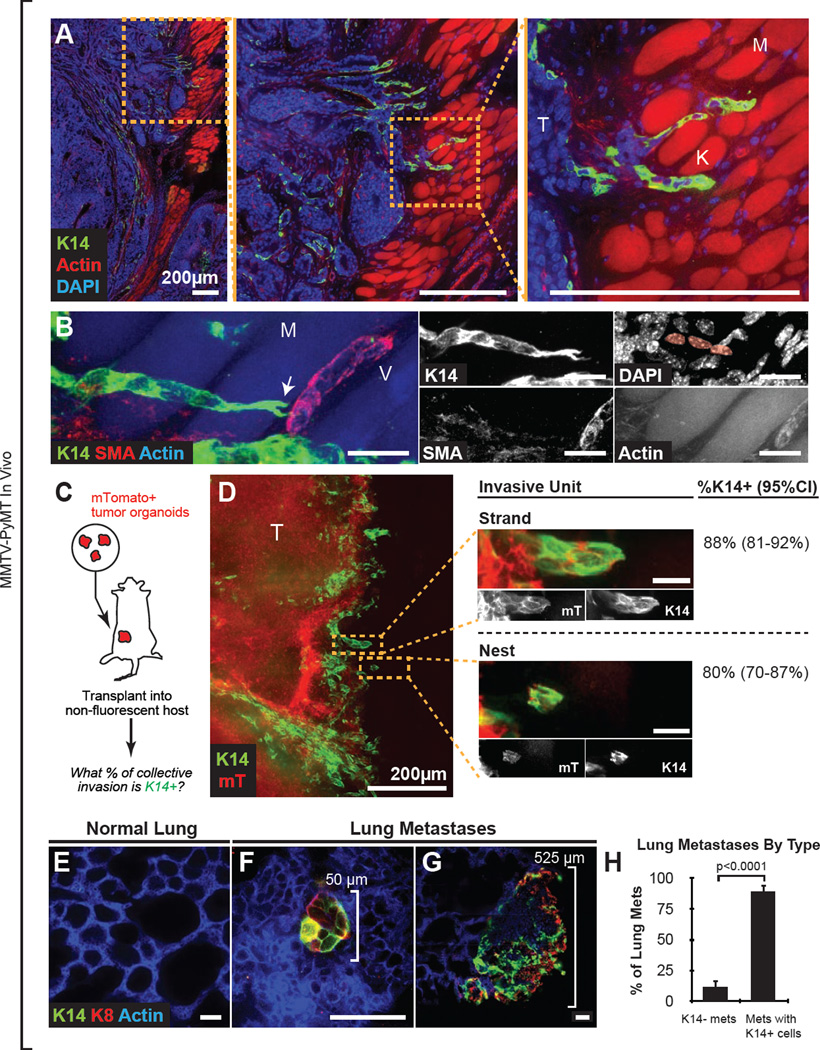
We next asked whether K14+ cells also led collective invasion in vivo in this luminal model. To address this question, we optically reconstructed large regions of the tumor-stromal border from fixed sections of intact tumors. K14+ cells were concentrated at tumor-stromal borders (n=9 reconstructed tumors from 8 mice, and Figure 2A) and led multicellular invasive strands into the adjacent muscle (Figure 2A,B and Movie S2). As in 3D culture, K14+ cells were negative for SMA and co-expressed luminal K8 (Figures 2B and S2A). Thus, K14+ cells lead collective invasion in 3D culture and in vivo.
Figure 2. K14+ Cells are Enriched at the Tumor Invasive Border and in Lung Metastases.
(A) Area of tumor invasion into muscle in 100-µm thick mammary tumor sections, stained with K14, DAPI, and phalloidin. T, tumor; K, K14+ leader cells; M, phalloidin+ muscle fibers.
(B) Collectively invading K14+ leaders at high magnification stained with K14, phalloidin, SMA, and DAPI. M, muscle; V, SMA+ vessel; arrow, leading K14+ cell with forked protrusions. Inset highlights the chain of three K14+ cells. This micrograph represents a z-projection of over 40 µm to capture the organization of the invasive strand. See Movie S2.
(C) Schema to quantify the percentage of collective invasion that is led by K14+ cells. mTomato+ tumor organoids were isolated from MMTV-PyMT;mT/mG mice. Organoids were transplanted orthotopically into non-fluorescent congenic hosts. Transplanted tumors >1cm in size were harvested to generate montages of the tumor-stromal border.
(D) Micrograph of a representative mTomato+ tumor-stromal border stained with K14. mTomato+ regions (in red) denote tumor-derived cells. Insets denote multicellular groups of cells invading into the adjacent stroma. An invasive strand was defined as a protrusive group of cells connected to the main tumor. A nest was defined as an isolated group of tumor cells. N=145 invasive strands and N=83 nests were counted from 13 sections from 5 mice. 95% confidence intervals denoted in parentheses. The micrograph represents a 40 µm z-projection.
(E–G) Reconstructed metastases in thick sections of lungs from MMTV-PyMT mice, stained with K14, K8, and phalloidin. Normal lung parenchyma was K14 negative (E). Both micro-metastatic and larger metastatic lesions had K14+ and K8+ cells (F–G).
(H) Metastatic lung lesions were identified by K8 positivity and classified based on their K14 status. Data are presented as mean ± sd. n=226 metastases, from 5 tumor mice. P-value determined by two-sided t-test.
Scale bars are 20 µm in (B,D) and 50 µm in (E–G). See also Figure S2.
We next asked what fraction of collective invasion in vivo was led by K14+ cells. We employed an orthotopic transplant model to mark donor-derived tumor cells unambiguously. We transplanted MMTV-PyMT mammary tumor organoids co-expressing a red fluorescent reporter (mTomato) into the cleared mammary fat pads of non-fluorescent congenic hosts (FVB), and identified invasive units at the tumor-stromal border, blind to K14 expression (Figure 2C–D). These invasive units included multicellular groups of cells connected to the main tumor mass, which we defined as invasive strands, and groups of cells unconnected from the main tumor mass, which we defined as tumor nests (Figure 2D, middle panel). We then classified each invasive unit: in the case of strands, we asked whether the leader cells were K14+ and in the case of nests, whether any cell in the group was K14+. By this measure, we observed that 88% of invasive strands and 80% of tumor nests were K14+ (Figure 2D, right panel). Next, we systematically quantified the distance of K14+ and K14− cells from the tumor-stromal border in large optically reconstructed montages (Figure S2E–G). We observed strong enrichment for K14+ cells in the first 100 µm from the stromal border, while K14− cells were relatively uniformly distributed (n=5 montages from 4 tumors, Figure S2H). We conclude that K14+ cancer cells are strongly enriched at the tumor-stromal interface and lead the majority of collective invasion in vivo.
Since MMTV-PyMT tumors develop metastases with high penetrance, we next quantified the K14 status of their lung metastases. Although K14 was absent in normal lung parenchyma, we observed K14 staining in >88% of metastatic lesions, from micrometastases up to large, 500 µm lesions (Figures 2E–H). In contrast, K14+ cells constituted only a small percentage of the total cell population in primary tumor (1.4 ± 0.2% by FACS, n=3 mice and Figure S2D). Consistent with their phenotype in the primary tumor, K14+ metastases were negative for SMA in both micrometastatic and large metastatic lesions (Figures S2B–C). We conclude that although K14+ cells are rare in the primary tumors of this luminal breast cancer model, they are strongly enriched in lung metastases.
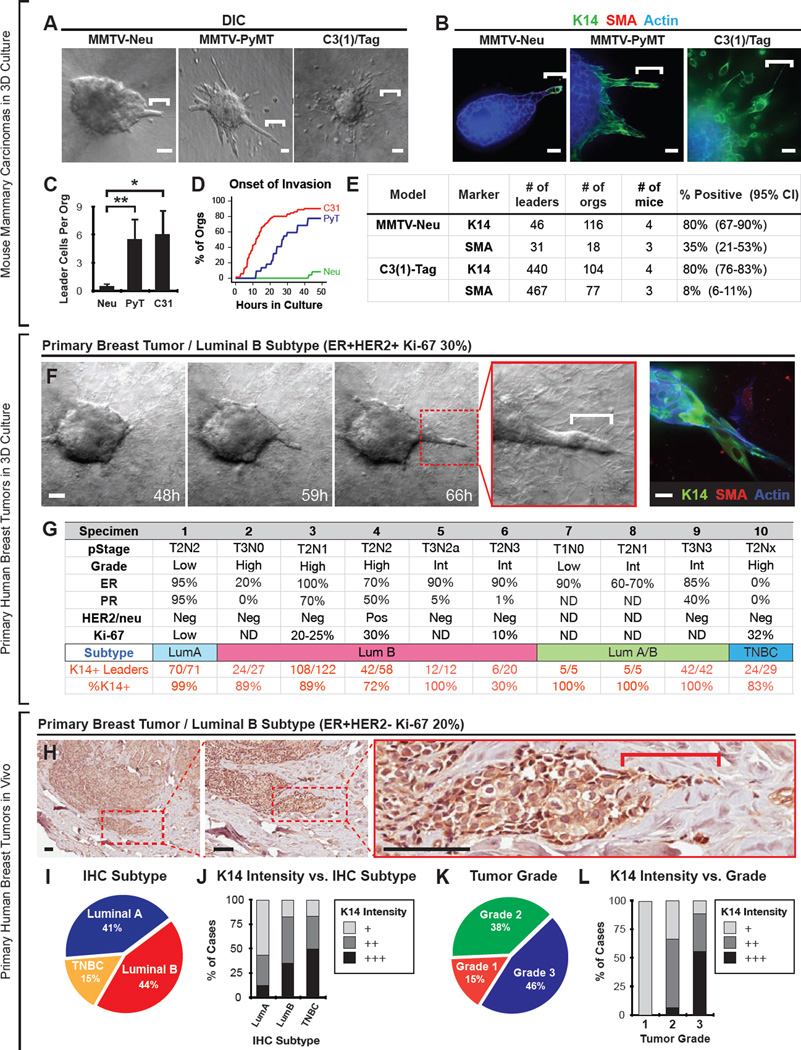
K14+ Cells Lead Collective Invasion Across Multiple Mouse Models of Breast Cancer
To test the generality of our observations, we next characterized invasion in two additional genetically engineered mouse models of breast cancer: MMTV-Neu, a model of HER2+ luminal breast cancer and C3(1)/Tag, a model of basal breast cancer (Guy et al., 1992b; Maroulakou et al., 1994). Tumor organoids isolated from these models displayed pronounced differences in the morphologic patterns and timing of invasion in 3D culture (Figure 3A–B and Movie S3). MMTV-Neu tumor organoids had the fewest leader cells, MMTV-PyMT organoids had an intermediate number, and C3(1)/Tag tumor organoids had the most leader cells (Figure 3C–D). Despite these differences in the frequency of invasion, >80% of leader cells in all three models were K14+ (Figure 3E). Similarly, the majority of K14+ leader cells co-expressed luminal K8, but did not express SMA in all three models (Figure 3B,E and S3A–B).
Figure 3. K14+ Cells Lead Collective Invasion Across Mouse Models of Breast Cancer and in Primary Human Breast Cancers.
(A) Time-lapse DIC microscopy of collagen I embedded organoids derived from MMTV-Neu, MMTV-PyMT, or C3(1)/Tag mammary tumors. White bars, leader cells. Also see Movie S3.
(B) Micrographs of leader cells from the three mouse models in (A) stained with K14, SMA, and phalloidin.
(C) Quantification of the number of invasive leaders per tumor organoid in (A). For MMTV-Neu, N=116 organoids from 4 mice. For MMTV-PyMT, N=245 organoids from 10 mice. For C3(1)/Tag, N=104 organoids from 4 mice. Data are presented as mean ± sd. *, P-value < 0.05. **, P-value < 0.01. P-value determined by two-sided t-test.
(D) The percentage of invasive tumor organoids from the three mouse models in (A) as a function of time in culture (in hrs). Onset of invasion was defined as the first instance of protrusive cell motility into collagen I matrix. N=22–70 tumor organoids per mouse model.
(E) Frequency of leader cells expressing K14 or SMA in MMTV-Neu or C3(1)/Tag tumor organoids. 95% confidence intervals for each proportion denoted in parentheses.
(F) Time-lapse DIC microscopy of a human luminal breast tumor organoid embedded in collagen I matrix (sample S4). White bar, leader cell. At right, micrograph of tumor organoid from same tumor specimen, stained for K14, SMA, and F-actin. Also see Movie S3.
(G) Pathologic characteristics of harvested human breast tumors, including stage, grade, ER status, PR status, HER2/Neu status, Ki-67 percentage, and the measured frequency of K14+ leaders. 9 of 10 tumors were ER positive luminal breast tumors, with ER positivity ranging from 20–100%. The breast cancer subtype was determined using surrogate immunohistochemistry definitions (Goldhirsch et al., 2013).
(H) Representative micrograph of a K14+ collective invasion front from an archival specimen classified as luminal B.
(I) The distribution of cases according to breast cancer subtype assigned using surrogate IHC subtype definitions. N=39 cases in total.
(J) The frequency of cases with low (+), medium (++), or high (+++) K14 staining stratified by IHC-defined breast cancer subtype. K14 intensity was scored as in Figure S4A.
(K) The distribution of cases according to histologic tumor grade.
(L) The frequency of cases with low (+), medium (++), or high (+++) K14 staining stratified by tumor grade.
Scale bar represents 50 µm in (A,F: left movie series), 20 µm in (B,F: right panel), and 100 µm in H. See also Figures S3 and S4.
K14+ Cells Lead Collective Invasion in Primary Human Breast Tumors in 3D Culture and In Vivo
To validate our observations from mouse models, we next characterized invasive leader cells in fresh human breast tumor specimens. We isolated organoids from primary tumor tissue obtained at initial breast surgery (n=10, Figure 3F), using established protocols (Nguyen-Ngoc et al., 2012). We classified each specimen using surrogate immunohistochemistry definitions (Goldhirsch et al. 2013). In total, there were 9 luminal breast cancers, 5 of which were luminal B. In each case, human tumor organoids invaded into collagen I matrix (Figure 3F and Movie S3). Although the numbers of invasive strands and disseminated cells varied among tumor samples, >86% of leaders expressed K14 across all samples (Figure 3F–G and S3E). K14+ cells typically co-expressed nuclear p63 (Figure S3C) and typically did not express SMA (Figure 3F and S3E). A subset of K14+ cells were also positive for luminal K8 and K18 (Figure S3D). Together, these data demonstrate that the cells that lead invasion in 3D culture express basal epithelial markers across multiple mouse models of breast cancer and in primary human breast tumors.
We therefore hypothesized that K14+ cells would lead collective invasion in primary human breast tumors in vivo. We first examined fixed sections from intact tumor specimens corresponding to three cases from 3D culture. We observed that K14+ cells led collective invasion in each case (Figure S3F). We next assayed for K14+ cells in archival specimens of primary breast tumors (n=39 specimens) from multiple subtypes of human breast cancer. Using consensus guidelines, we then assigned these specimens to breast cancer subtypes based on their ER, PR, HER2/Neu, and Ki-67 status, as assayed by immunohistochemistry (Goldhirsch et al., 2013). There were 16 luminal A cases, 17 luminal B cases (including 4 cases with HER2+ disease), and 6 triple-negative breast carcinomas (Figure 3I). We stained these samples for K14 using immunohistochemistry and classified their intensity to 3 levels: low (+), medium (++), and high (+++) (Figure S4A).
We observed strong K14 staining (CK3) in collective invasive fronts in luminal A, luminal B (both HER2− and HER2+), and triple negative breast cancers (Figure 3H,J and S4D–G). Breast cancer subtype was not statistically significantly associated with K14 staining intensity (Fisher exact test p-value = 0.11). We also stratified samples by histologic grade (Figure 3K). In general, higher histologic grade was associated with higher K14 staining intensity (Figure 3L, Fisher exact test p-value < 1 × 10^-4). However, even among luminal B tumors with high grade (grade 3), we identified cases with low, medium, and high K14 staining (Figure S4A). We observed three distinct patterns of K14 staining in human tumors: cytoplasmic staining, cytoskeletal staining with intense staining bundles of K14, and a border-enriched pattern, in which staining appeared specific to cells at the border of the collective invasion front (Figure S4B). The triple negative breast cancers showed a statistically significant increase in cytoskeletal staining pattern relative to luminal tumors (Figure S4C, Fisher-exact test p-value < 0.01). Together, these data demonstrate that K14 is expressed in collective invasion fronts across the major subtypes of human breast cancer.
Luminal Tumor Cells Acquire Markers of Basal Differentiation
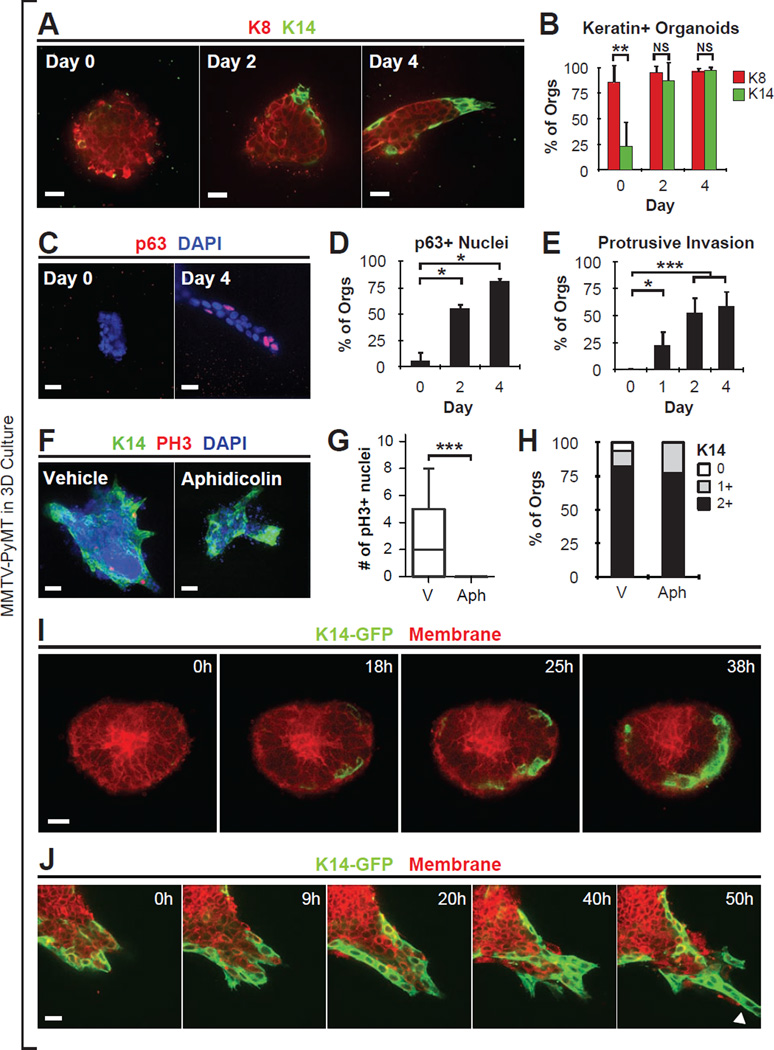
To address how K14+ leaders are generated, we asked when K14 was expressed in 3D culture of MMTV-PyMT tumor organoids. Consistent with the low in vivo frequency of K14+ cells in this luminal mouse model, tumor organoids were initially K8+, with few K14+ cells. During the first 48 hr of culture, the number of K14+ cells and p63+ cells in tumor organoids both increased significantly (Figure 4A–D and S5A–B). This expansion in basal marker positive cells coincided closely in time with the onset of collective invasion (Figure 4E and S5C). K14+ and p63+ cells were most frequently observed on the basal surfaces of tumor organoids, in contact with ECM. Thus, tumor organoids start 3D culture primarily luminal in character and subsequently expand a population of cells with basal differentiation markers.
Figure 4. Luminal Tumor Cells Acquire Markers of Basal Differentiation.
(A) Mouse mammary tumor organoids grown in 3D collagen matrix, stained with K8 and K14.
(B) Bar-graph of percentage of tumor organoids with K8 or K14 intensity equal to or greater than 1+ as a function of time. K14 and K8 intensity was quantified into 0 (no or few), 1 (intermediate), 2 (bright) K14 signal. Data presented as mean ± sd. For K14: N=719 organoids, from 5–8 mice per day; For K8: N=269 organoids, from 3 mice per day.
(C) Mammary tumor organoids grown in 3D collagen matrix, stained with p63 and DAPI.
(D) The percentage of organoids with nuclei positive for p63 was counted. Data presented as mean ± sd. N=114 organoids, from 2 mice per day.
(E) Invasion was quantified by scoring protrusive morphology of cancer cells in contact with the ECM. Data presented as mean ± sd. N=916 organoids, from 4–7 mice per day.
(F) Tumor organoids treated from day 0 with DMSO vehicle or a mitosis inhibitor (aphidicolin 10 µM), stained with K14, pH3, and DAPI.
(G) The number of pH3+ nuclei per tumor organoid in vehicle or mitosis inhibitor (aphidicolin) treated conditions. Data presented as boxplots. N=79 organoids, from 2 mice per condition.
(H) K14 intensity scored in vehicle or aphidicolin. K14 intensity was quantified into 0 (no or few), 1 (intermediate), 2 (bright) K14 signal. N=110 organoids, from 2 mice per condition.
(I) Time-lapse microscopy of MMTV-PyMT tumor organoids expressing GFP under the control of the K14 promoter (K14-GFP), beginning at the start of culture. See also Movie S4.
(J) K14-GFP+ cells migrate collectively and lead trailing K14-GFP- cells. Arrowhead, K14+ leader cell invasion. See Movie S4.
All p-values determined by two-sided t-test. * denotes P-value < 0.05. ** denotes P-value < 0.01. *** denotes P-value < 0.001. Scale bars are 40 µm in (F), and 20 µm in (A,C,I–J). See also Figure S5.
One possible cellular mechanism for the expansion of K14+ cells is cell proliferation. However, the number of mitotically active (pH3+) K14− cells was significantly greater than the number of mitotically active K14+ cells during the first 2 days of culture (Figure S5D–E). Furthermore, treatment with aphidicolin blocked mitosis completely but did not prevent an increase in K14+ cells (Figure 4F–H). We conclude that proliferation is not required for acquisition of a K14+ cell phenotype.
Alternatively, luminal tumor cells may convert directly from K14− to K14+ states. To test this hypothesis directly, we generated MMTV-PyMT mice carrying a K14 promoter biosensor in which GFP reports on K14 gene expression (Vaezi et al., 2002). In these tumor organoids, we observed individual cells at the basal surface transition from K14− to K14+ states (Figure 4I, cross-sectional view and S5F, en-face and Movie S4. N=22 movies). Following phenotypic conversion, the newly K14+ cells dynamically extended and retracted membrane protrusions into the matrix (N=149 movies) and led migration of trailing GFP− cells (N=70 movies, Figure 4J and Movie S4). Consistent with these data in mouse, we observed that organoids derived from multiple primary human breast tumors similarly expanded their fraction of K14+ and p63+ cells (Figure S5G–I). Taken together, these data reveal that the invasive leader phenotype is a differentiation state exhibited by cancer cells, not a fixed lineage.
K14+ Cells Acquire Leader Cell Behaviors Specifically in Collagen I Rich Local Microenvironments
We previously demonstrated that extracellular matrix (ECM) composition strongly regulates tumor invasion (Nguyen-Ngoc et al., 2012). Organoids isolated from the same tumor but allocated to different ECMs robustly invaded when cultured in collagen I, but not when cultured in basement membrane gels (Matrigel). We therefore hypothesized that different ECM environments might have a differential capacity to induce the basal gene expression characteristic of invasive leader cells.
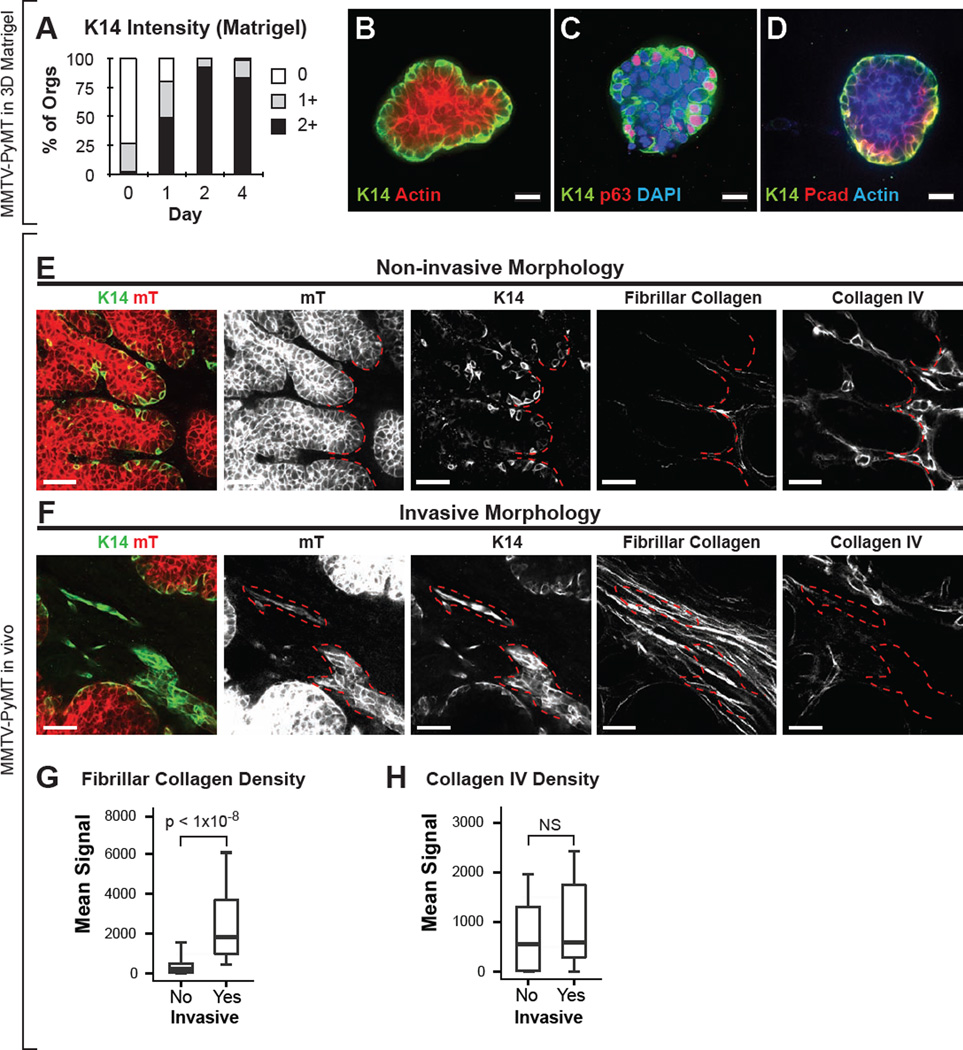
To test this hypothesis, we cultured MMTV-PyMT tumor organoids in 3D Matrigel. Although tumor organoids were non-invasive in Matrigel (Figure S6A–C), we still observed a robust increase in the number of K14+ tumor cells. At day 0, there were few K14+ cells but by day 4, 99% of tumor organoids had basal K14+ staining (Figure 5A–B). As in collagen I, Matrigel-embedded tumor organoids expressed nuclear p63 and P-cadherin specifically in basally located cells in contact with ECM, and did not express SMA (Figure 5C–D and Figure S6D). Consistent with these results, we also observed phenotypic conversion in K14-GFP;PyMT tumor organoids cultured in Matrigel (Figure S6E and Movie S5. N=5 movies). Taken together, these data demonstrate that induction of the basal epithelial program does not require collagen I and occurs in diverse ECM microenvironments.
Figure 5. K14+ Cells Acquire Leader Cell Behaviors Specifically in Collagen I Rich Local Microenvironments.
(A) K14 intensity of tumor organoids grown in 3D Matrigel. K14 intensity was quantified into 0 (no or few), 1 (intermediate), 2 (bright) K14 signal. N=471 organoids, 3–5 tumors per day.
(B–D) Micrographs of tumor organoids cultured in 3D Matrigel stained with K14 and phalloidin (B), K14, p63, and DAPI (C), or K14, P-cadherin, and phalloidin (D).
(E–F) Thick sections from in vivo primary MMTV-PyMT tumor were stained for K14, collagen type IV, and assayed for fibrillar collagen by SHG. Regions with K14+ cells were identified and classified as having either non-invasive or invasive morphology. Non-invasive morphology was defined as K14+ cells with smooth membrane borders (E, left-most panel). Invasive morphology was defined as protrusive strands of K14+ cells (F, left-most panel). The same sections were assayed for fibrillar collagen (second panels from right) and for collagen IV (right-most panels). Red hash marks outline the border of non-invasive (E) and invasive structures (F).
(G–H). The correlation of invasive morphology of K14+ cells with the abundance of fibrillar collagen or collagen IV density in vivo in MMTV-PyMT tumors. Data presented as boxplots. N=37–53 sections per condition, across 8 mice. P-values determined by two-sided t-test. Scale bars represent 20 µm in (B–D), and 40 µm in (E–F). See also Figure S6.
If phenotypic conversion induces a stable change in invasive potential, then switching to an invasive microenvironment should induce an acute change in K14+ cell behavior. To test this hypothesis, we conducted a matrix-swapping experiment. We cultured PyMT tumor organoids in 3D Matrigel for 4 days, liberated organoids from the gel, and re-embedded them in collagen I. Under these conditions, there was rapid collective invasion from the basal layer within 12 hr (Figure S6F–G, n=14 movies, and Movie S6). In response to collagen I matrix, individual K14+ cells at the basal surface became protrusive and subsequently initiated collective invasion (Figure S6G, insets). These findings show that phenotypic conversion and collective invasion are distinct steps and that the latent invasive potential of K14+ cells is only converted to invasive cell behaviors in specific tumor microenvironments.
In vivo, we observed that although K14+ cells led collective invasion, there were also K14+ cells with non-invasive morphology (Figure 2A). These findings raised the question which factors in vivo distinguish K14+ cells that manifest invasive morphology from those that do not. Because the local ECM context within a primary tumor is heterogeneous, we hypothesized that differences in invasive morphology in vivo would correlate with differences in ECM context. Accordingly, we assayed fibrillar collagen density in fixed thick tumor sections by second harmonic generation (SHG) and collagen IV density by immunofluorescence. Areas were classified as having either invasive or non-invasive morphology based on the protrusive morphology of the K14+ cells in the region (Figure 5E–F). Invasive regions were associated with an ~8-fold increase in median collagen I fiber density relative to non-invasive regions (Figure 5G). In contrast, collagen IV, a component of basement membrane and Matrigel, was observed at comparable levels in both invasive and non-invasive regions (Figure 5H). Taken together, our data demonstrate that 3D Matrigel and 3D collagen I ECM microenvironments induce distinct cell morphologies similar to those observed in specific regions of primary tumors in vivo, and suggest that K14+ cells in vivo acquire leader cell behaviors specifically in collagen I rich local microenvironments.
Basal Epithelial Genes K14 and p63 Are Required for Collective Invasion in 3D Culture
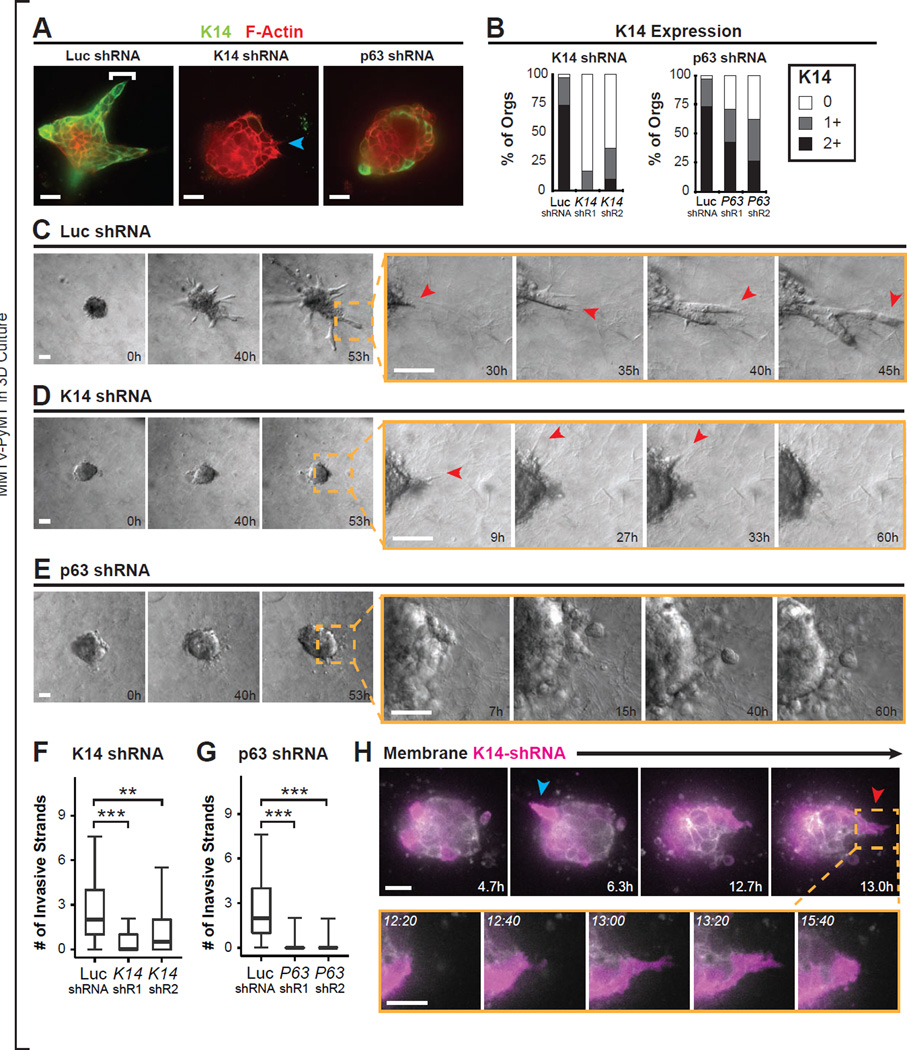
We next asked whether expression of basal epithelial genes by leader cells is functionally required for collective invasion. Accordingly, we developed protocols for efficient lentiviral transduction of primary tumor organoids. We tested five different hairpin shRNAs targeting K14 and p63 and determined the two hairpins with the highest knockdown efficiency by Western blotting (Figure S7A–B). As a control, we used a hairpin targeting luciferase (Luc-kd).
To assess the requirement for K14 in collective invasion, we cultured Luc-kd, K14-kd1, and K14-kd2 tumor organoids in 3D collagen I and observed them by time-lapse DIC microscopy. Control Luc-kd organoids strongly expressed K14 and invaded vigorously into collagen I matrix (Figure 6A–C and Movie S7). In contrast, both K14 expression and collective invasion were markedly reduced in K14-kd organoids (Figure 6A,B,D,F). Although K14-kd tumor organoids generally lacked multicellular invasive strands, we still observed subcellular protrusions extending from K14-kd organoids (Figure 6D, red arrows and Figure S7C). We next infected organoids with lentiviruses encoding both K14-shRNA and GFP to label K14-kd cells in real-time. By tracking infected organoids at single cell resolution, we observed individual K14-kd cells that were protrusive and motile (Figure 6H). Thus, we conclude that K14 is dispensable for subcellular protrusions but is required for the transition to persistent collective invasion.
Figure 6. Basal Epithelial Genes K14 and p63 Are Required for Collective Invasion in 3D Culture.
(A) Micrographs of MMTV-PyMT tumor organoids transduced with lentiviral particles encoding shRNAs against luciferase control (Luc shRNA), K14 (K14 shRNA), or p63 (p63 shRNA), and then embedded in collagen I matrix and stained for K14 and Phalloidin. Left panel: bracket, leader cell. Middle panel: blue arrow, K14- protrusive cell.
(B) K14 intensity was quantified into 0 (no or few), 1 (intermediate), or 2 (bright) K14 signal for organoids from (A). N=61–93 organoids per condition, from 3 independent experiments.
(C–E) Time-lapse DIC microscopy of transduced organoids as in (A). In Luc shRNA transduced organoids, the tumor organoid invades collectively (C). Inset shows protrusive cells that extend and expand into a collective invasive strand. In K14 shRNA transduced organoids, tumor organoids do not invade collectively (D). Inset shows protrusive behavior is intact. In p63 shRNA transduced organoids, tumor organoids do not invade collectively (E). Inset shows lack of protrusions and rounded cell borders in p63-kd1 tumor organoids. See Movie S7.
(F–G) The number of collective invasive strands in Luc shRNA, K14-shRNA kd1 and kd2, and p63-shRNA kd1 and kd2 transduced organoids was determined from timelapse movies. The maximal number of invasive strands at any time was determined. N=170 movies for Luc shRNA from 7 independent experiments. N=133 movies for K14 shRNA kd1 from 7 independent experiments. N=36 movies for K14 shRNA kd2 from 3 independent experiments. N=90 movies for p63 shRNA kd1 from 5 independent experiments. N=42 movies for p63 shRNA kd2 from 3 independent experiments. Data presented as boxplots. P-values determined by two-sided t-test. **, p-value < than 0.001. ***, p-value < 1 × 10^-10.
(H) Time-lapse sequence of a representative tumor organoid transduced with lentiviral particles encoding for dual expression of K14-shRNA and GFP. GFP is shown in false color purple. Arrows, individual GFP+ (and K14-shRNA expressing) protrusive cells.
Scale bars represent 20 µm in (A), 50 µm in (C–E), and 10 µm in (H). See also Figure S7.
We next examined the phenotypic consequences of p63 knockdown (p63-kd1 and p63-kd2) in PyMT primary tumor organoids cultured in collagen I (Figure 6A,B,E,G). We observed a strong reduction in p63 protein levels and in collective invasion in p63-kd organoids (Figure 6E,G and S7D). However, compared with K14-kd organoids, p63-kd organoids had fewer protrusive cells and more rounded cell borders (Figure 6E, S7C, and Movie S7). Because knockdown with p63 hairpin abrogated collective invasion but yielded only a partial reduction in K14, these data suggest that p63 has essential K14-independent functions in collective invasion. In aggregate, these data demonstrate that multiple basal epithelial genes expressed by leader cells are individually required for collective invasion in 3D culture. Furthermore, these data show that basal epithelial genes are required for collective invasion in a luminal mouse model of breast cancer, despite being expressed in only a small minority of the tumor cells.
In Vivo Knockdown of K14 Disrupts Collective Invasion at the Tumor-Stromal Border
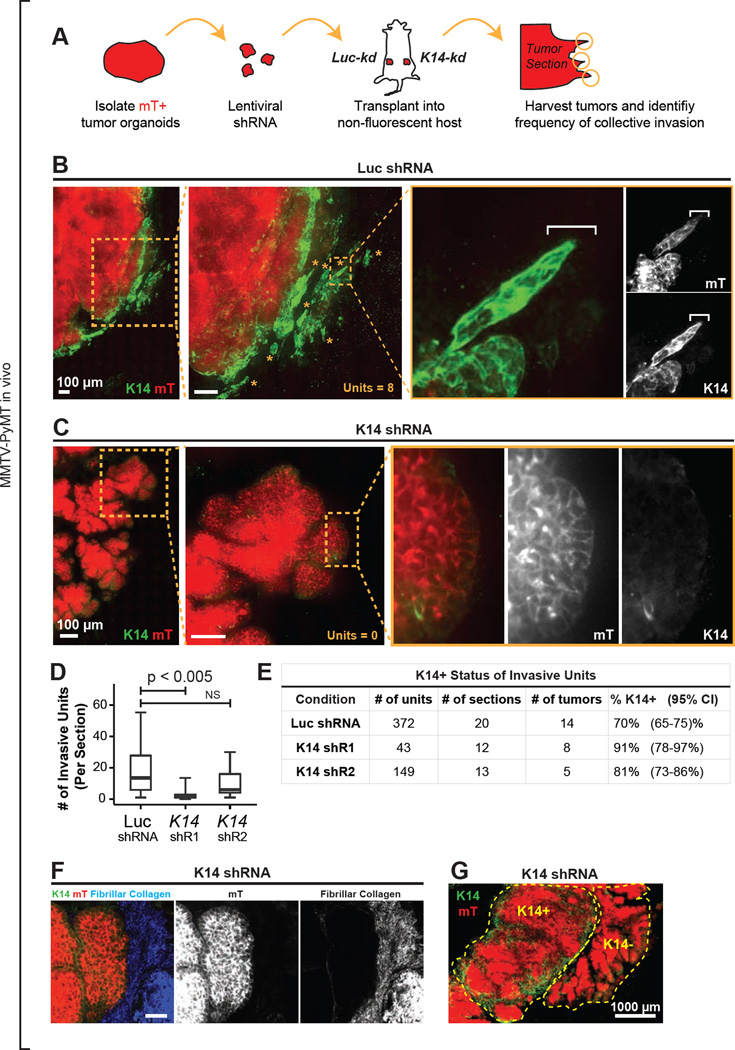
We next sought to test the requirement for K14 in collective invasion in vivo. We isolated tumor organoids from fluorescently labeled advanced carcinomas (mTomato+; MMTV-PyMT), transduced them with Luc-kd, K14-kd1, or K14-kd2 lentivirus, and then orthotopically transplanted the transduced organoids into the cleared mammary fat pads of non-fluorescent congenic hosts (Figure 7A). Although tumor growth appeared similar, we observed strong differences in collective invasion at the tumor stromal border. In Luc-kd tumors, cells at the tumor stromal border strongly expressed K14 and organized many K14+ infiltrating invasive strands and nests of cells (Figure 7B). In contrast, in K14-kd1 tumors, we observed large regions in which the tumor stromal border was K14- and lacked either invasive strands or nests (Figure 7C,D). The K14- regions instead displayed borders with a rounded morphology (Figure 7C,D). Furthermore, whereas fibrillar collagen was associated with collective invasion in control tumors (Figure 5E), in K14-kd1 tumors, this association was disrupted. Within K14-kd1 tumors we observed non-invasive borders that were both K14- and surrounded by dense fibrillar collagen (Figure 7F). To quantify the differences in collective invasion between knock-down conditions, we determined the number of invasive strands and nests per tumor section. The number of collective invasive units per section was significantly reduced in K14-kd1 tumors relative to Luc-kd control tumors (Figure 7D). Consistent with the less efficient knockdown of K14-kd2 observed by immunofluorescence and western blotting, we observed a qualitatively similar reduction in invasive units in K14-kd2 tumors (Figure 7D–E). The residual invasion observed in both K14-kd1 and K14-kd2 tumors was predominantly K14+ and was concentrated in discrete K14+ tumor foci (Figure 7E,G). These data demonstrate the presence of residual tumor cells competent to express K14, and suggest that despite puromycin selection prior to transplantation, K14-competent cells preferentially expanded in vivo. Together these data demonstrate: (1) that K14 is required in vivo for collective invasion at the tumor-stromal border; (2) that elevated collagen I is not sufficient to induce collective invasion in vivo in the absence of K14 expression; and (3) that targeting a basal invasive program expressed in a small minority of tumor cells is sufficient to disrupt the invasive process in an advanced carcinoma.
Figure 7. In Vivo Knockdown of K14 Disrupts Collective Invasion at the Tumor-Stromal Border.
(A) Schema to test the in vivo requirement for K14 in collective invasion. Fluorescent mTomato+ (mT+) tumor organoids were transduced with either Luc shRNA (Luc-kd) or K14-shRNA (K14-kd), selected with puromycin, and transplanted into the cleared mammary fat pads of non-fluorescent congenic FVB hosts. Tumor was isolated at ~1cm and montages were assembled of the tumor-stromal border.
(B) Micrograph of the tumor-stromal border from a representative Luc-kd tumor stained with K14. mT+ regions (in red), tumor-derived cells. Insets, collective invasive units in the Luc-kd tumor. The micrograph represents a z-projection of over 40 µm.
(C) Micrograph of the tumor-stromal border from a representative K14-kd1 tumor stained with K14. mT+ regions (in red), tumor-derived cells. Insets, representative K14- non-invasive border.
(D) Median number of invasive units per section in Luc-kd and K14-kd tumors, with data presented as a boxplot. Invasive units included strands and nests as described in Figure 2E. P-values determined by 2-sided t-test.
(E) Frequency of invasive units expressing K14 in Luc-kd and K14-kd tumor sections. 95% confidence intervals for each proportion denoted in parentheses.
(F) Representative micrograph of K14-kd1 tumor stromal border stained for K14 and assayed for fibrillar collagen by SHG. mT+ regions (in red), tumor-derived cells.
(G) Micrograph of a K14-kd1 tumor stained with K14, demonstrating K14+ and K14-tumor foci. The micrograph represents a z-projection of over 40 µm. Scale bars represent 100 µm in (B–C), 40 µm in (F) and 1000 µm in (G).
DISCUSSION
The transition to invasive behavior in epithelial cancer cells is a critical step in metastasis; yet both the cellular and molecular basis of this transition is incompletely understood. An important challenge is that primary tumors are complex heterogeneous tissues containing phenotypically and genetically distinct cancer cell subpopulations interacting with an altered extracellular matrix and diverse stromal cells (Almendro et al., 2013; Egeblad et al., 2010a; Egeblad et al., 2010b; Fidler, 2003; Weigelt and Bissell, 2008). It is important to reduce this complexity to specific molecular regulators of tumor cell behavior in order to identify targets for therapeutic intervention.
To accomplish this goal, we developed a general strategy for identifying the most invasive cells in a primary epithelial tumor. We then applied this strategy and discovered that the most invasive cancer cells are behaviorally and molecularly distinct from bulk tumor cells. Importantly, the invasive leader cells expressed basal epithelial genes, including K14 and p63, across mouse models of breast cancer and in diverse human breast tumors. Cells with this phenotype led the majority of collective invasion in vivo and were strongly overrepresented in lung metastases. We therefore have exploited our assays to resolve the heterogeneous cancer cell populations within the primary tumor into functional classes based on their invasive behavior. We then leveraged this understanding to establish the requirement for K14 and p63 for collective invasion in primary cells derived from poorly differentiated carcinomas.
Previous studies have shown that heterotypic epithelial-stromal interactions contribute to cancer invasion in diverse malignancies (Condeelis and Pollard, 2006; Gaggioli et al., 2007). Our data establish the importance of heterotypic epithelial-epithelial interactions between different cancer cell subpopulations in collective invasion. Importantly, while invasive leader cells were highly protrusive and migratory, they retained characteristically epithelial expression of cell adhesion proteins (e.g. E-cadherin) and intermediate filament proteins (e.g. K14, K8) during invasion.
K14 expression also marks other highly migratory epithelial cell populations. During development, the migratory cells of the normal mammary embryonic placode are K14+K8+SMA- (Moumen et al., 2011; Sun et al., 2010). Similarly, the highly invasive rat breast cancer MTLn3 cell line is K14+K8+, whereas non-invasive MTCs do not express these markers (Lichtner et al., 1991). Likewise, highly invasive human breast cancer cell lines MDA-MB-468, MDA-MB-436, and BPLER are K14+K18+, while cell lines with low invasive potential, such as MCF-7, T47D, and ZR75 express a pure luminal phenotype (Gordon et al., 2003; Petrocca et al., 2013). Experimental models suggest that basal breast cancers arise from luminal progenitors, and that mutations in genetic drivers of breast cancer, such as BRCA1, can shift luminal progenitors towards basal differentiation (Lim et al., 2009; Molyneux et al., 2010). Consistent with these observations, a number of pathologic studies have shown that basal cytokeratins, including K14, are associated with poor patient prognosis, early relapse, and reduced overall survival (de Silva Rudland et al., 2011; Gusterson et al., 2005; Laakso et al., 2006; Malzahn et al., 1998). Our study provides a biological basis for these findings, namely that K14 marks a subpopulation of tumor cells capable of initiating collective invasion, a critical step in metastatic progression.
Importantly, we directly observed phenotypic conversion of K14- luminal tumor cells to K14+ cells with invasive potential. We previously demonstrated that collective invasion is strongly regulated by the composition of the local extracellular matrix (Nguyen-Ngoc et al., 2012). In the present study, we demonstrated that the basal invasive program can be induced in diverse ECM microenvironments. However, consistent with our prior study, basal gene expression was only associated with invasive cell behaviors and morphologies in collagen I rich microenvironments in 3D culture and in vivo.
In our study we established that the basal epithelial intermediate filament cytoskeleton is specifically required for collective invasion in cancer. Our data suggest that actin-based sub-cellular protrusions and multicellular collective invasion can be uncoupled and have distinct cytoskeletal requirements. The precise mechanism by which cytokeratin 14 regulates collective invasion, be it directly modifying the tensile strength and viscoelastic properties of the cell (Yamada et al., 2002), regulating cell-cell adhesion (Weber et al., 2012), or by signal transduction mechanisms independent of their mechanical properties (Kim et al., 2006), remains unclear and warrants further study. Because cells with basal differentiation are found in many epithelial organs, our findings may also have implications for invasion in other solid tumors.
EXPERIMENTAL PROCEDURES
Mouse Lines and Breeding
Mice used in this study were backcrossed onto and maintained on the FVB/n background in a specific pathogen-free facility. Animal protocols were approved by the Johns Hopkins IACUC. FVB/N-Tg(MMTV-PyVT)634Mul/J (MMTV-PyMT) (Guy et al., 1992a), FVB/N-Tg(MMTVneu)202Mul/J (MMTV-Neu) (Guy et al., 1992b), FVB-Tg(C3-1-TAg)cJeg/JegJ (C3(1)/Tag) (Maroulakou et al., 1994) and B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) (Muzumdar et al., 2007) mice were obtained from the Jackson Lab. K14- GFP-actin mice (Vaezi et al., 2002) were a generous gift from Elaine Fuchs (Rockefeller University). For confocal time-lapse experiments, MMTV-PyMT mice were crossed with mT/mG mice and/or K14-GFP-actin mice.
Analyses of Fresh Primary Human Tumors
Primary breast cancers were acquired in accordance with IRB exempt protocols NA_00052607 and NA_00077976 (Nguyen-Ngoc et al., 2012). In brief, each tumor sample was de-identified prior to receipt with limited pre-specified clinical information provided with each sample. The tumor sample was kept in cold DMEM media when in transit. The tumor sample was rinsed with antibiotic wash, minced, and then digested in collagenase with or without trypsin. Tumor organoids were treated with DNase, and separated out by differential centrifugation at 1500 rpm × 4–6 spins. Tumor organoids were then allocated to either Matrigel or 3D collagen I, and cultured in human mammary epithelium medium containing insulin, EGF, hydrocortisone, and cholera toxin. Medium was replaced every 3–4 days.
Statistics
Statistical analysis was conducted using R. For all boxplots, the whiskers represent the 5th and 95th percentiles. All tests used and P values are specified in the figure legends. P < 0.05 was considered significant.
Additional Materials and Methods, including isolation of tumor organoids, antibodies, and staining procedures used, are described in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Molecularly distinct tumor cells lead collective invasion in breast cancer.
Keratin-14+ cells lead collective invasion across major subtypes of breast cancer.
Invasive leader cells are generated by induction of a basal epithelial program.
Basal epithelial gene expression is required for collective invasion.
ACKNOWLEDGEMENTS
We thank Koen Schipper, Jen Beck, and Audrey Brenot for technical support. We thank Ellen Tully for performing K14 immunohistochemistry on archival tumors. Human tissue samples were provided by the Johns Hopkins Hospital and by the Cooperative Human Tissue Network, which is funded by the NCI. We also thank William C. Hines, Paul Yaswen, Pedram Argani and Lisa Jacobs for assistance with acquisition of primary human tumor specimens. We thank Mikala Egeblad, William Matsui, Vered Stearns, Sara Sukumar, Tamara Lotan, and Charles Rudin for scientific discussions. This study was funded by grants from the NCI (R01 CA056721 to Z.W.; U01 CA155758, P50 CA103175, and P50 CA88843 to A.J.E.), the NIEHS (U01 ES019458 to Z.W.), by a Safeway Foundation Award for Breast Cancer Research (to A.J.E.), by funds from the Avon Foundation for Women (to A.J.E.), by funds from the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research (to A.J.E.), and from the U.S. Department of Defense (W81XWH-12-1-0018 to K.J.C). A.J.E. was funded by a Research Scholar Grant, RSG-12-141- 01-CSM from the American Cancer Society. This study benefited from a Zeiss 710 NLO 2- photon microscope purchased through funds from S10RR024550.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests. The authors declare no competing financial interests.
REFERENCES
- 1.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 2.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A, Keely PJ. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Silva Rudland S, Platt-Higgins A, Winstanley JH, Jones NJ, Barraclough R, West C, Carroll J, Rudland PS. Statistical association of basal cell keratins with metastasis-inducing proteins in a prognostically unfavorable group of sporadic breast cancers. Am. J. Pathol. 2011;179:1061–1072. doi: 10.1016/j.ajpath.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell. 2010a;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010b;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 10.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 12.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 13.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL. Breast cell invasive potential relates to the myoepithelial phenotype. Int. J. Cancer. 2003;106:8–16. doi: 10.1002/ijc.11172. [DOI] [PubMed] [Google Scholar]
- 15.Gray RS, Cheung KJ, Ewald AJ. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr. Opin. Cell Biol. 2010;22:640–650. doi: 10.1016/j.ceb.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusterson BA, Ross DT, Heath VJ, Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 2005;7:143–148. doi: 10.1186/bcr1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992a;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. U.S.A. 1992b;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 22.Laakso M, Tanner M, Nilsson J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, Malmstrom P, Wilking N, Bergh J, Isola J. Basoluminal carcinoma: a new biologically and prognostically distinct entity between basal and luminal breast cancer. Clin. Cancer Res. 2006;12:4185–4191. doi: 10.1158/1078-0432.CCR-06-0353. [DOI] [PubMed] [Google Scholar]
- 23.Leighton J, Kalla RL, Turner JM, Jr, Fennell RH., Jr Pathogenesis of tumor invasion. II. Aggregate replication. Cancer Res. 1960;20:575–586. [PubMed] [Google Scholar]
- 24.Lichtner RB, Julian JA, North SM, Glasser SR, Nicolson GL. Coexpression of cytokeratins characteristic for myoepithelial and luminal cell lineages in rat 13762NF mammary adenocarcinoma tumors and their spontaneous metastases. Cancer Res. 1991;51:5943–5950. [PubMed] [Google Scholar]
- 25.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 26.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malzahn K, Mitze M, Thoenes M, Moll R. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Arch. 1998;433:119–129. doi: 10.1007/s004280050226. [DOI] [PubMed] [Google Scholar]
- 28.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Moumen M, Chiche A, Cagnet S, Petit V, Raymond K, Faraldo MM, Deugnier MA, Glukhova MA. The mammary myoepithelial cell. Int. J. Dev. Biol. 2011;55:763–771. doi: 10.1387/ijdb.113385mm. [DOI] [PubMed] [Google Scholar]
- 31.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 32.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen-Ngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, Ewald AJ. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2595–E2604. doi: 10.1073/pnas.1212834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, Kung AL, Hide W, Ince TA, Lieberman J. A Genome-wide siRNA Screen Identifies Proteasome Addiction as a Vulnerability of Basal-like Triple-Negative Breast Cancer Cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J. Natl. Cancer Inst. Monogr. 2010;2010:210–213. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 38.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun P, Yuan Y, Li A, Li B, Dai X. Cytokeratin expression during mouse embryonic and early postnatal mammary gland development. Histochem. Cell Biol. 2010;133:213–221. doi: 10.1007/s00418-009-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 41.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev. Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin. Cancer Biol. 2008;18:311–321. doi: 10.1016/j.semcancer.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada S, Wirtz D, Coulombe PA. Pairwise assembly determines the intrinsic potential for self-organization and mechanical properties of keratin filaments. Mol. Biol. Cell. 2002;13:382–391. doi: 10.1091/mbc.01-10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.