Abstract
Biomotors have been classified into linear and rotational motors. For 35 years, it has been popularly believed that viral dsDNA-packaging apparatuses are pentameric rotation motors. Recently, a third class of hexameric motor has been found in bacteriophage phi29 that utilizes a mechanism of revolution without rotation, friction, coiling, or torque. This review addresses how packaging motors control dsDNA one-way traffic; how four electropositive layers in the channel interact with the electronegative phosphate backbone to generate four steps in translocating one dsDNA helix; how motors resolve the mismatch between 10.5 bases and 12 connector subunits per cycle of revolution; and how ATP regulates sequential action of motor ATPase. Since motors with all number of subunits can utilize the revolution mechanism, this finding helps resolve puzzles and debates concerning the oligomeric nature of packaging motors in many phage systems. This revolution mechanism helps to solve the undesirable dsDNA supercoiling issue involved in rotation.
Keywords: Bionanomotor, AAA+ ATPase Superfamily, One-way Traffic Mechanism, DNA packaging, Virus Assembly, Nanobiotechnology
Viral DNA packaging motors—a marvelous field with disputes
The importance of nanomotors for cells or nanotechnology is akin to that of mechanical motors to daily life. Mechanical motors power cars to drive us to destinations, and nanobiomotors translocate DNA, RNA, and other cargo to facilitate biological functions. Extensive studies on nanobiomotors have resulted in many fabulous and marvelous findings, but also much wonderment and conjecture, as well as puzzles and mystery, even fervent debates and disputes. Historically, nanobiomotors have been found to use two types of motor mechanisms: linear and rotational (Grigoriev et al., 2004; Vale, 1993). For 35 years, it has been popularly believed that the DNA packaging machines of dsDNA viruses are pentameric rotation motors (Hendrix, 1978; Aathavan et al., 2009; Ding et al., 2011; Moffitt et al., 2009; Caspar and Klug, 1962; Morais et al., 2001; Sun et al., 2008; Simpson et al., 2000). Recently, the bacteriophage phi29 DNA packaging motor has been described as a handicapped pentamer with one non-functional subunit and four rotating subunits (Chistol et al., 2012; Moffitt et al., 2009; Yu et al., 2010). Many other models have also been proposed for the motor action of dsDNA viruses (Guo et al., 1998; Chen and Guo, 1997; Black and Silverman, 1978; Maluf et al., 2006; Moffitt et al., 2009; Aathavan et al., 2009) (for review, see (Guo, 1994; Guo and Lee, 2007; Rao and Feiss, 2008; Zhang et al., 2012; Serwer, 2003)). However, the latest thorough investigation has unexpectedly revealed that the phi29 DNA packaging motor is a hexameric revolution motor that does not rotate (Guo et al., 1998; Shu et al., 2007; Zhang et al., 2012; Schwartz et al., 2013b; Schwartz et al., 2013a; Zhao et al., in press), a discovery disparate to previous models.
In living organisms, a common fundamental process is the transportation of dsDNA from one cellular compartment to another. The AAA+ (ATPases associated with a variety of cellular activities) superfamily includes a class of nanomotors that facilitate a wide range of functions (Zhang and Wigley, 2008; Snider and Houry, 2008; Snider et al., 2008; Ammelburg et al., 2006), many of which are involved in dsDNA riding, tracking, packaging, and translocation that are critical to DNA repair, replication, recombination, chromosome segregation, DNA/RNA transportation, membrane sorting, cellular reorganization, and other processes (Martin et al., 2005; Ammelburg et al., 2006). One common feature of many nanomotors is a hexameric arrangement of subunits (Mueller-Cajar et al., 2011; Wang et al., 2011; Aker et al., 2007; Willows et al., 2004; Chen et al., 2002). Despite their functional diversity, the common characteristic of this family is their ability to convert energy obtained from the binding or hydrolysis of ATP γ-phosphate bond into mechanical force, usually involving a conformational change of the ATPase. The change in conformation generates a gain or a loss of affinity for its substrate inducing a mechanical movement that either makes or breaks contacts between macromolecules. This results in local/global protein unfolding, complex assembly/disassembly, or grabbing/pushing of dsDNA for translocation (McNally et al., 2010; Guenther et al., 1997; Schwartz et al., 2012). The hexagonal shape of the motor facilitates bottom-up assembly in nanomachine manufacturing and produces stable structures, arrangements, and robust machines that may be functionalized in human cells to remedy functional defects that will advance the emerging field of RNA nanotechnology (Guo, 2010; Zhang et al., 2012; Shu et al., 2013a).
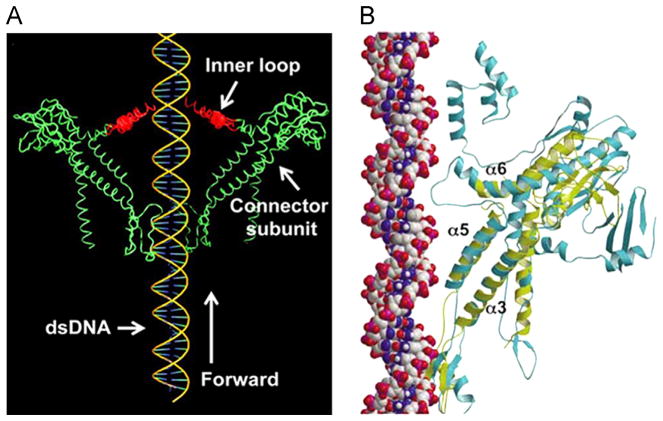
DsDNA (double-stranded DNA) viruses translocate their genomic DNA into preformed protein shells, termed procapsids, during replication (see reviews (Guo and Lee, 2007; Rao and Feiss, 2008; Zhang et al., 2012; Serwer, 2010)). This entropically unfavorable process is accomplished by a nanomotor that also uses ATP as an energy source (Guo et al., 1987c; Chemla et al., 2005; Hwang et al., 1996; Sabanayagam et al., 2007; Lee et al., 2008). The dsDNA packaging motor consists of a protein channel and two packaging molecules that carry out its activities. A discovery 25 years ago showed that the larger molecule serves as part of the ATPase complex, and that the smaller one is responsible for dsDNA binding and cleaving (Guo et al., 1987c); this idea now is well-established for many different viruses. Besides the well-characterized connector channel functioning as a portal (Bazinet and King, 1985), the motor of phage phi29 involves an ATPase protein gp16 (Fig. 1) (Guo et al., 1987c; Guo et al., 1987b; Huang and Guo, 2003a; Huang and Guo, 2003b; Lee and Guo, 2006; Lee et al., 2008; Ibarra et al., 2001; Grimes and Anderson, 1990) and a hexameric packaging RNA ring (Guo et al., 1987a; Guo et al., 1998; Shu et al., 2007; Zhang et al., in press). The motor connector portal contains a center channel created by 12 circling copies of the protein gp10 that provides a pathway for dsDNA translocation (Jimenez et al., 1986; Guasch et al., 2002; Badasso et al., 2000).
Fig. 1.
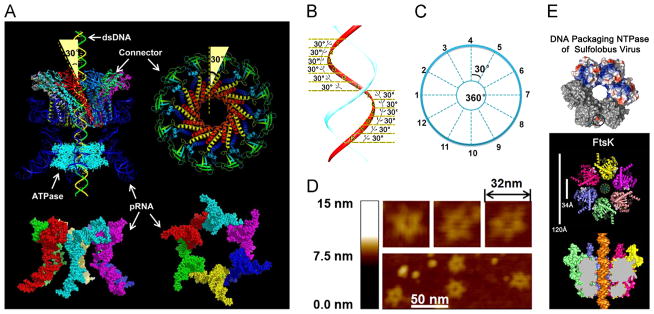
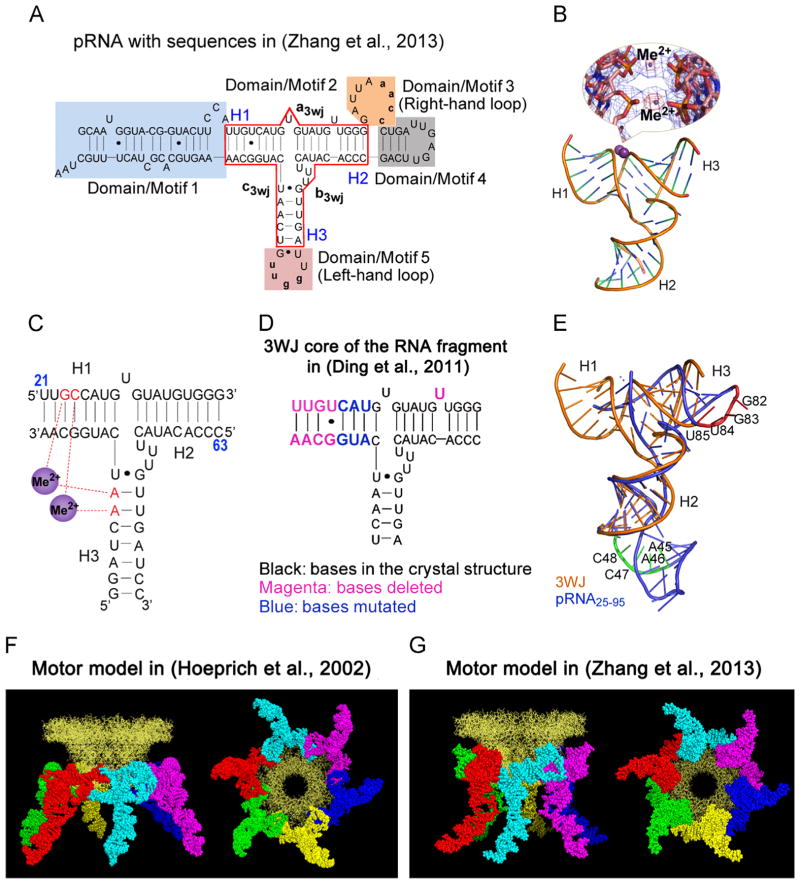
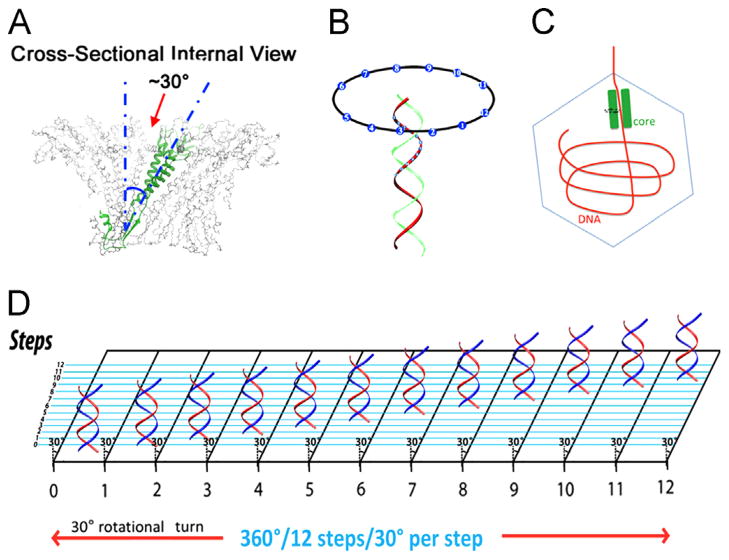
Illustration of the phi29 DNA-packaging motor hexameric structure. (A) Side view (left) and top view (right) of the phi29 packaging motor and pRNA hexamer. The 30° tilt of the helix of the channel subunit is depicted in both external and internal views. (Bottom) Side and bottom views of hexameric pRNA derived from 3WJ crystal structures (Zhang et al., in press). (B, C) The contact at every 30° for twelve 30° transitions translocates dsDNA one helical turn through the connector. (D) AFM images of pRNA hexamer. (E) Hexameric motor models of helicase FtsK and B204 NTPase. (A) and (C), adapted from Zhao et al. (2013), © 2013 with permission from American Chemical Society; (B) and (D), adapted from Schwartz et al. (2013b), © 2013 with permission from Elsevier; (E) upper panel, adapted from Happonen et al. (2013), © 2013 with permission from American Society for Microbiology, and lower panel, adapted from Massey et al. (2006), © 2006 with permission from Elsevier.
Cellular proteins that show a strong similarity to the phi29 viral DNA packaging motor include FtsK, a hexameric motor that transports DNA and separates intertwined chromosomes during cell division, and members of the SpoIIIE family (Lowe et al., 2008; Demarre et al., 2013; Barre, 2007), other hexamers responsible for transportation of DNA from mother cell to pre-spore during Bacillus subtilis cell division (Bath et al., 2000). FtsK contains three components: one for DNA translocation, one for orientation control, and one for anchoring to the substrate. Extensive studies suggest that FtsK employs a “rotary inchworm” mechanism to transport DNA. During each cycle of ATP binding and hydrolysis within each FtsK subunit, one motif acts to tightly bind to the helix while the other progresses forward along the dsDNA. This process causes translational movement and is repeated by handing off of the helix to an adjacent subunit (Massey et al., 2006).
Many mysteries have been encountered during the course of study of dsDNA translocation motors. It was found that the FtsK protein transport closed circular dsDNA without covalent bond breakage or DNA topology changes (Grainge, 2013; Demarre et al., 2013), and other motors were found to operate as a one-way traffic machine to control DNA movement, despite the intrinsic symmetry of the double-stranded DNA helix, and some allow dsDNA to cross cell membranes without affecting the hydrophobic or hydrophilic nature of the membrane (Demarre et al., 2013). Some motor channel walls display a swirl or pinwheel shaped structure, but no physical rotation components have been confirmed. This mini-review focuses on addressing the following puzzles, primarily focusing on the phi29 packaging motor: 1. Is the motor pRNA and ATPase a hexamer or a pentamer? 2. How can the revolution motors transport a DNA helix with such an unusual strong force without rotating, coiling, or torsion? 3. How does the motor execute unidirectional transportation? 4. Do positively charged amino acids facilitate movement of negatively charged dsDNA, and how does the motor resolve the mismatch between a dsDNA helix of 10.5 bases/360° and the 12 subunit connector? 5. Why, in certain cases, does the motor display physical motion without ATP hydrolysis?
Puzzle 1. is the motor pRNA a hexamer or a pentamer?
In 1987, an RNA component was discovered in the phi29 DNA packaging motor (Guo et al., 1987a) and the RNA was shown to be a hexameric ring (Guo et al., 1998; Zhang et al., 1998). AFM imaging and an RNA crystal structure at 3.05 Å resolution confirmed that phi29 pRNA form a hexamer ring on the DNA packaging motor (Fig. 1) (Zhang et al., in press; Schwartz et al., 2013a). These findings end the long-term debate regarding whether the pRNA is a pentamer or a hexamer. This part of the review serves to comment on why a hexamer has also been reported as a pentamer to shed light on future structural studies of RNA.
Why has the hexameric pRNA structure been reported as a pentamer in other crystal structure computation?
Phi29 pRNA contain a three-way junction (3WJ) core that displays an unusual thermodynamic stability (Shu et al., 2011). This core has a primary role in driving the folding of the entire pRNA to produce its mature global structure. The thermodynamically stable pRNA 3WJ scaffold can be assembled from three RNA strands; and various functional small RNA motifs or fragments, when fused to the three arms of this pRNA 3WJ, fold into their authentic structures (Shu et al., 2011; Haque et al., 2012; Reif et al., 2013; Shu et al., 2013b) in vitro and in vivo, supporting the assumption that the fragments used to make the pRNA 3WJ core adopts the same structure as within the full pRNA (Shu et al., 2011). The 3WJ displays an unusual thermodynamic stability, is stable in serum, remains intact at ultra-low concentrations, and is resistant to 8 M urea. The crystal structure by Zhang et al. revealed two divalent metal ions that coordinate four nucleotides of the pRNA 3WJ (Fig. 2). The global structure of the resulting pRNA/connector complex is similar to a hexamer model derived from biochemical and biophysical data (Hoeprich and Guo, 2002). However, different from the authentically folded RNA of Zhang et al., in the RNA crystal structure by Ding et al. (2011) the RNA is different, likely because two of the four nucleotides responsible for the coordination of Mg2+ are missing. In addition, several nucleotides in the 3WJ core region and the procapsid binding domain were mutated (Fig. 2D). These differences result in an RNA structure that is different from the pRNA folding driven by the 3WJ core. Our results suggest that an RNA core with the lowest Gibbs free energy (ΔG) is critical for RNA folding, and alteration of nucleotides in the core region will change the RNA global folding. It is important to note that the pRNA model derived from computational and biochemical methods 11 years ago (Fig. 2F) is very close to the most recent one derived from the crystal structure (Fig. 2G).
Fig. 2.
Comparison of the two RNA molecules used in crystallization to produce hexamer and pentamer. (A) The sequence of the hexameric pRNA used in (Zhang et al., in press). (B) Crystal structure of pRNA 3WJ with two metal binding sites (Me2+, magenta), shown as sphere. A close-up of the metal binding sites superposed on the 2Fo-Fc electron density map (blue mesh contoured at 1.0 σ) and the anomalous difference map (red mesh contoured at 4.5 σ) are shown in the inset. (C) Schematic representation of the 3WJ structure with metal coordinating nucleotides in red. Numbers in blue represent the nucleotide locations in the wild-type pRNA sequence. (D) The 3WJ core in the RNA fragment sequence used in (Ding et al., 2011), the mutated and deleted bases are shown in different colors as indicated. (E) Superposition of the pRNA25–95 (Ding et al., 2011) (blue) and 3WJ domain (Zhang et al., in press) (gold) crystal structures with the left- (red) and right-hand (green) loops highlighted. The motor model with the pRNA derived from computational and biochemical methods in (Hoeprich and Guo, 2002) (F) and the one derived from crystallization in (Zhang et al., in press) (G) are shown as side view (left) and bottom view (right). (A)–(E) and (G), adapted from Zhang et al. (in press), © 2013 with permission from Cold Spring Harbor Laboratory Press; (F), adapted from Hoeprich and Guo, (2002), © 2002 with permission from The American Society for Biochemistry and Molecular Biology. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Why has cryo-EM imaging of the same pRNA complex resulted in a hexamer structure in one lab and a pentamer in another lab?
Using similar cryo-EM approaches for reconstructions, both hexameric (Ibarra et al., 2000) and pentameric (Morais et al., 2001; Simpson et al., 2000) pRNA images have been reported. The possible cause for this discrepancy could be due to the special property of RNA that differs from proteins. Application of cryo-EM for RNA structural studies remains challenging due to the sensitivity of RNA to RNase degradation during sample preparation, as well as structural flexibility resulting from different energy landscape in RNA folding (Hofacker et al., 2010; Li et al., 2007). Cryo-EM reconstructions depend on computation to average the copy number and structure of the complex. Degradation by RNase can result in an apparent lower copy number of pRNA per procapsid than found in vivo. RNase degraded during single molecule photobleaching studies resulting in an underestimation of the copy number (Shu et al., 2007), and a hexamer perceived as pentamer (Chistol et al., 2012; Yu et al., 2010; Morais et al., 2008; Comolli et al., 2008). Different environments can also make the computation averaging of RNA structure difficult because of the structural flexibility that RNA gains from different energy landscapes.
The presence of hexameric folds in the motor have been revealed by biochemical, structural studies, nano-fabrication approaches (Guo et al., 1998; Zhang et al., 1998; Hendrix, 1998; Bourassa and Major, 2002; Shu et al., 2007; Xiao et al., 2008; Moll and Guo, 2007; Ibarra et al., 2000; Xiao et al., 2010; Zhang et al., in press; Fang et al., 2005), and activity assays (Chen et al., 1999; Chen et al., 2000). Single molecule imaging revealed that a fully active ring can be formed by either a pure dimer or pure trimer (Xiao et al., 2008). The least common multiple of two and three is six, a stoichiometry comparable to the phi29 motor portal vertex that contains 12 subunits (Badasso et al., 2000; Guasch et al., 1998). Proponents of a pentameric or hexameric pRNA disagree on important aspects of phage and motor structure. Hexamer supporters believe that pRNA binds to the dodecameric connector, a shape that exhibits 6-fold symmetry. Pentamer supporters suggest that pRNA binds the 5-fold procapsid shell, but cross-linking approaches have shown that pRNA binds the connector protein gp10 and not the procapsid protein (Garver and Guo, 2000; Garver and Guo, 1997). Furthermore, pRNA binds to three basic amino acids, Arg–Lys–Arg, near the gp10 N-terminus (Xiao et al., 2005; Atz et al., 2007). Proponents of a pentamer have also suggested that pRNA hexamers are formed initially, but after binding, one monomer dissociates from the procapsid due to a conformational change and leaves behind a bound pentamer (Morais et al., 2008; Morais et al., 2001; Ding et al., 2011). However, single molecule photobleaching assays show that an active motor still contains six copies of pRNA (Shu et al., 2007).
Fig. 6.
DNA revolves and transports through 30° tilted connector subunits. Translocation is facilitated by the anti-parallel arrangement between right-handed dsDNA helices and the left-handed 30° tilted connector channel (A, B). (C) The offset of tilted T7 dsDNA core around the DNA packaging channel revealed by cryo-EM. When viewed from top to bottom in this illustration, the core stack will process counterclockwise. (D) A planar view showing how DNA advances and travels along the circular wall of the connector channel with no torsion or coiling force and continues through the connector channel, touching each subunit, translocating 12 discrete steps at 30° integrals per subunit. (A), (B) and (D), adapted from Schwartz et al. (2013b), © 2013 with permission from Elsevier; (C), adapted from Guo et al. (2013), © 2013 with permission from National Academy of Sciences.
Puzzle 2: is the motor ATPase a hexamer or a pentamer?
Many AAA+ superfamily members are found to be hexamers (Mueller-Cajar et al., 2011; Wang et al., 2011; Aker et al., 2007; Willows et al., 2004; Chen et al., 2002; Happonen et al., 2013; Snider et al., 2008), some are assembled from three dimers (Sim et al., 2008; Skordalakes and Berger, 2006; Ziegelin et al., 2003). However, there is disagreement as to the oligomeric state of the phi29 DNA packaging motor. Phi29 gp16 ATPase was reported to be a pentamer by cryo-EM (Morais et al., 2008), but single molecule packaging experiments have led to the contradictory finding that four steps, rather than the expected five, of motor action are sufficient to package a single helical turn of DNA (Chistol et al., 2012; Moffitt et al., 2009) (Fig. 3). In contrast, multiple approaches have concluded gp16 to be hexameric, including native gel shift assays, capillary electrophoresis (CE), Hill constant determination, and titration of mutant subunits using computational binomial distribution (Schwartz et al., 2013a). All these assays revealed that assembly of gp16 followed a monomer→dimer→tetramer→hexamer pathway.
Fig. 3.
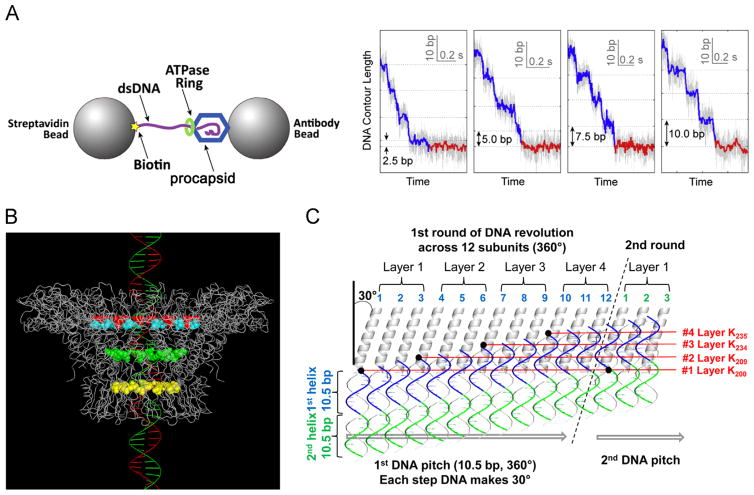
Single-molecule experiment revealing the four pauses during DNA packaging and the illustration of the effect of lysine layers resulting in the pauses. (A) Design and data showing four transition steps of 2.5 bp, 5.0 bp, 7.5 bp, and 10 bp. (B) Structure of the phi29 DNA packaging motor showing the four lysine rings lining the inner wall of the phi29 connector channel: lysine rings K200 (yellow) and K209 (green), with 229 (cyan) and 246 (red) representing the boundary of the connector inner flexible loops that harboring the other two lysine rings K234 and K235. (C) A detailed schema of DNA revolution through the connector channel showing that the negatively-charged phosphate backbone of dsDNA contacts the four lysine layers leading to 4 discrete pauses during packaging of 10.5 bp of genomic dsDNA. See text for details. (A), adapted from Chistol et al. (2012), © 2012 with permission from Elsevier; (B) and (C), adapted from Zhao et al. (2013), © 2013 with permission from American Chemical Society. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Natural DNA or RNA translocation motors contain P-loop NTPase components for contacting DNA or RNA that mostly display a hexameric configuration. Structural computation for P-loop ATPases identified a FtsK–HerA superfamily that is predominantly involved in translocation of DNA and peptides through membrane pores (Iyer et al., 2004; Burroughs et al., 2007). Crystal structures revealed that members of this subfamily, such as FtsK and SpoIIIE, display a hexameric arrangement (Lowe et al., 2008; Demarre et al., 2013; Barre, 2007; Bath et al., 2000; Massey et al., 2006) (Fig. 1E). Structural and sequence computations have led to the conclusion that the DNA-packaging ATPases of various dsDNA viruses belong to this hexameric FtsK–HerA superfamily (Iyer et al., 2004; Burroughs et al., 2007). Crystal structures of the DNA packaging motors from Sulfolobus virus B204 (Happonen et al., 2013) reveal a hexameric structure. These studies lead to a conclusion that all ATPase of viral DNA packaging motors assemble into hexameric but not pentameric structure.
Puzzle 3: how can the revolution motors transport a DNA helix with such an unusual strong force without rotating, coiling, or torsion?
Packaging has been found to proceed even when the connector is covalently linked to the capsid protein of the procapsid (Baumann et al., 2006; Maluf and Feiss, 2006) (Fig. 4). Single-molecule force spectroscopy has demonstrated that the connector does not rotate (Hugel et al., 2007). Since the phi29 connector does not rotate, there is no reason to believe that the ATPase motor rotates because the gp16 ring is tightly bound to the pRNA ring (Lee and Guo, 2006) that is immobilized at the external end of the stationary connector. Single molecule studies also revealed that when the end of the phi29 dsDNA was tethered to the beads, the motor was still active in packaging (Shu et al., 2007; Chang et al., 2008), indicating that dsDNA does not rotate during packaging. If the motor components do not rotate, then what is the mechanism of a motor that translocates a dsDNA helix? Unidirectional movement by a “push through a one-way valve” model was recently proposed (Schwartz et al., 2012; Fang et al., 2012; Zhao et al., 2013; Zhang et al., 2012; Jing et al., 2010) and solves this puzzle. Pushing DNA through the connector channel by the ATPase is entropically unfavorable, requiring the connector to function like a valve to prevent DNA from slipping out during DNA packaging (Black, 1989; Casjens, 2011; Guo and Lee, 2007). This model is compatible with the previously proposed ratchet (Serwer, 2003) and compression (Ray et al., 2010b; Ray et al., 2010a; Dixit et al., 2012) models of DNA packaging, but cannot be easily reconciled with the T4 gp17 ATPase structure (Sun et al., 2008). The recent findings of a revolution without rotation mechanism (Zhao et al., 2013; Schwartz et al., 2013b; Schwartz et al., 2013a) solves many puzzles pondered for many years.
Fig. 4.
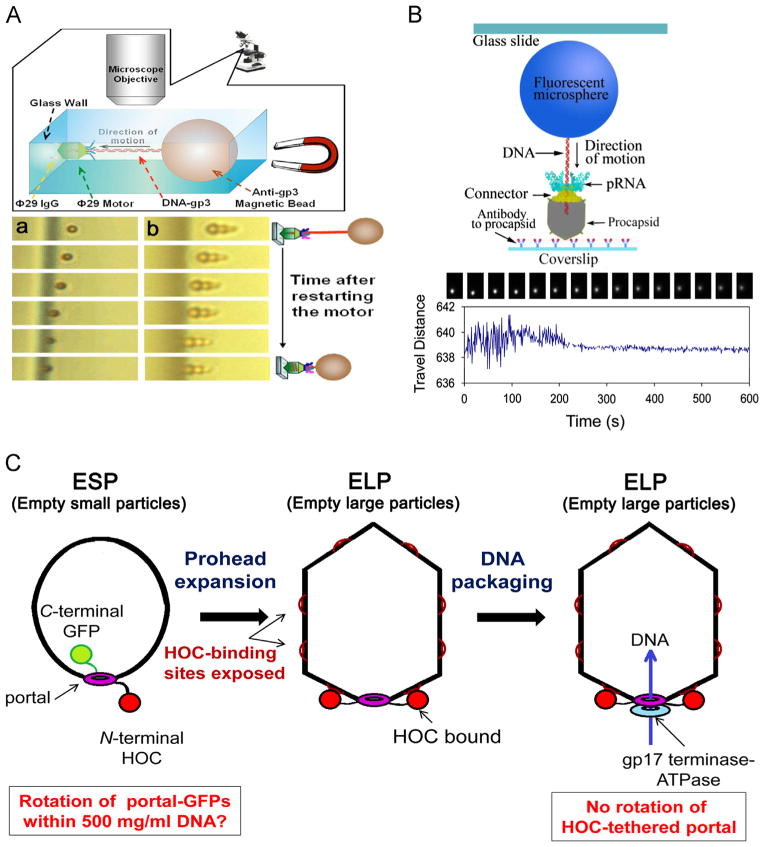
Data showing that no DNA (A and B) or connector (C) rotates during active packaging of viral DNA. (A) Design (upper) and temporal micrographs (lower) of tethered DNA being packaged. The phi29 genome ends are covalently bound to the terminal protein gp3. Only one bead is bound to DNA in (a), while a cluster of beads are bound in (b). (B) Experimental design and sequential images of the direct observation of DNA packaging. Both (A) and (B) show that the motor is still active in packaging with the end of the DNA tethered with beads. (C) The T4 portal does not rotate during packaging is shown by normal DNA packaging into T4 phage heads with half the portal dodecamers assembled as C-terminal GFP fusion proteins, and with N-terminal portal fusions to T4 Hoc (Highly antigenic outer capsid binding protein) tethered to its protease immune binding site on the expanded capsid without blocking packaging in vitro or in vivo. (A), adapted from Chang et al. (2008), © 2008 with permission from American Institute of Physics; (B), adapted from Shu et al. (2007), © 2007 with permission from Nature Publishing Group; (C), adapted from Baumann et al. (2006), © 2006 with permission from John Wiley and Sons.
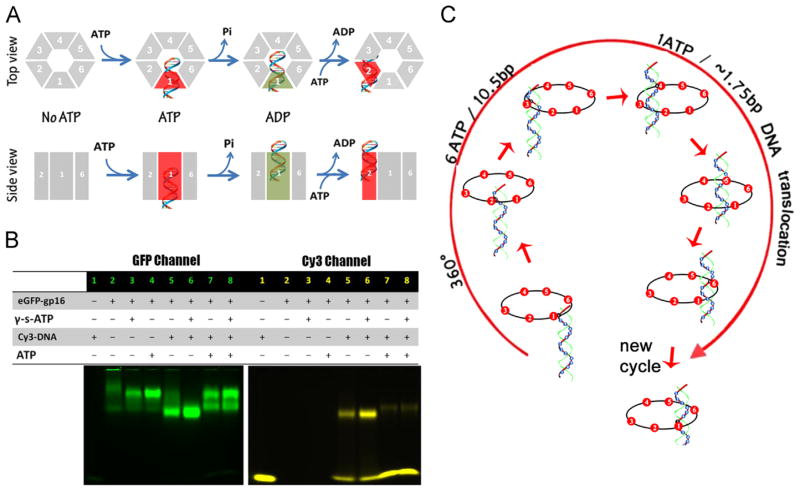
It has long been revealed that the motor subunits work sequentially (Chen and Guo, 1997). The phi29 packaging motor is one of the strongest biomotors, providing forces up to 57 pN (Smith et al., 2001). The phi29 motor contains six copies of the ATPase gp16 (Puzzle 2), and the binding of ATP to one ATPase subunit stimulates it to adopt a conformational change with an associated 40-fold enhancement in affinity for dsDNA (Schwartz et al., 2013b). Following ATP hydrolysis, the ATPase subunit assumes a new conformation with higher entropy, but lower affinity for dsDNA, thereby pushing dsDNA away and transferring it to an adjacent subunit (Schwartz et al., 2013b; Schwartz et al., 2013a). Communication from one subunit to the adjacent subunit is mediated by Arginine Finger domain (unpublished data) where the Arginine Finger of the subunit will turn to the adjacent subunit after the hydrolysis of ATP and the release of DNA to promote another round of ATP hydrolysis. DNA revolves unidirectionally around both the channel wall of the connector and the gp16 ATPase, but neither the dsDNA nor the ATPase rotates. It has been shown that the binding of only one bound ATP is sufficient to induce strong DNA-binding as one ATP is hydrolyzed in each step and six ATPs are consumed in one cycle to translocate the dsDNA through one helical turn. Sequential binding of the same phosphodiester backbone strand to the hexameric ATPase requires dsDNA to move 1.8 bp (base pairs) each step (10.5 bp per turn ÷ 6 ATP=1.8 bp/ATP) (Fig. 5). This agrees with the empirical data demonstrating that one ATP is used to package 2 bp (Guo et al., 1987c) or 1.8 bp (Morita et al., 1993) of DNA. The cooperativity and sequential action among hexameric ATPase subunits (Chen and Guo, 1997; Moffitt et al., 2009) also promotes the revolution of dsDNA around the connector channel (Fig. 6). The contact between the connector and the dsDNA chain is transferred from one point on the phosphate backbone to another on the same strand in a 5′ to 3′ direction (Moffitt et al., 2009; Aathavan et al., 2009; Zhao et al., 2013).
Fig. 5.
Mechanism of sequential revolution in translocating genomic dsDNA within ATPase ring. (A) Binding of ATP to one gp16 subunit stimulates it to adapt a conformation with higher affinity for dsDNA. ATP hydrolysis forces gp16 to assume a new conformation with lower affinity for dsDNA, thus pushing dsDNA away from this subunit and transferring it to an adjacent subunit. (B) Gel images showing interaction of eGFP-gp16 with 40bp Cy3-dsDNA in the presence or absence of ATP or γ-S-ATP. (C) The revolution of dsDNA along the ATPase hexameric ring. (A) and (C), adapted from Schwartz et al. (2013b), © 2013 with permission from Elsevier; (B), adapted from Schwartz et al. (2013a), © 2013 with permission from Elsevier.
The anti-parallel arrangement between the dsDNA helix and the channel subunits of the connector dodecamer (Fig. 6) also contributes to the revolution of DNA without rotating, coiling, or torsional strain. The connector helices that interact with DNA are tilted as 30° (Jing et al., 2010; Zhao et al., 2013; Schwartz et al., 2013b), relative to the long axis of the dsDNA. The dsDNA shifts +30° each time it goes through one of the 12 subunits (360° ÷ 12=30°), is perfectly compensated by the −30° tilting of the helices lining the connector channel wall, and results in a net change of “0°”. The “+” and the “−” result from the anti-parallel arrangement of DNA and connector helices. This model explains how the motor can transport dsDNA without involving rotation, coiling, or a torsional force. Revolution mechanism might reconcile the stoichiometric inconsistency among many bacteriophages whose ATPases have been reported to be tetrameric (Chang et al., 2012; Medina et al., 2011; Fuller et al., 2007; Ortega and Catalano, 2006), pentameric (see above), hexameric (Guo et al., 1998; Zhang et al., 1998; Hendrix, 1998; Shu et al., 2007; Xiao et al., 2008; Moll and Guo, 2007; Shu et al., 2007; Xiao et al., 2010; Zhang et al., 2012), and nonameric (Roy et al., 2011).
The viral DNA packaging motor rotation mechanism has been proposed long time ago (Hendrix, 1978). However, the long quest for motor mechanism to overcome the super coiling issue of the lengthy DNA during packaging has been in vain. Fortunately, motors with any number of subunits can utilize this revolution mechanism that solves the supercoiling issue regarding the transportation of dsDNA helices. During the revolution of dsDNA through the channel, dsDNA will advance by touching the channel wall instead of proceeding through the center of the channel (Guo et al., 2013). This is in agreement with the recent finding by cryo-EM-imaging showing that the T7 dsDNA core is tilted from its central axis (Fig. 6C). The clear pattern indicates that the DNA core stack does have a small offset and tilt that is only detected clearly when the gp15 layer of the core stack is truly resolved (Guo et al., 2013). The authors also found evidence of a counterclockwise motion of the dsDNA viewed from the T7 top of the connector, as observed from the N-terminus shown in fig. 6C (Guo et al., 2013). This is in agreement with the clockwise revolution of dsDNA viewed from the wider end to the narrower end of the phi29 connector, as observed from the C-terminus (Zhao et al., 2013; Zhang et al., 2012; Jing et al., 2010; Schwartz et al., 2013b).
Puzzle 4: how does the motor execute unidirectional transportation?
The phi29 motor has been shown to use a “push through one-way valve” mechanism to translocate dsDNA (Jing et al., 2010; Zhang et al., 2012). The direction of translocation is controlled by five actions (Schwartz et al., 2013; Zhao et al., 2013; Fang et al., 2012): (1) motor ATPase undergoing a series of entropy transition and conformational changes when binding ATP or dsDNA; (2) the 30° angle of each subunit of the dodecameric connector channel, that run anti-parallel to the dsDNA helix, facilitating the one-way traffic of the dsDNA and coinciding with the 12 subunits of the connector; (3) internal channel loops of the connector protein serving as a ratchet valve to prevent dsDNA reversal; (4) the 5′→3′ movement of one strand of dsDNA through the phi29 motor connector channel wall ensuring unidirectional movement of the DNA; and (5) four lysine layers interacting with the DNA phosphate backbone, resulting in four steps of forward transition and pausing during DNA translocation.
The anti-parallel arrangement between the α-helices and the dsDNA argues against a nut and bolt rotation model, since the corresponding whorl would display a parallel arrangement in this scenario. A mutant connector in which the internal loop was deleted allowed packaged dsDNA to reverse direction and slip out of the mutant procapsid (Fang et al., 2012; Geng et al., 2011; Grimes et al., 2011; Serwer, 2010; Isidro et al., 2004), lending support to the theory that the channel loops may act as a ratchet, preventing packaged DNA from leaking (Fig. 7) and supporting the “push through one-way valve” model (Fang et al., 2012). When the internal loops were deleted, the one way channel became two-way, allowing both ssRNA or ssDNA to pass through (Zhao et al., 2013; Geng et al., 2013).
Fig. 7.
The role of the flexible inner channel loop in DNA one-way traffic. Illustration of the ratchet model in phi29 bacteriophage (A) and the superimposition of the connector subunit of SPP1 (cyan) and phi29 (yellow) (B). The conservation of the three-helical substructure (α3, α5 and α6) in the portal proteins of bacteriophages SPP1 and phi29 strongly suggest that this substructure is important for DNA translocation. (A), adapted from Zhao et al. (2013), © 2013 with permission from American Chemical Society; (B), adapted from Lebedev et al. (2007), © 2007 with permission from Nature Publishing Group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Gaps created in single-stranded Phi29 and T4 DNA have been found to halt packaging (Moll and Guo, 2005; Oram et al., 2008). A 3′ end extension with a 12-base overhang had little effect, but a 20 base extension blocked T4 DNA packaging (Oram et al., 2008). Chemical modification of the phosphate backbone of the 3′-5′ strand did not inhibit DNA translocation, and modification of 10 bases of the 5′-3′ strand being packaged was found to be tolerated, but a change of 11 bases of the 5′-3′ strand resulted in severe reduction in packaging activity (Aathavan et al., 2009). These results support the notions that the motor can complete one turn of 360° with single-stranded DNA and that DNA revolves along the motor using the single strand of the dsDNA that is packaged in the 5′→3′ direction. If the connector is a one-way valve that does not allow dsDNA to reverse direction during DNA packaging, how does dsDNA come out of the capsid from the one-way-inward motor to enter the host cell? Significant conformational changes in the motor channel occur after genomic dsDNA been packaged, as evidenced in different bacteriophages including SPP1 (Cuervo et al., 2007; Lhuillier et al., 2009), phi29 (Geng et al., 2011; Tao et al., 1998), λ (Petrov and Harvey, 2011), and T7 (Hu et al., 2013; Kemp et al., 2004; Chang et al., 2010). These conformational changes will alter the one-way inward property of the motor channel and allow ejection of the genome during infection (Molineux and Panja, 2013).
Puzzle 5: do positively charged amino acids facilitate movement of negatively charged dsDNA, and how does the motor resolve the mismatch between a dsDNA helix of 10.5 bases/360° and the 12 subunit connector?(Fig. 3)
The negatively charged phi29 connector interior channel surface is decorated with four rings of positively charged lysine residues that have been proposed to have a role in DNA translocation (Guasch et al., 2002; Badasso et al., 2000). The mismatch between 10.5 bp and 12 subunits per turn suggest that there are only four perfect contacts for the positively charged lysine to meet the negatively charge phosphate at each cycle (Fig. 3B). The transition from a mismatch to four static contacts will generate four steps of change in DNA packaging speed. Examination of the phi29 crystal structure reveals that the length of the connector channel is 7 nm and the four lysine layers span half the channel length, ~3.5 nm. Thus, the average distance between each lysine layer is about ~0.9 nm (Fig. 3B). Since the axial rise of each bp of B DNA is about 0.34 nm, 2.6 bp is the necessary step size to cross the lysine layer (0.9 ÷ 0.34=2.6), and four steps of transition will occur during packaging one helical turn (Schwartz et al., 2013a; Schwartz et al., 2013b; Zhao et al., 2013). This number agrees with the finding of four discrete steps per cycle and translocation of 2.5 bp per step in single molecule studies using optical tweezers (Fig. 3A).
The 2.6 bp per lysine layer agrees with calculations those based on the pitch size and connector stoichiometry. During DNA revolution, the negatively charged phosphate backbone of dsDNA makes contact with the positively charged lysine layer located inside the channel (Guasch et al., 2002; Guasch et al., 2002). A 360° revolution equals one pitch of dsDNA with 10.5 bp, and one revolution cycle goes around 12 connector subunits. Thus, 0.875 bp will pass one connector subunit (10.5 bp per cycle). On average, each of the four lysine layers is responsible for contact with three subunits (12 subunits ÷ 4 layers=3 subunits). Thus, for each lysine layer or three connector subunits, 2.6 bp will translocate through the connector (Fig. 3C) (Schwartz et al., 2013a; Schwartz et al., 2013b; Zhao et al., 2013). This number also agrees with the finding of four discrete steps per cycle and translocation of 2.5 bp per step in single molecule studies using optical tweezers, although the authors interpreted the data differently using a handicapped pentameric motor as mentioned earlier in this review (Chistol et al., 2012; Moffitt et al., 2009). It has been reported that mutation of one lysine ring does not significantly affect motor packaging activity (Fang et al., 2012) and the mutation of two or more lysines results in a motor that packages, but cannot hold onto the dsDNA, resulting in DNA reversal (Fang et al., 2012; Grimes et al., 2011). Mutating more lysines inhibits phage production. Since the location and the number of lysine are not critical, the data suggest that the lysine layers are not essential to the mechanism, but are auxiliary components that may improve efficiency.
Puzzle 6: why, in certain cases, does the motor display physical motion without ATP hydrolysis?
Classical dogma posits that nanobiomotors are powered by the energy released from ATP hydrolysis. Although the process of converting the chemical energy stored in ATP to physical motion is still unknown, several reports have indicated that the onset of motion can be linked to ATP binding, rather than ATP hydrolysis (Kinosita et al., 2000; Chang et al., 2012; Acharya et al., 2003; Gradia et al., 1997; Gradia et al., 1999). Similar to AAA+ motor proteins that undergo a cycle of conformational changes between two distinct states during their interaction with ATP, the phi29 motor ATPase gp16 (Guo et al., 1987c; Ibarra et al., 2001; Lee et al., 2008) also displays high and low affinity states for DNA (Fig. 5). Recently, it has been qualitatively demonstrated via EMSA (Electrophoretic Mobility Shift Assay) that gp16 exhibits binding to dsDNA and exhibiting stronger binding to DNA in the presence of γ-S-ATP, a non-hydrolysable ATP analog (Schwartz et al., 2012). This finding was further validated by Förster Resonance Energy Transfer (FRET) analysis and sucrose gradient sedimentation, both demonstrated how a motor promotes motion without ATP hydrolysis.
The motor undergoes a cycle of conformational changes between two distinct states during its interaction with ATP; the initial step is the binding of ATP that results in a reduction of entropy in the ATPase by a conformational change (Guo et al., 1987c; Ibarra et al., 2001; Lee et al., 2008). The entropy lost is compensated by a subsequent step of ATP hydrolysis resulting in entropy increase with another conformational change. Thus, coupling motion to a change in entropy is an intrinsic property of the protein. Both a DNA binding domain and a Walker-A motif are present in ATPase gp16 (Guo et al., 1987c; Schwartz et al., 2012). Generally, the Walker A motif of AAA+ ATPases is responsible for ATP binding, while the Walker B motif initiates ATP hydrolysis (Story and Steitz, 1992). This conformational change induced by ATP binding disappeared when a mutation was introduced to the Walker A motif.
Conclusion and Perspectives
The model of action of the phi29 DNA packaging motor revolves without rotating, coiling, or generating torque (Schwartz et al., 2013b), offers a model for revolution motors of differing stoichiometry. The topics discussed in this review offer a series of applications in nanotechnology. The riding system along one strand of dsDNA relates to cargo transportation at the nanoscale level and a tool for studying force generation mechanisms in a moving world.
Acknowledgments
The authors would like to thank Lei Lin for her EMSAs and thank Lindsay Black for providing Fig. 4C. The work was supported by NIH grants R01 EB003730, R01 EB012135, and U01 CA151648 to PG who is a co-founder of Kylin Therapeutics, Inc. and Biomotor and Nucleic Acids Nanotech Development, Ltd. Funding to Peixuan Guo’s Endowed Chair in Nanobiotechnology position is by the William Fairish Endowment Fund.
References
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- Aker J, Hesselink R, Engel R, Karlova R, Borst JW, Visser AJWG, de Vries SC. In vivo hexamerization and characterization of the Arabidopsis AAA ATPase CDC48A complex using forster resonance energy transfer-fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Plant Physiol. 2007;145:339–350. doi: 10.1104/pp.107.103986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the Bacteriophage phi29 prohead RNA-connector interaction. J Mol Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badasso MO, Leiman PG, Tao Y, He Y, Ohlendorf DH, Rossmann MG, Anderson D. Purification, crystallization and initial X-ray analysis of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2000;56:1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- Barre FX. FtsK and SpoIIIE: the tale of the conserved tails. Mol Microbiol. 2007;66:1051–1055. doi: 10.1111/j.1365-2958.2007.05981.x. [DOI] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. The DNA translocation vertex of dsDNA bacteriophages. Ann Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- Black LW. DNA Packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Black LW, Silverman D. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28:643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa N, Major F. Implication of the prohead RNA in phage phi29 DNA packaging. Biochimie. 2002;84:945–951. doi: 10.1016/s0300-9084(02)01444-x. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. In: Volff J-N, editor. Gene and Protein Evlolution. Genome Dyn; 2007. pp. 48–65. [DOI] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symposium on Quantitative Biology. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Zhang H, Shu D, Guo P, Savran C. Bright-field analysis of phi29 DNA packaging motor using a magnetomechanical system. Appl Phys Lett. 2008;93 doi: 10.1063/1.3000606. 153902–153902–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JR, Andrews BT, Catalano CE. Energy-independent helicase activity of a viral genome packaging motor. Biochemistry. 2012;51:391–400. doi: 10.1021/bi201604b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage phi29 genomic DNA translocation. J Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA: A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem. 2000;275 (23):17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Yu X, Egelman EH. The hexameric ring structure of the Escherichia coli RuvB branch migration protein. J Mol Biol. 2002;319:587–591. doi: 10.1016/S0022-2836(02)00353-4. [DOI] [PubMed] [Google Scholar]
- Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, Jardine PJ, Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi 29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- Demarre G, Galli E, Barre FX. The FtsK Family of DNA Pumps. Adv Exp Med Biol. 2013;767:245–262. doi: 10.1007/978-1-4614-5037-5_12. [DOI] [PubMed] [Google Scholar]
- Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci USA. 2011;108:7357–7362. doi: 10.1073/pnas.1016690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci USA. 2012;109:20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Jing P, Haque F, Guo P. Role of channel lysines and push through a one-way valve mechanism of viral DNA packaging motor. Biophys J. 2012;102:127–135. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Cai Q, Qin PZ. The procapsid binding domain of phi29 packaging RNA has a modular architecture and requires 2′-hydroxyl groups in packaging RNA interaction. Biochemistry. 2005;44:9348–9358. doi: 10.1021/bi0475020. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver K, Guo P. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage phi29. RNA. 1997;3:1068–1079. [PMC free article] [PubMed] [Google Scholar]
- Garver K, Guo P. Mapping the inter-RNA interaction of phage phi29 by site-specific photoaffinity crosslinking. J Biol Chem. 2000;275 (4):2817–2824. doi: 10.1074/jbc.275.4.2817. [DOI] [PubMed] [Google Scholar]
- Geng J, shaoying W, Huaming F, Peixuan G. Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded DNA and RNA. ACS Nano. 2013;7 (4):3315–3323. doi: 10.1021/nn400020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Fang H, Haque F, Zhang L, Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011;32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Grainge I. Simple topology: FtsK-directed recombination at the dif site. Biochem Soc Trans. 2013;41:595–600. doi: 10.1042/BST20120299. [DOI] [PubMed] [Google Scholar]
- Grigoriev DN, Moll W, Hall J, Guo P. Bionanomotor. In: Nalwa HS, editor. Encyclopedia of Nanoscience and Nanotechnology. American Scientific Publishers; 2004. pp. 361–374. [Google Scholar]
- Grimes S, Anderson D. RNA Dependence of the Bateriophage phi29 DNA Packaging ATPase. J Mol Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of phi29 connector channel loops in late-stage DNA packaging. J Mol Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Parraga A, Valpuesta JM, Carrascosa JL, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage phi29 connector particle. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Crystal structure of the delta subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci USA. 2013;110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. Introduction: principles, perspectives, and potential applications in viral assembly. Seminars in Virology (Editor’s Introduction) 1994;5 (1):1–3. [Google Scholar]
- Guo P. The emerging field of RNA nanotechnology. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987a;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage φ29 DNA-gp3. J Mol Biol. 1987b;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J Mol Biol. 1987c;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Guo PX, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happonen LJ, Oksanen E, Liljeroos L, Goldman A, Kajander T, Butcher SJ. The structure of the NTPase that powers DNA packaging into Sulfolobus turreted icosahedral virus 2. J Virol. 2013;87:8388–8398. doi: 10.1128/JVI.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Ultrastable Synergistic Tetravalent RNA Nanoparticles For Targeting To Cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage DNA packaging: RNA gears in a DNA transport machine (Minireview) Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA(pRNA) monomer, dimer, and hexamer of phi29 DNA Packaging motor. J Biol Chem. 2002;277 (23):20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Flamm C, Heine C, Wolfinger MT, Scheuermann G, Stadler PF. BarMap: RNA folding on dynamic energy landscapes. RNA. 2010;16:1308–1316. doi: 10.1261/rna.2093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LP, Guo P. Use of acetone to attain highly active and soluble DNA packaging protein gp16 of phi29 for ATPase assay. Virology. 2003a;312:449–457. doi: 10.1016/s0042-6822(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Huang LP, Guo P. Use of PEG to acquire highly soluble DNA-packaging enzyme gp16 of bacterial virus phi29 for stoichiometry quantification. J Virol Methods. 2003b;109:235–244. doi: 10.1016/s0166-0934(03)00077-6. [DOI] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Faik W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:558–567. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage lambda. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- Ibarra B, Valpuesta JM, Carrascosa JL. Purification and functional characterization of p16, the ATPase of the bacteriophage phi29 packaging machinary. Nucleic Acids Res. 2001;29:4264–4273. doi: 10.1093/nar/29.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–263. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Santisteban A, Carazo JM, Carrascosa JL. Computer graphic display method for visualizing three-dimensional biological structures. Science. 1986;232:1113–1115. doi: 10.1126/science.3754654. [DOI] [PubMed] [Google Scholar]
- Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–3627. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- Kinosita K, Jr, Yasuda R, Noji H, Adachi K. A rotary molecular motor that can work at near 100% efficiency. Philos Trans R Soc London B Biol Sci. 2000;355:473–489. doi: 10.1098/rstb.2000.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J Mol Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- Lee TJ, Zhang H, Liang D, Guo P. Strand and nucleotide-dependent ATPase activity of gp16 of bacterial virus phi29 DNA packaging motor. Virology. 2008;380:69–74. doi: 10.1016/j.virol.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, Gilles M, Dethan G, Orlova EV, Couprie J, Tavares P, Zinn-Justin S. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci USA. 2009;106:8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PT, Bustamante C, Tinoco I., Jr Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf NK, Feiss M. Virus DNA translocation: progress towards a first ascent of mount pretty difficult. Mol Microbiol. 2006;61:1–4. doi: 10.1111/j.1365-2958.2006.05214.x. [DOI] [PubMed] [Google Scholar]
- Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina EM, Andrews BT, Nakatani E, Catalano CE. The bacteriophage lambda gpNu3 scaffolding protein is an intrinsically disordered and biologically functional procapsid assembly catalyst. J Mol Biol. 2011;412:723–736. doi: 10.1016/j.jmb.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- Moll D, Guo P. Grouping of ferritin and gold nanoparticles conjugated to pRNA of the phage phi29 DNA-packaging motor. J Nanosci Nanotechnol (JNN) 2007;7:3257–3267. doi: 10.1166/jnn.2007.914. [DOI] [PubMed] [Google Scholar]
- Moll WD, Guo P. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J Mol Biol. 2005;351:100–107. doi: 10.1016/j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson D, Rossman MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais MC, Tao Y, Olsen NH, Grimes S, Jardine PJ, Anderson D, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage-T3. Virology. 1993;193:748–752. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature. 2011;479:194–199. doi: 10.1038/nature10568. [DOI] [PubMed] [Google Scholar]
- Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 Terminase by DNA Structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Harvey SC. Role of DNA–DNA interactions on the structure and thermodynamics of bacteriophages Lambda and P4. J Struct Biol. 2011;174:137–146. doi: 10.1016/j.jsb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Ray K, Ma J, Oram M, Lakowicz JR, Black LW. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J Mol Biol. 2010a;395:1102–1113. doi: 10.1016/j.jmb.2009.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010b;398:224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif R, Haque F, Guo P. Fluorogenic RNA nanoparticles for monitoring RNA folding and degradation in real time in living cells. Nucleic Acid Ther. 2013;22 (6):428–437. doi: 10.1089/nat.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Cingolani G. Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta Crystallogr Sec F-Struct Biol Crystallization Commun. 2011;67:104–110. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, De Donatis GM, Fang H, Guo P. The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA+ superfamily. Virology. 2013a;443:20–27. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology. 2013b;443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Fang H, Huang L, Guo P. Sequential action of ATPase, ATP, ADP, Pi and dsDNA in procapsid-free system to enlighten mechanism in viral dsDNA packaging. Nucleic Acids Res. 2012;40:2577–2586. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P. A hypothesis for bacteriophage DNA packaging motors. Viruses. 2010;2:1821–1843. doi: 10.3390/v2091821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P. Models of bacteriophage DNA packaging motors. J Struct Biol. 2003;141:179–188. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Shu D, Haque F, Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nature Protocols. 2013a;8:1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA. 2013b;19:766–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J, Ozgur S, Lin BY, Yu JH, Broker TR, Chow LT, Griffith J. Remodeling of the human papillomavirus Type 11 replication origin into discrete nucleoprotein particles and looped structures by the E2 Protein. J Mol Biol. 2008;375:1165–1177. doi: 10.1016/j.jmb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase Activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Snider J, Houry WA. AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans. 2008;36:72–77. doi: 10.1042/BST0360072. [DOI] [PubMed] [Google Scholar]
- Snider J, Thibault G, Houry WA. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008;9:216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the rec-A protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. Motor proteins. In: Kreis T, Vale R, editors. Guidebook to the Cytoskeletal and Motor Proteins. Oxford University Press; 1993. pp. 175–211. [Google Scholar]
- Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- Willows RD, Hansson A, Birch D, Al-Karadaghi S, Hansson M. EM single particle analysis of the ATP-dependent BchI complex of magnesium chelatase: an AAA(+) hexamer. J Struct Biol. 2004;146:227–233. doi: 10.1016/j.jsb.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Xiao F, Moll D, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Demeler B, Guo P. Assembly mechanism of the sixty-subunit nanoparticles via interaction of RNA with the reengineered protein connector of phi29 DNA-Packaging motor. ACS Nano. 2010;4:3293–3301. doi: 10.1021/nn100158k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J Mol Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud S, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage phi29 DNA. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schwartz C, De Donatis GM, Guo P. Push through one-way valve mechanism of viral DNA packaging. Adv Virus Res. 2012;83:415–465. doi: 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA. RNA. doi: 10.1261/rna.037077.112. http://dx.doi.org/10.1261/rna.037077.112, in press. [DOI] [PMC free article] [PubMed]
- Zhang X, Wigley DB. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol. 2008;15:1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Khisamutdinov E, Schwartz C, Guo P. Mechanism of one-way traffic of hexameric Phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano. 2013;7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelin G, Niedenzu T, Lurz R, Saenger W, Lanka E. Hexameric RSF1010 helicase RepA: the structural and functional importance of single amino acid residues. Nucleic Acids Res. 2003;31:5917–5929. doi: 10.1093/nar/gkg790. [DOI] [PMC free article] [PubMed] [Google Scholar]