Abstract
Neuropeptides are cell to cell signaling molecules that modulate a wide range of physiological processes. Neuropeptide release has been studied in samples sizes ranging from single cells and neuronal clusters, to defined brain nuclei and large brain regions. We have developed and optimized cell stimulation and collection approaches for the efficient measurement of neuropeptide release from neuronal samples using a dual capillary system. The defining feature is a capillary that contains octadecyl-modified silica nanoparticles on its inner wall to capture and extract releasates. This collection capillary is inserted into another capillary used to deliver solutions that chemically stimulate the cells, with solution flow up the inner capillary to facilitate peptide collection. The efficiency of peptide collection was evaluated using six peptide standards mixed in physiological saline. The extracted peptides eluted from these capillaries were characterized via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) with low femtomole detection limits. Using the capillary collection system in small custom-fabricated culturing chambers, individual cultured neurons and neuronal clusters from the model animal Aplysia californica were stimulated with distinct neuronal secretagogues and the releasates collected and characterized using MALDI-TOF MS.
Keywords: neuropeptides, solid phase extraction, silica nanoparticle deposited capillary, neuronal release, single neuron, MALDI MS
Introduction
The ability to monitor the release of peptides during neuronal network activity allows one to study a broad variety of physiological processes, including neuromodulation, neurotransmission, cell outgrowth, cell survival, and hormonal signaling between organs.1,2 Depending on the specific system under investigation—individual neurons located in chemically heterogeneous tissue, well-defined brain nuclei, or larger brain regions—the scale of the neuropeptide release varies greatly. Due to the diversity of peptide sequences and properties, and their wide range of endogenous concentrations in a chemically complex extracellular environment that contains high levels of inorganic salts, the characterization of cell to cell signaling neuropeptides is challenging. Here, a spatially defined release is correlated to specific environmental perturbations—the addition of distinct cellular secretagogues.
Successful means of characterizing neuronal release include microelectrodes, biosensors, liquid chromatography (LC) and capillary electrophoresis (CE) separations, and mass spectrometry (MS).3 When monitoring complex suites of neuropeptides, MS-based approaches are capable of simultaneously monitoring multiple peptides without analyte preselection or pretagging,4–7 making MS well suited for peptide discovery. Among the available MS methods, electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) have been used in many peptide studies.8–10
Here we use MALDI-time-of-flight (TOF) MS as it is compatible with small sample sizes and tolerant of salts and other impurities.9,11–13 Regardless of the MS figures of merit, a sample approach that can efficiently collect peptides from well-defined locations while removing cell media components is desirable. Pipette tips packed with extraction media have been used for collecting, concentrating, and desalting biological samples,14,15 including releasates from rat brain slices before MALDI MS detection.16 Recently, we described a mixed mode extraction media consisting of a particle embedded polymer monolithic capillary that was successfully used for monitoring release from single cultured neurons.17 The embedded extraction particles provided enhanced extraction capacity and the monolithic materials increased the phase ratio, which improved the overall efficiency of peptide extraction from a complex extracellular matrix, and allowed us to access individual cultured neurons. Generally, the collection probe was placed close to the neuronal structure under investigation. After the addition of the appropriate secretagogue solutions, released peptides were collected from the neuron of interest and perhaps those adjacent to the target neuron, and then assayed via MALDI-TOF MS.
Several other methods have been reported for sampling peptide release from organs, tissues, and cell culture media. For example, the push-pull perfusion sampling technique with dual-tubing (capillary) has been used for studying release events in various biological systems; one tube is used to deliver stimulants/reactants and the other to transport the releasates out of the system for further processing.18,19 The two tubes can be placed near each other or in a concentric arrangement for sampling extracellular fluids. Recent modifications have focused on reducing the tissue damage by miniaturizing the probe, using a low flow rate,20,21 using electroosmotic sampling,22 and improving temporal and spatial resolution.21,23 Slaney et al.24 used segmented flow for push-pull sampling and achieved high temporal resolution at 7 s with volumes as small as 6 nL for in vivo monitoring of glutamate in the rat striatum. The microdialysis probe is an approach widely used for real-time monitoring.25,26 Microfluidic sampling methods that use the chemistrode27 and other devices28 allow stimulation and release to be integrated into a single device. A microfabricated two-channel device has been used to monitor released compounds from cells in a culture dish.29 By controlling the ratio of incoming and outgoing flows, this microfluidic probe obtained spatially resolved activity information from a geometrically open space without sealing the area/target. Recently, the technique was used for lysis of single adherent cells in a cell network to collect messenger RNA.30
Most of the previously reported dual-capillary systems normally act as sample collection approaches and do not clean up or process the samples. As a result, the releasate is collected along with physiological levels of salts that often require separation and/or extraction steps before further analysis. Here we describe several arrangements of the dual-capillary collection system where the outer capillary delivers the compounds and the inner capillary collects the release. Hence, the outer capillary helps to define the area where the release activity is monitored while the inner capillary, containing octadecyl-modified silica nanoparticles, collects and concentrates the peptides as they pass by the extraction media. Using the well-characterized animal model, Aplysia californica, neuropeptides from single cultured neurons upon KCl stimulation, and from bag cell clusters upon insulin stimulation, have been collected and detected by MALDI MS. Next, we used this approach to collect peptides released in a spatially resolved manner from the A. californica abdominal ganglion.
Materials and methods
Chemicals
Octadecyltrimethoxysilane (ODTS, 90%), fluosilicic acid (H2SiF6, 35%), silica (amorphous powder), boric acid (H2BO3, 99.8%,), and anhydrite toluene were purchased from Sigma-Aldrich (St. Louis, MO). Methanol (MeOH), acetonitrile (ACN), and formic acid (FA) were obtained from Fisher Scientific (Fairlawn, NJ).
Artificial sea water (ASW) consisting of 460 mM NaCl, 10 mM KCl, 10 mM CaCl2, 22 mM MgCl2, 26 mM MgSO4, and 10 mM HEPES, pH 7.8, was used as the sample matrix for extraction of the peptide standards. Six peptide standards—angiotensin II (3.8 mM), angiotensin I (3.1 mM), substance P (3.0 mM), bombesin (2.5 mM), somatostatin (2.4 mM), and adrenocorticotropic hormone (18–39) (ACTH) (3.5 mM)—were obtained from Sigma-Aldrich and prepared in water as stock solutions at the concentrations indicated. They were mixed together and diluted 1000-fold to 2.4 to 3.8 μM in ASW for evaluating the binding capacity of the octadecyl-modified silica nanoparticle deposited (OSND) capillary. Water used throughout the experiments was obtained from a Milli-Q water purification system (Millipore, Bedford, MA).
Fabrication of the OSND capillaries
Fused silica capillaries (100 μm inner diameter (i.d.) × 170 μm outer diameter (o.d.), Polymicro Technologies, Phoenix, AZ) were rinsed with 1.0 M of aqueous sodium hydroxide for 2.5 h, 30 min with water, 2 h with 1.0 M of hydrochloric acid, and again with water for 30 min. The next step, depositing the silica nanoparticles, was performed according to a previously published method.31,32 Briefly, a precursor solution was prepared by adding 0.6 g of the silica into 8 mL of H2SiF6. After stirring at 35 °C for 16 h, the mixture was centrifuged at 14,000 rpm for 10 min. Next, 1 mL of the supernatant was added to 0.5 mL of water and 0.6 mL of 0.1 M H3BO3. The mixture was vortexed, filled into the capillary, and both ends sealed. The deposition was allowed to proceed for 16 h at 40 °C. After deposition, the capillary was washed with water and dried at 120 °C for 4 h under constant N2 flow. Then the entire procedure, starting with the precursor solution preparation, was repeated for a second deposition. Finally, the silica nanoparticle-deposited capillaries were aged at 250 °C for 6 h. The final steps were to modify the silica nanoparticles with octadecyl groups. Here, 1 mL of dried toluene was added to 0.2 mL of ODTS, the capillary filled, and the ends sealed. The reaction proceeded at 105 °C for 24 h, followed by thorough washing steps with toluene, isopropanol, and methanol in sequence. The prepared OSND capillaries were stored until use.
Scanning electron microscopy (SEM) characterization
The silica nanoparticles deposited on the inner wall of the capillary were characterized with SEM using a Philips XL30 field-emission environmental scanning electron microscope (FEI Company, Hillsboro, OR) using a secondary electron detector. The OSND capillaries were broken and several pieces were mounted on the microscope sample stage with the inner surface facing up and then coated with Pd/Au using a Desk II turbo sputter coater (Denton Vacuum, Moorestown, NJ).
Dual-capillary collection system
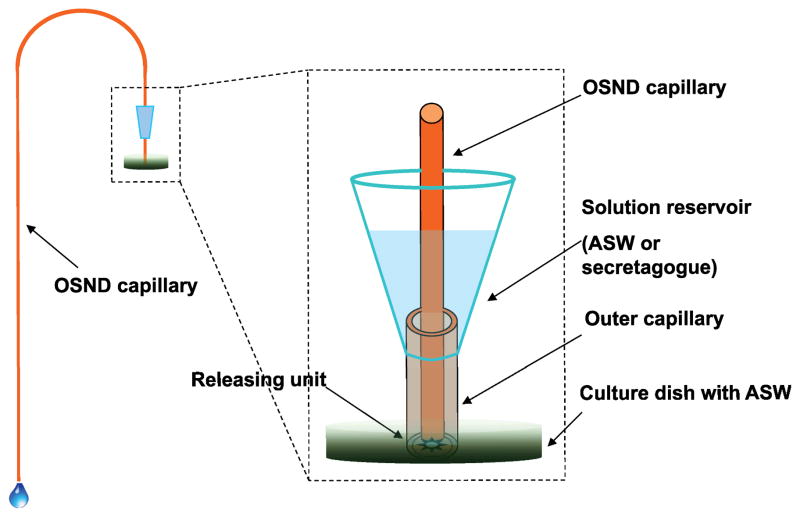
The dual-capillary collection system used in this work (Figure 1) consisted of two capillaries with different diameters positioned in a centric format, the outer capillary, 536 μm i.d × 665 μm o.d. and 2 cm long, was used to define the collection area; the inner OSND capillary was used for collecting the released peptides at 100 μm i.d.× 170 μm o.d. The outer capillary was connected to a solution reservoir that was created from a modified pipette tip capable of holding 0.5 mL of ASW or secretagogue and glued to the outer capillary. A one meter long OSND capillary was used as the inner collection capillary. When collection began, the open end of the outer capillary was positioned to enclose the releasing targets, such as isolated neuron clusters or a single cultured neuron. Then the inner capillary was inserted into the outer capillary with the end positioned as close as possible to the sample target and the capillary outlet placed at a lower height to generate the desired sampling flow ranging from 0.1 to 1.5 μL/min. Once the collection was completed, the inner collection capillary was replaced with a new one to start the next collection.
Figure 1.
Schematic illustration of the dual-capillary collection setup with an OSND capillary used as the peptide collection approach. Other arrangements of cell stimulation are also explored including confined cell culturing chambers and delivery of chemicals through blood vessels. In all cases, the efficient collection, concentration and desalting of the peptides is required prior to MS characterization.
Extraction efficiency of OSND capillaries
The extraction efficiency of the OSND capillaries was evaluated by extracting standard peptides prepared in ASW. The capillaries were connected to a syringe mounted on a KDS270 syringe pump (KD Scientific, Holliston, MA) through PEEK tubing (1/16 × 0.007″) (IDEX, Oak Habor, WA), with the other end positioned in the peptide sample solution. The capillaries were first conditioned with methanol and equilibrated with water. Then the sample solution was loaded onto the columns at 0.3 μL/min by setting the syringe pump to withdraw mode. After extraction, the columns were washed with 12 μL of 20% MeOH in water, followed by an air flush to remove all of the liquid. Next the analytes were eluted with 90% methanol containing 0.5% FA, and further analyzed by MALDI-TOF MS/capillary liquid chromatography (CapLC)-ultraviolet (UV) detection, as described below.
CapLC-UV/ESI-MS assays
The eluents from the OSND columns were first dried using a SpeedVac concentrator (Thermo Scientific, San Jose, CA) and redissolved in 25 μL of a solution containing 99% H2O, 1% ACN, and 0.05% trifluoroacetic acid (TFA). Liquid chromatographic separation was performed with a U3000 CapLC system (Dionex, Sunnyvale, CA) equipped with an Acclaim® PepMap RSLC C18 column (300 μm, 15 cm, 2 μm 100 Å) using an elution gradient of 0–80% solvent B (80% ACN with 0.05% TFA) and solvent A (100% H2O with 0.05% TFA) in 35 min. Detection was performed with a diode array detector set at 214 nm, coupled to an HCTultra™ ESI mass spectrometer (Bruker Daltonics, Billerica, MA) for peak assignment.
MALDI-TOF MS analysis
MALDI-TOF MS was performed with an ultrafleXtreme™ MALDI TOF/TOF mass spectrometer using flexControl software (Bruker Daltonics). The extracted peptides were eluted at 1 μL/min with the flow controlled by a syringe pump for 15 s. Eluent fractions (0.25 μL) were directly applied onto targets prespotted with matrix (AnchorChip, Bruker Daltonics). Mass spectra were acquired with external calibration in reflectron mode with typical mass accuracies within 100 ppm. The resulting spectra represent the accumulation of 2000 laser shots without baseline correction or data filtering; flexAnalysis software (Bruker Daltonics) was used to process the spectra.
Release assays
A. californica, weighing 100–120 g, were obtained from the University of Miami/NIH National Resource for Aplysia and kept in an aquarium containing aerated and filtered artificial sea water (Instant Ocean, Aquarium Systems Inc., Mentor, OH) maintained at 14 °C. Before dissection, animals were anesthetized by injection of isotonic MgCl2 into the body cavity at 50% of body weight.
Sample preparation
For the ganglion release experiments, abdominal ganglia were surgically dissected from the CNS of A. californica without further treatment and pinned on an elastomer-filled (Sylgard 184, Dow Corning, Midland, MI) Petri dish. For the neuron cluster experiments, the bag cell clusters were isolated from the abdominal ganglia without enzyme treatment. For the single bag cell neuron assays, special sample wells were fabricated in order to facilitate the single neuron culture and release. A polydimethylsiloxane (PDMS) film of 200 μm thickness with punched pores (0.686 mm i.d.) was placed on the inside bottom of a Petri dish containing 4 mL of ASW. To isolate the bag cell neurons, the abdominal ganglia were surgically dissected, followed by treatment with 1% protease (type IX Bacterial, Sigma-Aldrich) prepared in ASW antibiotic solution (ASW containing penicillin G, gentamycin and streptomycin) for 100 min at 34 °C to help remove connective tissues and reduce adherence between cells. After rinsing the ganglia with ASW to remove extra protease, bag cell neurons from the abdominal ganglia were isolated manually under the guidance of a Leica MZ 7.5 high-performance stereomicroscope (Leica Microsystems Inc., Bannockburn, IL) and transported into the sample well by micropipette. The cells were cultured overnight at room temperature before the release studies.
Releasate collection
For the spatially resolved ganglion releasate collection, a perfusion method combined with an OSND capillary was used. In order to mimic the delivery of compounds to neurons, we did not use the concentric dual capillary setup but instead inserted a stainless steel tube (305 μm i.d.×140 μm o.d.) connected to a blood vessel of the abdominal ganglion to deliver the secretagogue. Through Teflon tubing, the stainless steel tube was connected to a glass sample vial containing 20 mL of either ASW (control) or KCl (secretagogue) solutions. The entire ganglion was stimulated with KCl and the releasate collected by positioning three OSND capillaries at different areas of the ganglion: the bag cell clusters, right upper quadrant (RUQ) R3–R8 neurons, and left upper quadrant (LUQ) L1–L4 neurons. The cultured neurons were stimulated with 50 mM elevated KCl solution prepared in ASW (the NaCl concentration was reduced accordingly in order to maintain the osmolarity). Insulin as the secretagogue was also prepared in ASW at 5 μM to stimulate the bag cell clusters.
After collecting the releasates, the OSND capillaries were rinsed with 12 μL of 20% MeOH in water to eliminate inorganic salts. The releasates were then eluted with 0.5% FA in 90% MeOH at 1 μL/min and directly spotted onto a MALDI target with the matrix prespotted, 0.25 μL for each spot, for further MALDI-TOF MS analysis. Releasates determined to be stimulation-dependent were distinguished from the chemical background by comparing the mass spectra from samples obtained before and during stimulation. Peptides were identified by their mass-to-charge ratio (m/z) and according to prior extensive in situ, immunohistochemical, MS-based, and tandem MS studies on these specific cells.16,33,34
Results and discussion
Dual-capillary peptide collection
Our goal was to develop a system to allow us to chemically stimulate neurons and measure peptide release from defined regions. This approach is distinct from traditional push-pull perfusion and other dual-tube systems; it allows secretagogue delivery and releasate collection from cultured neurons and brain slices, followed by analyte pretreatments, such as analyte concentration and desalting, prior to MALDI MS characterization. As shown in Figure 1, an outer capillary was used to enclose the collection area/releasing targets, and a concentric inner capillary was used for sample collection. We oftentimes work with ganglia and brain slices that are located within an open extracellular environment such as a tissue located within a dish. Under these conditions, peptides are mixed and thus diluted by orders of magnitude after release without such a physical confinement. Here we adjusted the flow rate ratio so that the incoming solution did not spread to adjacent areas. As a result, a larger sampling (collection) flow rate aided our collection and enhanced the spatial resolution. A ratio of 9 has been used to perform localized cell stimulation at a spatial resolution of ~100 μm.30 As the collection flow rate increases, the releasates are diluted by the greater volume of media, perhaps making detection of trace compounds more difficult. In addition, collection of released material in a larger volume may increase the likelihood of sampling of solutions from adjacent areas, thus raising the possibility of incorrect spatial release profile information. To reduce this possibility, we formed a soft “seal” to the tissue with the outer capillary, thus allowing a more defined collection region. To simplify sample handling, the inner OSND capillary was used to sample the releasates while acting as an extraction media to remove inorganic salts and small molecules; this allowed the collected material to be prepared for direct MS characterization.
The diameters of the outer and inner capillaries were optimized for our specific samples and our mass spectrometer. We selected an outer capillary dimension of 536 i.d. μm × 665 μm o.d., as this is suitable for enclosing a neuronal target (e.g., a neuronal cluster or single cultured neuron) while not being too large to compromise the spatial resolution of the collection. Since the flow is provided by gravity, an adequate difference between capillary diameters is required to allow the solution to flow. An inner capillary of 100 μm i.d.× 170 μm o.d. worked well. By filling the sample reservoir with 0.5 mL of solution and lowering the inner capillary outlet 0 to 90 cm lower than the inlet end, our flow rate was in the range of 0–1.5 μL/min (calculated assuming Poiseuille flow).35
Considering that a typical collection volume of releasates was around 10 μL and the sample reservoir contained 0.5 mL, the flow rate variation caused by the solution volume change at a flow rate of 0.3 μL/min was less than 0.2%; thus, no significant change in solution volume during the stimulation and collection process was expected. One of the advantages of this collection setup is that the inner capillary is replaceable and so collections of different releasate fractions are possible, such as pre-stimulation, stimulation, and perhaps also temporally resolved collection, at specific time points. A new capillary was used for each collection to minimize cross-contamination. The position of the outer capillary was fixed at the beginning of each experiment.
Column fabrication and characterization
An open capillary with a modified inner wall proved to be the most suitable option for our dual-capillary collection system. In prior efforts to create appropriate volume extraction media collection probes, approaches including sol-gel methods,36 silica film coating,37 and polymer monolithic film coating38 have been used to increase the sorbent volume, instead of direct chemical modification applied on the capillary inner wall with the desired functional groups. Nanoparticles have gained wide application in catalysis,39 biological assays,40 and sample pretreatment,41 where the significant increase of the surface area plays a critical role. Therefore, a convenient liquid deposition protocol was used here to introduce nanoparticles into the collection capillary for improving the extraction efficiency of the peptide releasates from the complex extracellular media.
The silica nanoparticles were deposited onto the inner wall of the capillary.31 Figure S1 shows an SEM image of the capillary inner wall before and after the deposition of the silica nanoparticles. The nanoparticle deposition process was repeated to ensure a reasonable coverage on the capillary inner wall while also increasing the roughness of the inner wall. The nanoparticles were measured to be ~60 nm in diameter. Some clustering of the nanoparticles can also be observed, similar to a previous report.31 The silica nanoparticle-deposited capillary was then activated and grafted with a C18 functional group for further peptide collection. In terms of the reproducibility of the deposition process, no obvious differences were found in SEM characterizations of capillaries from different batches of deposition.
Capillary collection efficiency
The extraction conditions when using the OSND capillary for peptides were optimized. The extraction flow rate was 0.3 μL/min, low enough to ensure that the neuronal targets were not disturbed during stimulation. Before extraction, the OSND capillary was equilibrated with methanol and water in sequence. No ion-pair reagent such as TFA was used to equilibrate the capillary, as TFA should not be used in the media when working with cells. After loading the sample, a washing step removed excess salts and thus facilitated the MALDI-TOF MS analysis. We found that even 30 μL of water used for rinsing the capillary did not create an adequately clean MS background. After optimization, we found that a solution of 12 μL of 20% methanol in water removed most of the media-related peaks from the MALDI MS spectra while at the same time, no detectable losses were observed for the collected peptides.
In order to characterize the extraction efficiency of the OSND capillary, we evaluated the collection of six standard peptides mixed in ASW. The eluents from the capillary were dried and redissolved in a loading solution for further CapLC-UV analysis, with the chromatogram shown in Figure S2A. For comparison, a capillary directly modified with octadecyl without the silica nanoparticle deposition was also used for extraction; that chromatogram is shown in Figure S2B. An obvious increase in peak height and peak number in the chromatogram obtained with the OSND capillary confirms that it provides improved extraction efficiency. Not surprisingly, different peptides showed distinct improvements in their ability to be collected. Similar phenomena were observed in extracting small molecules.31
We also evaluated the binding capacity of the OSND capillary using substance P and bombesin prepared in ASW at 3.0 and 2.5 μM, respectively. The binding curves are shown in Figure S3. When the sample volume increased from 5 to 25 μL, the amount of the peptides extracted increased accordingly, and a loading volume of 25 μL did not saturate the capillary. The binding capacity was calculated for substance P and bombesin as 42 and 32 pmol, respectively. When using 25 μL of a mixture sample containing six standard peptides from 2.4 to 3.8 μM, the binding capacity of the peptides extracted by the OSND capillary was calculated to be ~230 pmol by comparing the results to those obtained by direct injection of the peptide standard mixture. The recoveries of the six peptides ranged from 35 to 57%, within the range of microextraction using a coated open-tubular capillary.31,42
During experiments to monitor neuronal release, the OSND capillary used for peptide collection needs to be replaced after each collection. Therefore, the reproducibility of the extraction efficiency of the capillaries fabricated in the same batch, and from different batches, was evaluated. The extraction efficiency of substance P and bombesin from the peptide standard mixture was calibrated. We found that the relative standard deviation (RSD) of the extraction efficiency of substance P and bombesin using capillaries from the same batch was 3.2% and 4.3% (N = 4), respectively; the RSD for capillaries from different batches was 4.7 and 6.5% (N = 9), respectively. Therefore, our OSND capillary fabrication process delivers adequate reproducibility.
Detection limits
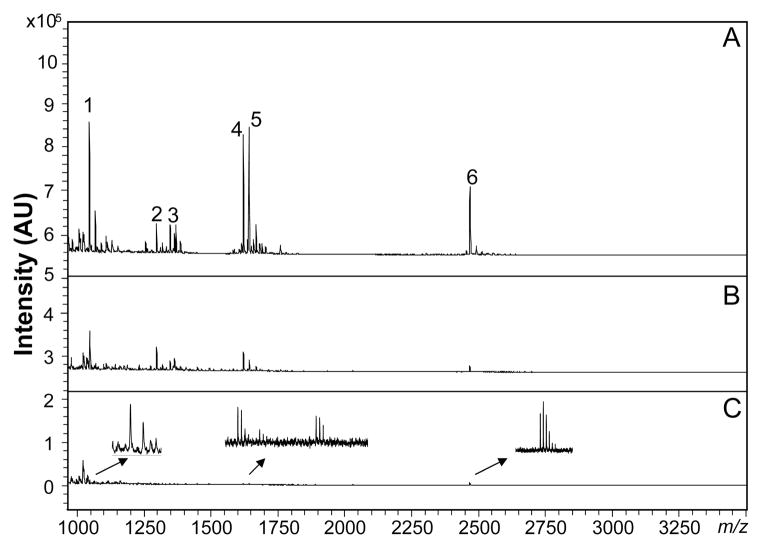
The detection limit of the OSND capillary collection method, followed by MALDI-TOF MS, was determined by extracting consecutively diluted peptide standards prepared in ASW. The resulting MALDI MS spectra can be seen in Figure 2. With a loading volume of 20 μL, angiotensin II at 0.38 nM, substance P at 0.3 nM, bombesin at 0.25 nM, and ACTH(18–29) at 0.35 nM are detectable (Figure 2C). The results were similar to previously reported particle-embedded polymer monolithic columns,17 and are low enough to follow release from individual cultured endocrine cells.
Figure 2.
Limits of detection for peptide standards collected using the OSND capillary, with the peptides consecutively diluted in seawater. Peptide concentrations: (A) 24–38 nM, (B) 2.4–3.8 nM, and (C) 240–380 pM. Peak identities: 1–angiotensin II, 2–angiotensin I, 3–substance P, 4– bombesin, 5–somatostatin, and 6–ACTH (18–39).
Neuronal release assays
In what follows, we highlight several arrangements of peptide collection using the extraction/collection capillary appropriate for different neuronal models, ranging from isolated cultured cells and clusters of cells, to measuring release from defined areas in a semi-intact ganglion preparation. In each case, secretagogue application and peptide collection is optimized for the amounts and dimensions of the cells/regions of interest. These approaches take advantage of the efficient collection, concentration and conditioning of the peptide samples prior to MALDI MS characterization.
Release from individually cultured cells
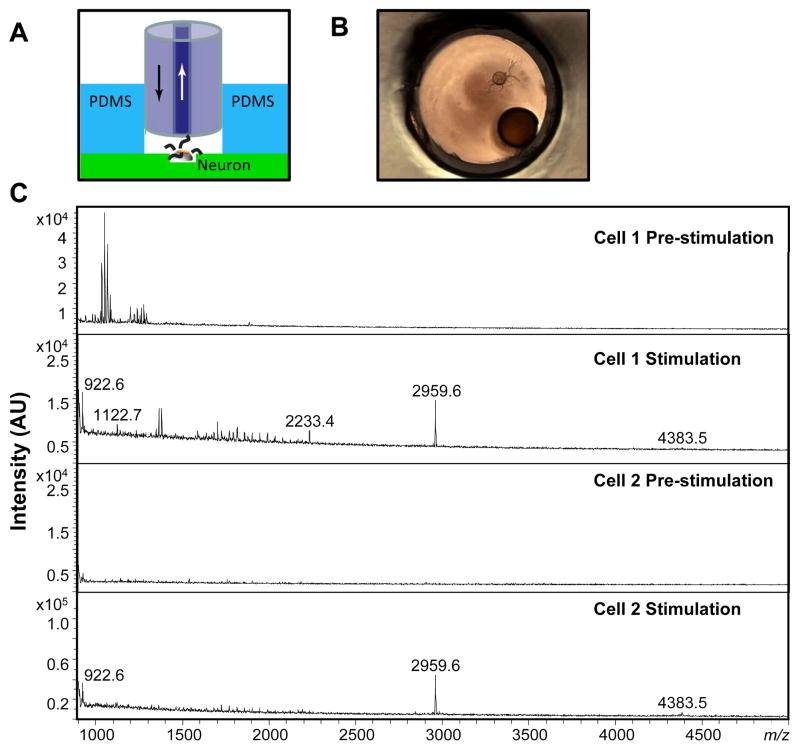
Monitoring the release of peptides from individual neurons helps us understand neuropeptide heterogeneity and even relate neuropeptide release to function and physiology. While we have measured release previously using a particle-embedded polymer monolithic capillary, that work was done by positioning the capillary to as close as possible to the cultured neuron and applying a secretagogue into the bath.17 Known peptides from several neurons were collected and characterized by MALDI MS. However, our collection efficiency was not optimized, leading to analyte losses and poorer detection sensitivity. Using the dual-capillary collection system, a greater fraction of releasate enters the collection capillary and we have better control of the extracellular media near the neuron. To further increase efficiency, we fabricated a sample well by punching holes in a thin film of PDMS having the same hole diameter as the outer diameter of the outer capillary, as can be seen in Figure 3A and B. The purpose is to encapsulate the neuron in a small volume. Bag cell neurons, which have been extensively studied under stimulation and their peptide profiles well-documented,33,34,43–45 were selected to validate the dual-capillary collection method. Using elevated KCl stimulation, releasates from a single bag cell neuron were collected and concentrated by the OSND capillary. Before collection, the outer capillary was inserted into the well, followed by insertion of the collection capillary as close as possible to the target neuron. Releasates from the individual bag cell neuron were collected and characterized by MALDI MS. The spectra of releasates from two individual neurons are shown in Figure 3C; acidic peptide (AP) at m/z 2959.6, and egg-laying hormone (ELH) at m/z 4383.5, were detected for both cells. In addition, α-BCP at m/z 1122.7 and α-BCP(1–7) at m/z 922.6 were observed, providing a more complete profile than previously reported.17
Figure 3.
(A) Schematic illustration of the single-neuron releasate collection approach using the dual-capillary collection system and the neuron sample well. Arrows indicate flow direction. (B) Microscopic view of a dual-capillary collection from a single cultured bag cell neuron in the well. (C) MALDI MS spectra from the releasates from two individual bag cell neurons collected both pre-stimulation (showing few peaks) and during/after KCl stimulation of the neuron showing α-BCP(1–7) at m/z 922.6, α-BCP at m/z 1122.7, AP(1–20) at m/z 2233.4, AP at m/z 2959.6, and ELH at m/z 4383.5.
Release from bag cell clusters
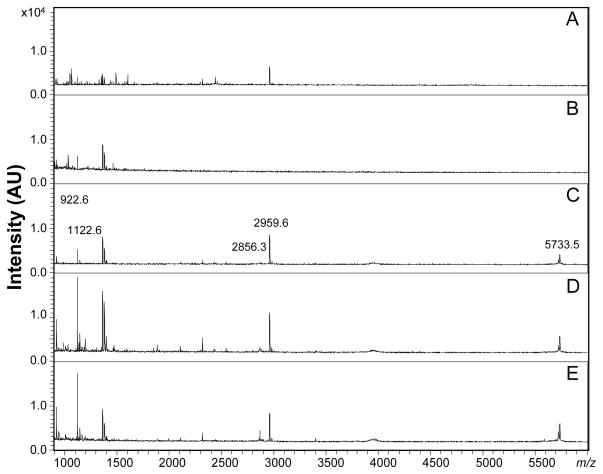
In addition to a selected cell, bag cell neuron clusters (each containing several hundred cells) located at the abdominal ganglion were selected for study. The outer capillary was able to encapsulate the whole cluster; when the end of the outer capillary attaches to the elastomer-filled Petri dish (where the cluster sits), a good seal is obtained. Under stimulation by 50 mM of KCl, the isolated bag cell cluster released known peptides (see Figure S4). In addition to the KCl, the release activity of the isolated bag cell cluster was also monitored under insulin stimulation. Insulin is able to simulate bag cell neurons and cause ELH release via perfusion and radioimmunoassay.46 With our dual-capillary collection approach, assays with different secretagogues can be used by replacing the secretagogue solution in the solution reservoir. As shown in Figure 4, 5 μM of insulin dissolved in ASW was used and the releasates collected and characterized by MALDI MS. We rinsed the apparatus using ASW. Two 30-min portions of efflux were collected, with the pre-stimulation MALDI spectra shown in Figure 4A and B. AP at low intensity was observed in the first 30 min of collection, generated during the placement of the capillary adjacent to the cluster. The second 30-min collection, using a new OSND capillary, showed clear pre-stimulation spectra, without specific peptides detected from the bag cell cluster.
Figure 4.
Bag cell cluster release after exposure to insulin. MALDI MS spectra showing ELH-related peptides, including α-BCP(1–7) observed at m/z 922.6, α-BCP observed at m/z 1122.6, and AP at m/z 2959.6. (A) Pre-stimulation 0–30 min, (B) pre-stimulation 30–60 min, (C) stimulation 0–30 min, (D) stimulation 30–60 min, and (E) stimulation 60–90 min.
Figure 5.
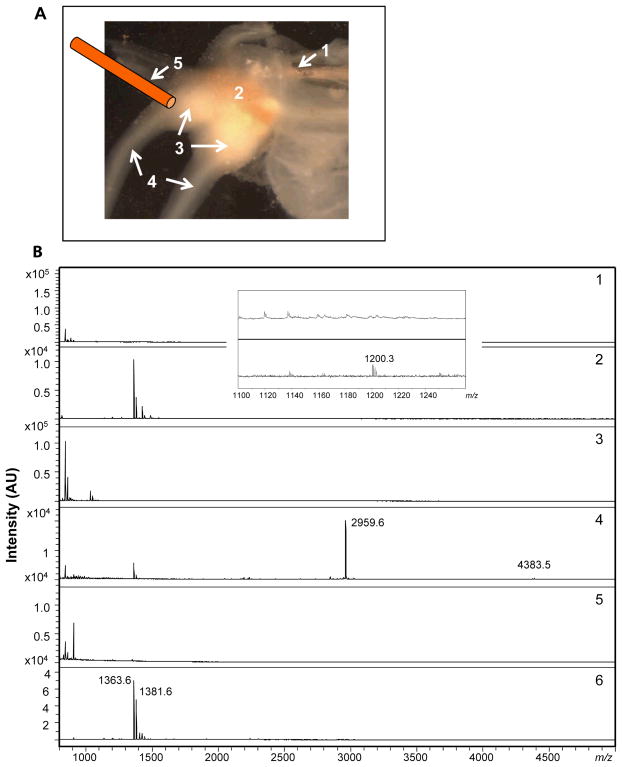
The assay of neuronal releasates collected pre-stimulation (showing few, if any peaks), during and after KCl chemical stimulation of the neurons from different areas of the abdominal ganglion. (A) Photomicrograph and schematic diagram showing the arrangement of perfusion and collection; in order to mimic the delivery of hormones through blood flow, we perfused the secretagogues through (1) a stainless steel capillary inserted into an artery, with (2) showing the abdominal ganglion, (3) the bag cell cluster, (4) the pleuro-abdominal connectives, and (5) the OSND capillary used to collect released compounds. (B) The resulting MALDI MS spectra of the releasates. (1) Left upper quadrant (LUQ) neuron pre-stimulation; (2) LUQ releasates showing the peptide LUQIN at m/z 1200.3. Insert shows a zoomed-in view of the peak at m/z 1200.3. (3) Bag cell cluster pre-stimulation, (4) bag cell cluster releasates with peaks at m/z 2959.6 (AP) and m/z 4383.5 (egg laying hormone, ELH), (5) R3–R8 neurons pre-stimulation, (6) R3–R8 neuron release showing R3–14-related peptides, including those observed at m/z 1363.6 and 1381.6.
After the insulin was applied, we observed bag cell peptide release, including α-BCP(1–7), α-BCP and AP; the peak at m/z 5733.5 was caused by co-extraction of insulin from the efflux and the peak at m/z 2866.3 belongs to insulin2+ (Figure 4C). By replacing the OSND capillary at desired time intervals, limited temporal information can be obtained. Here, two additional collections were made and peptide secretion continued during both collection periods, with the same peptides detected, as can be seen in Figure 4D and E. The secretion duration is found to be similar to a previous report.46
Comparing the two distinct solutions (elevated K and insulin), the same peptides were detected under insulin stimulation, except ELH at m/z 4383.5. The lack of ELH may be due to the lower intensity of this peak or differences in the peptide complement as a function of the stimulation paradigm. Intriguingly, while peptide release is observed after KCl stimulation of a single cultured bag cell neuron, we did not observe peptide release with insulin stimulation from a single bag cell neuron. While this may be because of lower amounts of peptide being released under insulin stimulation compared to KCl stimulation, it may be because the insulin receptors have a cellular distribution in fibers47 distinct from our isolated individual cell preparations. Two other potential secretagogues, glutamate and serotonin, were also tested without detectable signal obtained from either individual cultured bag cell neurons or from intact bag cell clusters.
Release from spatially resolved abdominal ganglion
A useful application of this approach is to collect release from spatially heterogeneous ganglia or brain slices, as peptide release is expected to be different at distinct locations. Our capillary collection system was used to obtain spatially resolved peptide release information from a single abdominal ganglion. Because of the irregular topology of the ganglia, direct contact of the outer capillary with the abdominal ganglion does not always provide a reasonable seal to the area of interest. In addition, the ganglion is covered with a thick sheath material and it takes time for the stimulants to permeate the sheath and reach the neurons underneath. More importantly, many compounds such as hormones are endogenously delivered through the blood vessel and not from direct application on top of the ganglion, and so if one wants to mimic the endogenous delivery approach, blood vessel perfusion is optimal. For these reasons, we modified the collection process; rather than using the concentric dual capillary collection setup to deliver chemicals, we used the blood vessel of the abdominal ganglion to deliver the secretagogues. As shown in Figure 5A, we connected a tube from a sample vial containing either ASW or KCl to the blood vessel entering the abdominal ganglion. After the ganglion was stimulated with KCl, three OSND capillaries were positioned at different releasing areas—the bag cell cluster, RUQ (R3–R14) neurons, and LUQ (L1–L4) neurons—to collect specific peptide release profiles. MALDI MS spectra of the respective releasates are shown in Figure 5B1–6. As can be seen in Figure 5B1 and 2, the LUQ neurons provided a small peak at m/z 1200.3, corresponding to LUQIN peptide.48,49 Whereas for the bag cell cluster, as shown in Figure 5B3 and 4, an intense peak at m/z 2959.6, and a small peak at m/z 4383.5, indicated the release of AP and ELH, respectively. R3–14 peptides exist in R3–R14 neurons at high concentration levels. Figure 5B5 and 6 show the release of RUQ neurons, as the signature R3–14 peptides50,51,52,53 at m/z 1363.6 and 1381.6 can be seen as intense peaks. Comparing the spectra for all of the releasates, we also observed two R3–14 peptides in the releasates from bag cell cluster and LUQ neurons. The R3–14 neurons send out axons into the brachial nerve where they branch so that small processes end in the abdominal sheath.50,54 Therefore, under stimulation of the whole ganglion, we expect that R3–14 peptides will be released from these processes over a wide area and thus be detected in the releasates collected from different neuronal areas.
Conclusions
Our delivery and peptide collection protocols allow us to collect peptide release profiles from defined locations at a ganglia and cluster, and even individual cells from Aplysia californica. In all cases, the OSND collection capillary is used for peptide collection, concentration and desalting to enable the released peptides to be characterized via MALDI-TOF MS over a broad concentration range. A number of stimulation delivery approaches have been highlighted that provide flexibility for a range of applications. Future enhancements will involve approaches for placing capillaries in a sequential series to achieve improved temporal information. This system allows us to examine activity-dependent changes in peptide release, even at the individual neuron level, and hence links information on single cell peptide content with functional information on neuropeptide release. While we have demonstrated the ability of this approach to follow neuropeptide release, its application to neurotransmitters and other small molecules is ongoing. Electrospray MS is well suited for single cell studies,55–57 and so this collection approach, when hyphenated to CE-ESI-MS, will be adaptable for following classical transmitter release.
Supplementary Material
Acknowledgments
We are grateful to Scott Robinson from the Image Technology Group, Beckman Institute at the University of Illinois at Urbana-Champaign for assistance in performing the SEM. The project described was supported by Award No. DA018310 from the National Institute for Drug Abuse, Award No. NS031609 from the National Institute of Neurological Disorders and Stroke, and the National Science Foundation Division of Chemistry under grant CHE-11-11705 (with co-funding from the Division of Biological Infrastructure). The content is solely the responsibility of the authors and does not necessarily represent the official views of the award agencies.
References
- 1.Strand FL, editor. Neuropeptides: Regulators of Physiological Processes. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- 2.Sandman CA, editor. Neuropeptides: Structure and Function in Biology and Behavior. New York Academy of Sciences; New York: 1999. [Google Scholar]
- 3.Perry M, Li Q, Kennedy RT. Anal Chim Acta. 2009;653:1–22. doi: 10.1016/j.aca.2009.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. J Sep Sci. 2008;31:427–445. doi: 10.1002/jssc.200700450. [DOI] [PubMed] [Google Scholar]
- 5.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Zubieta JK, Kennedy RT. Anal Chem. 2009;81:2242–2250. doi: 10.1021/ac802391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Sweedler JV. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 8.Yin P, Hou X, Romanova EV, Sweedler JV. Methods Mol Biol. 2011;789:223–236. doi: 10.1007/978-1-61779-310-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Nat Methods. 2011;8:S20–29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei H, Nolkrantz K, Parkin MC, Chisolm CN, O’Callaghan JP, Kennedy RT. Anal Chem. 2006;78:4342–4351. doi: 10.1021/ac052196x. [DOI] [PubMed] [Google Scholar]
- 11.Croushore CA, Supharoek SA, Lee CY, Jakmunee J, Sweedler JV. Anal Chem. 2012;84:9446–9452. doi: 10.1021/ac302283u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li LJ, Garden RW, Sweedler JV. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- 13.Rubakhin SS, Lanni EJ, Sweedler JV. Curr Opin Biotechnol. 2013;24:95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horning OB, Kjeldsen F, Theodorsen S, Vorm O, Jensen ON. J Proteome Res. 2008;7:3159–3167. doi: 10.1021/pr700865c. [DOI] [PubMed] [Google Scholar]
- 15.Kumazawa T, Hasegawa C, Uchigasaki S, Lee XP, Suzuki O, Sato K. J Chromatogr. 2011;1218:2521–2527. doi: 10.1016/j.chroma.2011.02.070. [DOI] [PubMed] [Google Scholar]
- 16.Hatcher NG, Atkins N, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Proc Natl Acad Sci U S A. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan Y, Rubakhin SS, Sweedler JV. Anal Chem. 2011;83:9557–9563. doi: 10.1021/ac202338e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers RD, Adell A, Lankford MF. Neurosci Biobehav Rev. 1998;22:371–387. doi: 10.1016/s0149-7634(97)00025-0. [DOI] [PubMed] [Google Scholar]
- 19.Gaddum JH. J Physiol (Lond) 1961;155:1p–2p. [Google Scholar]
- 20.Kottegoda S, Shaik I, Shippy SA. J Neurosci Methods. 2002;121:93–101. doi: 10.1016/s0165-0270(02)00245-5. [DOI] [PubMed] [Google Scholar]
- 21.Pritchett JS, Pulido JS, Shippy SA. Anal Chem. 2008;80:5342–5349. doi: 10.1021/ac800238d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamsher AE, Xu H, Guy Y, Sandberg M, Weber SG. Anal Chem. 2010:6370–6376. doi: 10.1021/ac101271r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson EE, Pritchett JS, Shippy SA. Analyst. 2009;134:401–406. doi: 10.1039/b813887g. [DOI] [PubMed] [Google Scholar]
- 24.Slaney TR, Nie J, Hershey ND, Thwar PK, Linderman J, Burns MA, Kennedy RT. Anal Chem. 2011;83:5207–5213. doi: 10.1021/ac2003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz KN, Kennedy RT. Annu Rev Anal Chem. 2008;1:627–661. doi: 10.1146/annurev.anchem.1.031207.113047. [DOI] [PubMed] [Google Scholar]
- 26.Nandia P, Lunte SM. Anal Chim Acta. 2009;651:1–14. doi: 10.1016/j.aca.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D, Du W, Liu Y, Liu W, Kuznetsov A, Mendez FE, Philipson LH, Ismagilov RF. Proc Natl Acad Sci U S A. 2008;105:16843–16848. doi: 10.1073/pnas.0807916105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croushore CA, Sweedler JV. Lab Chip. 2013;13:1666–1676. doi: 10.1039/c3lc41334a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juncker D, SH, Delamarche E. Nat Materials. 2005:622–628. doi: 10.1038/nmat1435. [DOI] [PubMed] [Google Scholar]
- 30.Shiku H, Yamakawa T, Nashimoto Y, Takahashi Y, Torisawa Y, Yasukawa T, Ito-Sasaki T, Yokoo M, Abe H, Kambara H, Matsue T. Anal Biochem. 2009:138–142. doi: 10.1016/j.ab.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 31.Li T, Xu J, Wu JH, Feng YQ. J Chromatogr. 2009:2989–2995. doi: 10.1016/j.chroma.2009.01.076. [DOI] [PubMed] [Google Scholar]
- 32.Chou JS, Lee SC. J Electrochem Soc. 1994;141:3214–3218. [Google Scholar]
- 33.Garden RW, Shippy SA, Li LJ, Moroz TP, Sweedler JV. Proc Natl Acad Sci U S A. 1998;95:3972–3977. doi: 10.1073/pnas.95.7.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatcher NG, Richmond TA, Rubakhin SS, Sweedler JV. Anal Chem. 2005;77:1580–1587. doi: 10.1021/ac0487909. [DOI] [PubMed] [Google Scholar]
- 35.Sutera SP, Skalak R. Annu Rev Fluid Mech. 1993;25:1–19. [Google Scholar]
- 36.Segro SS, Tran MP, Kesani S, Alhendal A, Turner EB, Malik A. J Sep Sci. 2010;33:3075–3096. doi: 10.1002/jssc.201000316. [DOI] [PubMed] [Google Scholar]
- 37.Fan Y, Feng YQ, Shi ZG, Wang JB. Anal Chim Acta. 2005;543:1–8. [Google Scholar]
- 38.Zaidi SA, Cheong WJ. J Chromatogr. 2009;1216:2947–2952. doi: 10.1016/j.chroma.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Astruc D. Nanoparticles and Catalysis. Wiley-VCH Verlag GmbH & Co. KGaA; 2008. [Google Scholar]
- 40.De M, Ghosh PS, RVM 2008;20:4225–4241. [Google Scholar]
- 41.Kaur A, Gupta U. J Mater Chem. 2009;19:8279–8289. [Google Scholar]
- 42.Fan Y, Feng YQ, Shi ZG, Wang JB. Anal Chim Acta. 2005;543:1–8. [Google Scholar]
- 43.Scheller RH, Jackson JF, McAllister LB, Schwartz JH, Kandel ER, Axel R. Cell Biochem Biophys. 1982;28:707–719. doi: 10.1016/0092-8674(82)90050-2. [DOI] [PubMed] [Google Scholar]
- 44.Kupfermann I. Nature. 1967;216:814–815. doi: 10.1038/216814a0. [DOI] [PubMed] [Google Scholar]
- 45.Hatcher NG, Sweedler JV. J Neurophysiol. 2008;99:333–343. doi: 10.1152/jn.00968.2007. [DOI] [PubMed] [Google Scholar]
- 46.Jonas EA, Knox RJ, Smith TCM, Wayne NL, Connor JA, Kaczmarek LK. Nat Materials. 1997;385:343–346. doi: 10.1038/385343a0. [DOI] [PubMed] [Google Scholar]
- 47.Jonas EA, Knox RJ, Kaczmarek LK, Schwartz JH, Solomon DH. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1996;16:1645–1658. doi: 10.1523/JNEUROSCI.16-05-01645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickhan L, DesGroseillers L. DNA Cell Biol. 1991;10:249–258. doi: 10.1089/dna.1991.10.249. [DOI] [PubMed] [Google Scholar]
- 49.Aloyz RS, DesGroseillers L. Peptides. 1995;16:331–338. doi: 10.1016/0196-9781(94)00140-5. [DOI] [PubMed] [Google Scholar]
- 50.Rothman BS, Sigvardt KA, Hawke DH, Brown RO, Shively JE, Mayeri E. Peptides. 1985;6:1113–1118. doi: 10.1016/0196-9781(85)90436-x. [DOI] [PubMed] [Google Scholar]
- 51.Newcomb R, Scheller RH. J Neurosci Methods. 1987;7:854–863. doi: 10.1523/JNEUROSCI.07-03-00854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagle GT, Knock SL, Painter SD, Blankenship JE, Fritz RR, Kurosky A. Peptides. 1989;10:849–857. doi: 10.1016/0196-9781(89)90124-1. [DOI] [PubMed] [Google Scholar]
- 53.Garden RW, Moroz TP, Gleeson JM, Floyd PD, Li L, Rubakhin SS, Sweedler JV. J Neurochem. 1999;72:676–681. doi: 10.1046/j.1471-4159.1999.0720676.x. [DOI] [PubMed] [Google Scholar]
- 54.Price CH, McAdoo DJ. J Comp Neurol. 1979;188:647–677. doi: 10.1002/cne.901880409. [DOI] [PubMed] [Google Scholar]
- 55.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. ACS Chem Neurosci. 2012;3:782–792. doi: 10.1021/cn300100u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Anal Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lapainis T, Rubakhin SS, Sweedler JV. Anal Chem. 2009;81:5858–5864. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.