Abstract
Peroxisome proliferator-activated receptors (PPARs) are transcription factors that belong to the superfamily of nuclear hormone receptors and regulate the expression of several genes involved in metabolic processes that are potentially linked to the development of some diseases such as hyperlipidemia, diabetes, and obesity. One type of PPAR, PPAR-α, is a transcription factor that regulates the metabolism of lipids, carbohydrates, and amino acids and is activated by ligands such as polyunsaturated fatty acids and drugs used to treat dyslipidemias. There is evidence that genetic variants within the PPARα gene have been associated with a risk of the development of dyslipidemia and cardiovascular disease by influencing fasting and postprandial lipid concentrations; the gene variants have also been associated with an acceleration of the progression of type 2 diabetes. The interactions between genetic PPARα variants and the response to dietary factors will help to identify individuals or populations who can benefit from specific dietary recommendations. Interestingly, certain nutritional conditions, such as the prolonged consumption of a protein-restricted diet, can produce long-lasting effects on PPARα gene expression through modifications in the methylation of a specific locus surrounding the PPARα gene. Thus, this review underlines our current knowledge about the important role of PPAR-α as a mediator of the metabolic response to nutritional and environmental factors.
Introduction
The prevalence of chronic diseases of metabolic origin such as hyperlipidemia, diabetes, and obesity has increased in recent decades worldwide. These disorders have a complex etiology involving genetic, environmental, and nutritional factors. There is evidence that a group of nuclear receptors, called PPARs, are involved in these diseases (1–5). PPARs are well-characterized type II nuclear receptors identified in vertebrates that contain a cysteine-rich Zn finger–motif DNA binding domain (6, 7). The PPAR family consists of 3 members: PPAR-α, PPAR-δ (also called PPAR-β), and PPAR-γ (NR1C1, NR1C2, and NR1C3, respectively). PPAR-α was first described as a receptor that is activated by peroxisome proliferators, hence its name (8–11).
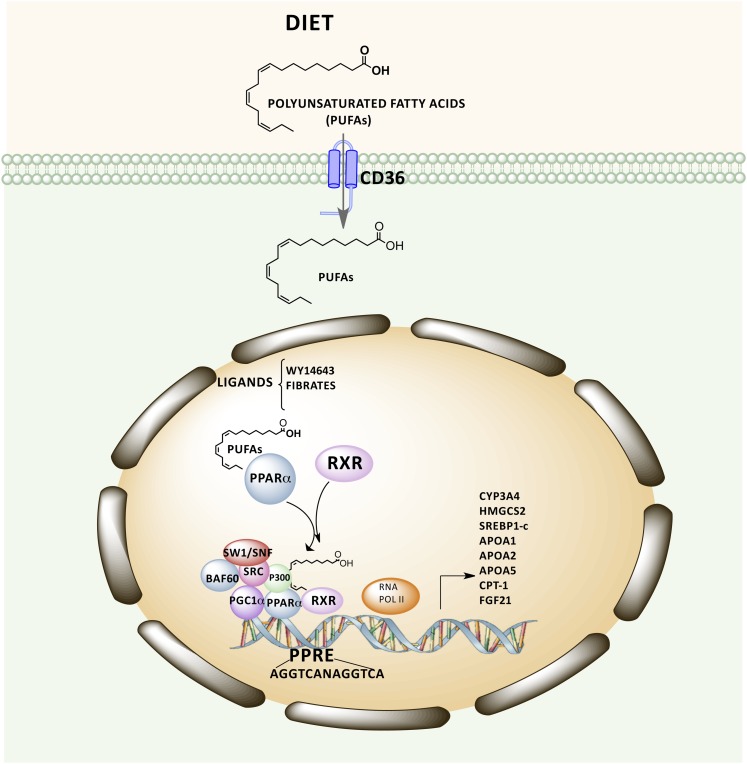
PPAR-α can be activated by certain natural and synthetic ligands such as PUFAs, eicosanoids, and hypolipidemic drugs (fibrates) and then modulates DNA transcription by binding to specific nucleotide sequences located in the regulatory regions of target genes known as peroxisome proliferator responsive elements (PPREs)6 (12, 13). DNA binding requires that a heterodimer containing PPAR-α and retinoic X receptor α interact with a PPRE that contains a consensus sequence (AGGTCA N AGGTCA) consisting of 2 direct repeating half-sites spaced by 1 nucleotide (14, 15). When ligands bind to PPARα, conformational changes in PPARα- retinoic X receptor α induce the active transcriptional complex to assemble with coactivator proteins either sequentially or into preassembled subcomplex modules. Therefore, coactivator complexes that acetylate (steroid receptor coactivators, p300) nucleosomes for chromatin remodeling and mediator components contact PPAR-α to facilitate the recruitment of the basal transcription machinery with RNA polymerase II for transcription of specific target genes (16, 17) (Fig. 1).
Figure 1.
Schematic representation of PPAR-α target gene transcription dependent on ligand binding and recruitment of coactivators. PPAR-α can be activated by certain ligands, such as PUFAs, 16:00/18:1-GPC, and fibrates, which modulate transcription of PPAR-α target genes. However, DNA binding requires the formation of a heterodimer containing PPAR-α and retinoic X receptor (RXR) α that is able to interact with peroxisome proliferator responsive elements. The assembly of this heterodimer with coactivator proteins facilitates the recruitment of the basal transcription machinery with RNA polymerase II for transcription of specific target genes. APO, apolipoprotein; BAF60, Brahma-related gene 1/Brahma -associated factor 60; CD36, Cluster of Differentiation 36; CPT-1, Carnitine palmitoyltransferase I; CYP, cytochrome P-450; FGF21, fibroblast growth factor 21; HMGCAS2, hydroxymethylglutaryl CoA synthase 2; p300, histone acetyltransferase p300; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-α PPRE, peroxisome proliferator responsive element; RNA POLII, RNA polymerase II; SRC, steroid receptor coactivator; SREBP1-c, sterol regulatory element binding protein 1c; SW1/SNF, switch/sucrose nonfermentable chromatin-modifying complex.
It is known that activation of PPAR-α by its ligands can modify multiple biological processes in the cell that are important, particularly in the mechanisms associated with body energy production, as well as in the inflammatory response, among others. Thus, in the current review, we focus on how PPAR-α can control these metabolic functions, showing an integrated understanding of 3 biological aspects of PPAR-α, its metabolic function, its most studied genetic variants, and the transgenerational reprogramming and how these aspects interact with environmental factors to carry out its metabolic role. In the past decade, genetic and genomic studies have provided evidence of the biological significance of PPAR-α genetic variants and their interactions with some diet components or hypolipidemic drugs, such as fenofibrate, to modulate metabolic phenotypes in humans because they are PPAR-α ligands. Finally, we consider the long-term consequences are in transgenerational environmental reprogramming of metabolism depending on nutritional status that may alter the epigenetic modifications of PPAR-α regulatory sequences that will influence the expression of its target genes.
Natural and synthetic PPAR-α ligands
PPAR-α is a receptor for a structurally diverse group of compounds, including natural and synthetic ligands. Within the PPAR-α ligand-binding domain, there is a large pocket ∼1400 μm3 into which the molecule ligand is bound. The ligand adopts a conformation within the receptor that allows formation of hydrogen bond interactions; these interactions stabilize the receptor in a configuration that leads to the transcriptional activation of PPAR-α via recruitment of coactivator proteins (18).
Among the natural PPAR-α ligands, some cells are able to generate an endogenous ligand, the phospholipid 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC), which is synthesized by the enzyme fatty acid synthase (19). In addition, other natural ligands such as PUFAs are provided by the diet (linoleic, α-linolenic, γ-linolenic, and arachidonic acids), which bind to PPAR-α at physiologic concentrations (12). It has been demonstrated that phytanic acid, a branched-chain fatty acid generated from phytol present in dairy products, is also a natural ligand of PPAR-α (27, 28). It is well-known that dietary PUFAs have effects on diverse biological processes such as insulin action, cardiovascular function, neural development, and immune function, some of them mediated via PPARα. Additionally, dietary PUFAs activate both directly and indirectly other transcription factors such as liver X receptor, hepatocyte nuclear factor-4, and sterol regulatory element binding protein, which mediate to some extent other biological processes affecting the expression of specific genes (Fig. 1) (20, 21).
Hostetler et al. (22), using direct fluorescence binding and fluorescence displacement assays, demonstrated that PPAR-α exhibits high affinity (1–4 nmol/L Kd values) for unsaturated long-chain fatty acyl-CoAs as well as unsaturated long-chain fatty acids commonly found in mammalian cells; these high-affinity ligands elicited conformational changes in PPAR-α structure that correlated functionally with coactivator binding. In contrast, saturated long-chain fatty acids were not or were only very weakly bound (22). In addition, Lin et al. (23), using a fluorescence-based method for measuring the dissociation constants (Kd) to characterize the interactions of PPAR-α with various natural ligands such as oleic acid, linoleic acid, linolenic acid, arachidonic acid, and leukotriene LTB4 (Kd values of 5.9, 4.8, 7.9, 17.3, and 60.8 nM, respectively), revealed that PPAR-α interacts with affinities in the nanomolar range with these ligands.
It has been suggested that activation of PPAR-α in the liver during fasting is due to serum FFAs that are released from adipose tissue; however, recent evidence indicates that circulating FFAs fail to activate hepatic PPAR-α, whereas it can be activated by dietary fatty acids and fatty acids generated via de novo lipogenesis (19, 20, 24, 25). In fact, it has been shown that the effects of dietary fatty acids on hepatic gene expression are quantitatively almost entirely mediated by PPAR-α (26).
PPAR-α also binds to eicosanoids such as 8-hydroxyeicosatetraenoic acid, with an affinity estimated in the range of 100 nM (12). Other natural compounds such as polyphenols have been described as ligands of PPAR-α (29, 30). Resveratrol, a natural polyphenol found in grapes, peanuts, and berries, and some of its derivatives and analogs, activate PPAR-α, resulting in brain protection against stroke (31–33); for instance, the derivate compound phosphate 15 has a potency higher than that of the drug ciprofibrate (33). Studies in HepG2 cells treated with genistein, another polyphenol that is the main soy isoflavone, induced the expression of PPAR-α at both messenger RNA (mRNA) and protein levels and enhanced expression of genes involved in fatty acid catabolism through activation of PPAR-α (34). Additional PPAR-α ligands from diet with hypolipidemic activity have been reported, such as the natural carotenoid abundant in seafood, astaxanthin, and the active compound extracted from the tomato, 9-oxo-10(E),12(E)-octadecadienoic acid (35, 36).
PPAR-α can be also activated by synthetic ligands such as hypolipidemic drugs that include bezafibrate, fenofibrate, clofibrate, and Wy14643 that induce up- and downregulation of expression of several genes involved in β-oxidation and lipid metabolism (10, 37). PPAR-α exhibits Kd values for bezafibrate and their CoA thioesters (bezafibroyl-CoA) in the same range (Kd = 13.1 and 2.7 nM, respectively) as for unsaturated long-chain fatty acids (22), whereas for fenofibrate is in the micromolar range (10–20 μM), indicating a lower affinity (38, 39). Wy14643 is a potent ligand that has a binding affinity to PPAR-α higher than that for the endogenous ligand 16:0/18:1-GPC (Ki = 11.06 vs. Ki = 33.20 nM, respectively); this may result in competition and a rapid displacement of the natural by the synthetic ligand (19). The piperidine synthetic agonists bind to PPAR-α very strongly with Ki values of 74 nM for CP-865529, 24.5 nM for CP-775146, and 10.8 nM for CP-868388. These compounds have been very useful in acute preclinical models for treating dyslipidemia (40).
Metabolic functions of PPAR-α
PPAR-α is highly expressed in brown adipose tissue, the liver, and, to a lesser extent, in the kidney, skeletal muscle, heart, and small and large intestines (41–43). PPAR-α functions as a lipid sensor in the liver and recognizes and responds to the influx of fatty acids by stimulating the transcription of specific genes (44). Studies performed using PPAR-α–null mice (knockout mice) have demonstrated that PPAR-α controls the expression of numerous genes related to lipid metabolism in the liver, including genes involved in mitochondrial β-oxidation, peroxisomal β-oxidation, fatty acid uptake and binding, and lipoprotein assembly and transport. In conditions such as starvation that lead to an increased demand for the oxidation of fatty acids, PPAR-α is essential for the upregulation of the expression of the genes coding for the enzymes necessary to fulfill the energy needs in these circumstances (45–49). However, if plasma FFAs do not activate hepatic PPAR-α during fasting and the production of the endogenous ligand (16:0/18:1 GPC) is controlled by fatty acid synthase during feeding, it remains unclear what the mechanism is for activation of PPAR-α–dependent gene regulation in a condition of high energy demand (50).
The genes involved in lipid and lipoprotein metabolism in humans and regulated by PPAR-α include the apolipoprotein (Apo) genes such as APOA1, APOA2, and APOA5, the genes involved in fatty acid oxidation (acyl-CoA oxidase, CPT-I, and CPT-II), those required for the desaturation of fatty acyl CoA (delta-6-desaturase), and genes involved in HDL metabolism (PLTP) and ketone body synthesis (HMGCS2) (51–54). PPAR-α does not merely serve as a transcriptional activator of fatty acid catabolism but plays a much broader role in lipid metabolism (24, 55). In this regard, PPAR-α agonists also enhance the activity of the sterol regulatory element binding protein 1c promoter because it contains a PPRE located at -453 bp in the human gene. Sterol regulatory element binding protein 1c is a transcription factor that plays a key role in the regulation of the gene expression of lipogenic enzymes and is essential for the genomic actions of insulin on the metabolism of both carbohydrates and lipids, depending on nutritional status (55).
Because PPAR-α is involved in ensuring energy availability during fasting and starvation, it plays a prominent role in the starvation response. This response is mediated in part by fibroblast growth factor 21, an endocrine regulator of the body’s adaptation to fasting that is produced in the liver in response to PPAR-α (56, 57). The function of fibroblast growth factor 21 is to both stimulate lipolysis of white adipose tissue to supply fatty acids to nonadipose tissue organs and to control ketogenesis in the liver (56) to obtain energy from fatty acids (Fig. 1).
In addition to its regulation of lipid metabolism, recent evidence shows that PPAR-α also regulates the metabolism of amino acids in the liver. It was found that PPAR-α decreases the mRNA expression of enzymes involved in the metabolism of amino acids when comparing the expression of mRNA from wild-type versus PPAR-α–null mice (58). Furthermore, in PPAR-α–null mice, fasting decreases the plasma levels of the gluconeogenic amino acid alanine and the ketogenic amino acid tyrosine. The amino acids linked to the urea cycle, which include aspartate, arginine, and citrulline, are increased in PPAR-α knockout mice compared with wild-type mice (59). In a model of dyslipidemia and insulin resistance, however, the PPAR-α agonist Wy14643 produces significant alterations in the plasma levels of both amino acids and nitrogen-containing metabolites, suggesting important effects on amino acid mobilization and catabolism (60).
On the other hand, PPAR-α is also known to modulate the transcription of genes involved in pathways of inflammatory response. This nuclear receptor modulates anti-inflammatory activity via downregulation of the activator protein-1 and nuclear factor-κB signaling pathways through a direct protein-protein interaction with p65 and c-Jun. In addition, experimental evidence suggests that PPAR-α activation attenuates or inhibits several mediators of vascular damage, including lipotoxicity, inflammation, reactive oxygen species, endothelial dysfunction, angiogenesis, and thrombosis (61–64). Moreover, activation of PPAR-α has been demonstrated to inhibit tumor growth and angiogenesis through suppressing hypoxia-inducible factor 1α signaling in cancer cells (65).
Recently, another role of PPAR-α was found by directly regulating the transcription of cytochrome P-450 3A4, the major human drug-metabolizing enzyme. Activation of PPAR-α by the synthetic and endogenous ligands Wy14643 and 16:0/18:1-GPC, respectively, increases the expression of a distinct set of cytochrome P-450 enzymes, including 3A4, 1A1, 1A2, 2B6, 2C8, and 7A1 in primary human hepatocytes (66).
PPAR-α genetic variants associated with metabolic phenotypes in humans
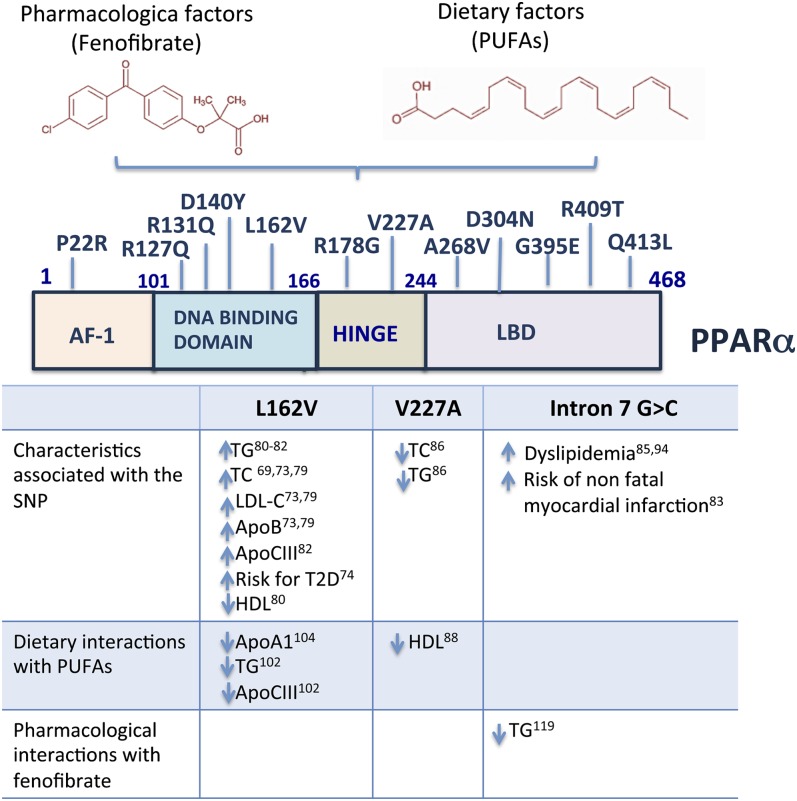
The human PPARα gene is located on the chromosome 22q13.3 and spans 93.15 kb. Genetic variants in both the DNA-binding and ligand-binding domains that influence the transcriptional activity of human PPAR-α were described previously (67–71). Single nucleotide polymorphisms (SNPs) within this gene are associated with metabolic features such as dyslipidemia and cardiovascular risk factors (72) and also influence both fasting and postprandial lipid concentrations (73) and the progression of type 2 diabetes (74, 75) (Fig. 2, Table 1).
Figure 2.
Location and associated effects of PPAR-α variants. Environmental factors such as diet and drugs may interact with PPARα genetic variants to modulate metabolic parameters. The L162V single nucleotide polymorphism (SNP) in the PPARα coding region has been associated with an increase in TG, total cholesterol (TC), LDL, ApoB, ApoC3, the risk of type 2 diabetes (T2D), and a decrease in HDL. There is an interaction between the SNP V227A or L162V with PUFA intake that is associated with a decrease in the levels of HDL, ApoA1, TGs, and ApoC3. In addition, the intron 7 G > C variant is associated with the fenofibrate response, dyslipidemia and risk of nonfatal myocardial infarction. The domains represented in the figure are activation function-1 (AF-1), DNA binding domain (DBD), and ligand binding domain (LBD). LDL-C, low density lipoprotein-cholesterol.
Table 1.
Common PPAR-α SNPs associated with metabolic phenotypes1
| SNP | Study population | Minor allele frequency, % | Metabolic effects | OR (95% CI) | P value | Ref. |
| L162V rs1800206 | 2373 subjects (1128 men and 1244 women) in the Framingham Offspring Study | 6.9 | Results observed in men | — | — | 79 |
| ↑ TC | — | 0.0012 | ||||
| ↑ LDL-C | — | 0.0004 | ||||
| ↑ ApoB | — | 0.009 | ||||
| In women | — | — | ||||
| ↑ ApoB | — | 0.03 | ||||
| 610 young healthy white subjects average age, 24 y) | 7.5 | Results observed in men | — | — | 80 | |
| ↑ TG | — | 0.0040 | ||||
| ↓ HDL | — | 0.0017 | ||||
| Danish study of middle-aged subjects (N = 5799; average age, 46 y) | 5.8 (95% CI: 5.4–6.2) | ↑ TG | — | 0.007 | 81 | |
| 59 healthy male students at the University of Cordoba (Spain), ranged from 18 to 49 y of age | 10.2 | ↑ TC | — | 0.003 | 73 | |
| ↑ LDL-C | — | 0.001 | ||||
| ↑ ApoB | — | 0.004 | ||||
| 2508 middle-age (50–61 y-old) healthy European men and 129 type 2 diabetic men and women (86 European, 43 Asian) | Europeans: 6.6 (95% CI: 3.0–10) | Results observed in subjects with type 2 diabetes | — | — | 69 | |
| Asians: 2.5 (95% CI: 0–6.0) | ↑ TC | — | 0.04 | |||
| ↑ ApoA1 | — | 0.003 | ||||
| 335 African Americans aged 30 y or older | 1.5 | ↑ ApoC2 | — | 0.0005 | 82 | |
| ↑ TG | — | 0.009 | ||||
| 570 subjects of an admixed population (147 males and 423 females) from Cuiaba City, Brazil | 5.2 | ↑ Dyslipidemia | 2.12 (1.0, 4.47) | 0.025, 0.0502 | 85 | |
| 767 subjects of the STOP-NIDDM trial (385 men and 382 women) | 7.0 | ↑ Development of type 2 diabetes | 1.93 (1.05, 3.58) | 0.035 | 74 | |
| 632 unrelated men from Quebec, Canada | 10.6 | Waist circumference (cm) is explain by PPAR-α–L162V × SFA interaction in 2.71% of variance | — | 0.01 | 78 | |
| 1003 men and 1103 women participating in the Framingham cohort | 6.98 | PPAR-α–L162V × low PUFA diet interaction | — | — | 102 | |
| ↑ TG | — | <0.05 | ||||
| ↑ ApoCIII | — | <0.05 | ||||
| PPAR-α–L162V × high PUFA diet interaction: | — | |||||
| ↓ TG | — | <0.05 | ||||
| ↓ ApoC3 | — | <0.05 | ||||
| 674 participants between 18 and 55 y of age; all subjects were recruited from the Quebec City metropolitan area | 8.8 | PPAR-α–L162V × high saturated fat intake interaction | — | — | 103 | |
| ↓ Peak particle diameters of LDL | — | 0.007 | ||||
| From 66 subjects from Quebec City metropolitan area, 20 healthy white men were selected, matched according to age and BMI | 10.6 | PPAR-α–162V × high PUFA diet interaction | — | — | 104 | |
| ↓ ApoA1 | — | 0.02 | ||||
| V227A rs1800234 | 401 apparently healthy subjects (207 men and 194 women), all Japanese | 5.1 | Results observed in women | — | — | 86 |
| ↓ TC | — | 0.046 | ||||
| ↓ TG | — | 0.038 | ||||
| 706 Japanese men | 5.0 | In nondrinker group | — | — | 70,90 | |
| ↓ TC | — | <0.05 | ||||
| ↓ TG | — | <0.05 | ||||
| 1964 men (1318 Chinese, 364 Malaysian, and 282 Asian Indians) and 2284 women (1581 Chinese, 397 Malaysian, and 306 Asian Indians) from the 1998 Singapore National Health Survey | 4.0 in Chinese 0.6 in Malays 0.3 in Asian Indians | PPAR-α–V227A × increasing dietary PUFA intake interaction in Chinese women | — | — | 88 | |
| ↓ HDL-C | — | 0.049 | ||||
| Intron 7G > C rs4253778 | 570 subjects from an admixed population (147 males and 423 females) from Cuiaba City, Brazil | 29.2 | ↑ Dyslipidemia | 1.56 (1.05, 2.33) | 0.027, 0.0292 | 85 |
| 1810 white subjects with type 2 diabetes from the prospective Go-DARTS | 18.2 (0.169–0.194) | ↑ TC | — | <0.05 | 83 | |
| ↑ HDL-C | — | <0.05 | ||||
| ↑ Risk of nonfatal myocardial infarction | 2.77,3 (1.34, 5.75), | 0.006 | ||||
| 358 white subjects (178 with a diagnosis of coronary artery disease and 180 without) | 15.5 | In the coronary artery disease group | — | — | 93 | |
| ↓ TC | — | 0.016 | ||||
| ↓ HDL-C | — | 0.029 | ||||
| 230 men from a hospital-based Indian population (110 patients with coronary artery disease and 120 healthy men) | 6.0 | ↑ Dyslipidemia | 2.95 (1.5, 4.39) | <0.05 | 94 | |
| 207 subjects with type 2 diabetes participating in the Diabetes Atherosclerosis Intervention Study (DAIS) | 11.9 | After fenofibrate treatment in patients with type 2 diabetes: | 3.10 (1.28, 7.52) | 0.012 | 119 | |
| ↓ TG | ||||||
| Haplotype of: rs135539, L162V, rs4253778 | 912 white subject with type 2 diabetes from 2 studies: UDACS and EDSC | 0.9 | ↑ onset of diabetes of type 2 diabetes, ∼10 y | 2.42 (0.70, 8.29) | 0.03 | 75 |
| 140+5435T > C rs135549 | 59 healthy male students at the University of Cordoba (Spain), age range,18–49 y e | 36.4 | Postprandial response with oral fat load (saturated fatty acids) | — | — | 73 |
| ↓ Small TRL-cholesterol | — | 0.039 | ||||
| ↓ Small TRL-TG | — | 0.008 | ||||
| 3′UTR G > A (rs6008259) | 10,134 white and 3480 African American subjects were selected from the ARIC study | 18.0 in whites, 67.0 in African Americans | PPAR-α 3′UTR G > A × linoleic acid intake interaction in white participants | — | — | 106 |
| ↓ TC | — | 0.03 | ||||
| ↓ LDL-C | — | 0.03 | ||||
| 3′UTR C > T (rs3892755) | 10,134 whites and 3480 African Americans were selected from the ARIC Study | 0.3 in whites, 28.5 in African Americans | PPAR-α-3′UTR C>T × long-chain n3 fatty acid intake in African Americans participants | — | — | 106 |
| ↓ TC | — | 0.03 | ||||
| ↓ LDL-C | — | 0.02 | ||||
| rs135550 | 861 subjects (white and nearly all of European ancestry) from GOLDN study | CEU4 20.0 | After fenofibrate treatment: | — | — | 121 |
| ↓ LDL-C | — | 0.001 | ||||
| rs9626730 | 861 subjects (white and nearly all of European ancestry) from GOLDN study | CEU4 14.2 | After fenofibrate treatment: | — | — | 121 |
| ↓ IL-2 | — | 0.0002 |
Apo, apolipoprotein; SNPs, single nucleotide polymorphisms; TC, total cholesterol; TG, triglyceride; TRL, triglyceride-rich lipoprotein.
P value adjusted for alcohol consumption and waist-to-hip ratio.
Hazard ratio.
White population from HapMap International Project (http://hapmap.ncbi.nlm.nih.gov/).
Metabolic phenotypes associated with the L162V genetic variant
A leucine-to-valine change in codon 162 (L162V, rs1800206), represented by a C to G substitution at the DNA-binding domain, encodes for a more active PPAR-α depending on the concentration of the ligand. In transfection assays performed in vitro, this variant is unresponsive to a low concentration of ligand compared with the wild type; however, in the presence of a high concentration of the synthetic ligand Wy14643 (>25 μM), the transcriptional activity of the 162V allele is higher compared with that of the 162L allele (67, 69). The L162V polymorphism has been associated with increased levels of TGs, total cholesterol (TC), LDL cholesterol, and apoA1 and apoB in whites (69, 73, 76–78).
The potential modulating the phenotype-associated effects of this polymorphism on lipid metabolism depends on several factors such as the disease state, advanced age, and sex. In 2373 subjects (1128 men and 1244 women) from the Framingham Offspring Study, the V162 allele was associated in men with increased serum concentrations of TC, LDL cholesterol, and apoB (P = 0.0012, P = 0.0004, and P = 0.009, respectively); in women, a similar trend was observed, but these findings did not reach statistical significance (P = 0.18 for TC and HDL cholesterol) with the exception of serum apoB (P = 0.03) (79). In 610 young healthy white subjects (average age, 24 y), men heterozygous for L162V showed 78% higher TG concentrations and 20% lower HDL levels than those homozygous for the common ancestral allele. However, no significant associations between PPAR-α genotype and lipids were shown in women (80). A Danish study of middle-aged (n = 5799; average age, 46 y) carriers of 162V showed 60% higher fasting serum TG concentrations (81). Moreover, in healthy men, the 162V allele was associated with higher fasting TC (P = 0.003), LDL cholesterol (P = 0.001), and apoB (P = 0.004), but not with postprandial parameters (73). In contrast, in subjects with type 2 diabetes (n = 129), but not in healthy men (n = 2508; 50–61 y old), carriers of the 162V allele had 9% higher plasma TC (P = 0.04) (69).
The minor allele frequency for L162V is lower in African Americans (1.5%) and Asians (2.5%) compared with whites (varies from 6.1% to 10.6%) (69, 78, 80, 82–84). In the African-American population (n = 335), carriers of the 162V allele have higher levels of plasma apoC3 (P = 0.0005) and TG (P = 0.009) than 162L homozygous individuals (82). A study of an admixed population of 570 subjects from Brazil showed an association with dyslipidemia when 2 SNPs, the 162V and intron 7G > C polymorphism, were present in the PPARα gene (rs4253778); carriers of the intron 7C allele showed a 1.56-fold increase in the risk of presenting with high blood lipids. In addition, haplotype analysis regarding L162V and intron 7G > C showed a significant association with dyslipidemia and LDL cholesterol levels. Carriers of the L-C haplotype were at an increased risk of dyslipidemia compared with carriers of the most common L-G haplotype (OR = 1.56, P = 0.021), and carriers of the V-C haplotype showed lower LDL cholesterol levels compared with L-G haplotype carriers (mean difference = −23.17 mg/dL, P = 0.044) (85).
In addition, some studies have reported that the presence of the 162V allele influences the progression of type 2 diabetes. A study of the STOP-NIDDM trial (N = 413) showed that L162V modulates the progression to type 2 diabetes. Subjects with impaired glucose tolerance carrying the 162V allele have a 1.9-fold (95% CI: 1.05, 3.58) increased risk of the development of diabetes (74). A haplotype that comprises L162V and 2 more PPAR-α SNPs (rs135539 A > C in intron 1 and rs4253778 G > C in intron 7) significantly influences the age at which type 2 diabetes develops in European subjects (n = 912). Individuals with the C-V-C haplotype (intron 1-L162V-intron 7) had an accelerated onset of diabetes of ∼10 y (P = 0.03) compared with subjects with the common A-L-G haplotype (75).
Metabolic phenotypes associated with the V227A genetic variant
The PPAR-α variant V227A (rs1800234) that is located in the hinge region between the DNA binding and ligand binding domains of the PPAR-α gene was identified in a Japanese population with a frequency of 5.0% (70, 86, 87) and in a Chinese population with a frequency of ∼4% (88). This polymorphism attenuates the transcription of cytochrome P-450 4A6 (∼35%–56%) and the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase genes in the presence of fibrate ligands. A 2-hybrid assay revealed that the variant 227A enhanced the recruitment of corepressors with an ∼2-fold greater avidity than the variant 227V and also induced a defective release of the nuclear corepressor 1 from PPAR-α in the presence of Wy14643 and α-linolenic acid (89).
Unlike the L162V variant, the effect of V227A seems to be more evident in women than in men (86, 88). Among women (n = 194), the mean serum TC and TG levels in carriers of the 227A allele were lower than in noncarriers (P = 0.046 and P = 0.038, respectively), and a stronger association between this SNP and TC concentration was observed in women younger than 45 y of age than in the total group of women (P = 0.023) (86). In addition, significant interactions between the V227A polymorphism and alcohol drinking habits were found for TC and TG when excluding subjects with possible familial hypertriglyceridemia (≥518 mg/dL), suggesting that the PPAR-α activity in the 227A carrier group with no alcohol drinking may be higher than in the wild-type group, but it may become reduced in subjects who consume alcohol (70, 90).
Metabolic phenotypes associated with the intron 7 G > C genetic variant
The intron 7 polymorphism (rs4253778) located in a noncoding region of PPARα has a reported allelic frequency of 13.4%–18.2% in European populations (83, 91, 92). The 7C allele is associated with a significantly earlier age at diagnosis of increased TC, LDL cholesterol, and risk of nonfatal myocardial infarction in white subjects with type 2 diabetes participating in the prospective population-based Genetics of Diabetes Audit and Research in Tayside, Scotland (Go-DARTS) study (N = 1810) (83). In contrast, in 358 subjects (178 with a confirmed diagnosis of coronary artery disease and 180 without a history of that disease), C allele carriers had significantly lower TC and LDL cholesterol levels (93). However, the presence of the C allele showed a positive association with dyslipidemia, defined as a plasma cholesterol concentration ≥200 mg/dL, a TG concentration ≥180 mg/dL, or the intake of lipid-lowering drugs in an Indian population (94). Inconsistencies in relation to lipid parameters associated with intron 7G > C polymorphism have been described; however, differences maybe associated with dietary habits, the proportion of ancestral components, and the disease status of the populations studied (72, 83, 85, 93, 95). In addition, it has speculated that this SNP could be in allelic association with an unidentified variant in a regulatory region of the PPARα gene that affects its transcriptional activity (91). Under this possibility, population stratification could alter the linkage disequilibrium between these genetic variants. Therefore, the discrepancy in results may be explained in part by differing environmental factors, genetic backgrounds, medications prescribed, and concentrations of saturated and polyunsaturated fats in the diet.
Other PPAR-α variants involved in metabolic responses
PPAR-α genetic variants also participate in developing adaptive metabolic responses. Populations living in extreme environments, such as Tibetans, exhibit physiological traits adapted to a high altitude: decreased arterial oxygen content, increased resting ventilation, lack of hypoxic pulmonary vasoconstriction, and reduced hemoglobin (96). PPAR-α is involved in hypoxia signaling via upregulation of its downstream target Pdk4, restricting entry of glycolytic intermediates into the tricarboxylic acid cycle (97). A recent genome-wide scan in Tibetans identified a haplotype of PPAR-α (consisting of 139 SNPs) that was significantly associated with a decreased hemoglobin concentration phenotype that is unique to this high-altitude population (P < 0.0009) (98). The putatively advantageous PPAR-α haplotype is correlated with increased serum FFA concentrations, suggesting a possible decrease in the activity of fatty acid oxidation (P < 0.01) (99).
In healthy men who were given a single oral fat load primarily composed of saturated fatty acids, lower postprandial TG (P = 0.008) and cholesterol (P = 0.039) responses were observed in the small triglyceride-rich lipoprotein fraction by the noncoding variant 140+5435T > C (rs135549) (73).
Interaction between the PPARα gene and diet
It is well-known that the variability in the interindividual response to any type of dietary intervention and many factors including age, sex, physical activity, alcohol, smoking, genetic factors, and others influence this response. This interaction between genetic variability and the response to dietary factors will help to identify individuals or populations who can benefit from specific dietary recommendations (100). Therefore, environmental factors such as diet interact with the genetic background to modulate metabolic parameters. The effect of PPAR-α polymorphisms on the relationship between diet and metabolic components has been evaluated (Fig. 2).
The interaction between the L162V polymorphism and the consumption of saturated fat in 632 unrelated men from Quebec explains 2.71% of the variance in waist circumference (P = 0.01) (78). PUFAs can interact directly with PPAR-α, acting on cis-regulatory elements of genes and turning mRNA synthesis on or off (101). A study of 1003 male and 1103 female participants in the Framingham Study consuming their habitual diets showed a significant gene-nutrient interaction between L162V and total PUFA intake with plasma TG (P < 0.05) and apoC3 (P < 0.05) concentrations. The 162V allele was associated with greater plasma concentrations of TG and apoC3 in subjects consuming a diet low in PUFAs (<6% of energy), whereas when PUFA intake was high, carriers of the 162V allele had lower TG and apoC3, indicating a significant dose-response relationship between PUFA intake and serum TG concentrations depending on the genotype (102). Additionally, L162V and dietary fat intake interaction has effects on the peak particle diameter of LDL, a risk factor for cardiovascular diseases. V162 carriers with higher saturated fat intakes had smaller peak particle diameters of LDL than those with lower intakes (103).
Furthermore, in healthy white men from Quebec, carriers of the 162V allele had lower apoA1 concentrations after a high PUFA diet (P = 0.02) (104). In addition, subjects that followed a low-fat diet for 8 wk and then were supplemented daily with 5 g of fish oil for 6 wk showed a significant genotype-diet interaction on the plasma C-reactive protein concentration (P = 0.01) (105). However, a statistically significant interaction was observed between the V227A polymorphism, dietary PUFA intake, and serum HDL cholesterol in Chinese women (n = 751, P = 0.049). In women who carried the 227A allele, increasing dietary PUFA intake was associated with lower serum HDL cholesterol concentrations (88).
The effect of PPAR-α genetic variants between PUFA intake and lipid measures shows differences among ethnic groups. Differences in allele frequencies of PPAR-α SNPs among populations and the variation of genetic structures of the PPAR-α locus in diverse ethnic groups may influence genotype-diet interactions. For instance, in the large biethnic (10,134 whites and 3480 African Americans) ARIC study, a significant interaction between the PPAR-α 3′-UTR (untranslated region) G > A (rs6008259) and n6 fatty acid (linoleic acid) intake with total serum cholesterol (P = 0.03) and LDL cholesterol concentrations (P = 0.03) in white participants was detected. Additionally, interactions by genotype between long-chain n3 fatty acid (EPA + DHA) intake with TC (P = 0.03) and LDL cholesterol (P = 0.02) were observed in African Americans for the 3′-UTR C > T (rs3892755) (106). The functional consequences of both UTR alleles are unknown.
In addition, an interaction has been demonstrated between the PPAR-γ Pro12Ala (rs1801282) and PPAR-α L162V genotypes with dietary intake of fatty acids on plasma lipids (107, 108). In the RISCK study (parallel-design, randomized, controlled trial) 466 subjects were genotyped; at baseline, there was no significant interaction between the PPAR-α L162V and PPAR-γ Pro12Ala genotypes on plasma lipid concentrations. However, after the dietary intervention, there was a genotype interaction that significantly influenced LDL cholesterol (P = 0.0002), particularly the small dense LDL proportion of total LDL (P = 0.005) (107).
PPAR-α and fenofibrate treatment response in metabolism
Fibrates, amphipathic carboxylic acids that include gemfibrozil, bezafibrate, clofibrate, and fenofibrate, are synthetic PPAR-α ligands that bind to and activate PPAR-α transcriptional function, leading to the modulation of the expression of its target genes. Fibrates are effective in reducing TG and raising HDL cholesterol levels, which reduce cardiovascular disease risk in people with diabetes (109). In fact, recent studies of fibrate trials (Bezafibrate Infarction Prevention study, Helsinki Heart Study, Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial, and Fenofibrate Intervention and Event Lowering in Diabetes) support the evidence that patients with insulin resistance (as occurs in diabetes and/or metabolic syndrome) benefit from therapy that includes fibrates (110–112). People with type 2 diabetes have low HDL cholesterol levels and high TG levels, both of which are associated with a higher risk of coronary heart disease (109, 112–114). Fenofibrate modifies lipid parameters by changing LDL particle morphology, increasing HDL cholesterol, and reducing TGs. In addition, fenofibrate reduces systemic inflammatory markers independent of its effects on lipid metabolism (114–116). Administration of fenofibrate to patients with hyperlipidemia decreases plasma concentrations of proinflammatory mediators such as IL-6, TNF-α, interferon-γ, fibrinogen, and C-reactive protein (117).
However, there is significant interindividual variation in response to fenofibrate (118). The association between PPAR-α 7G > C polymorphism and the reduction in plasma TGs in response to fenofibrate treatment (micronized fenofibrate, 200 mg/d for at least 3 y) was evaluated in subjects with type 2 diabetes participating in the Diabetes Atherosclerosis Intervention Study. The frequency of the 7G > G genotype was higher in high TG responders (relative reduction of TG levels >30% after treatment) than in low TG responders (85% vs. 69%, P < 0.05) (119).
Findings in families from the GOLDN (Genetics of Lipid-Lowering Drugs and Diet Network) study (861 participants who received 160 mg micronized fenofibrate once daily for 3 wk) showed rare variations (minor allele frequency >1%, 13 variants) in the PPARα gene that are associated with a reduced treatment response. After adjusting for baseline, fasting TG concentration when carrying at least 1 rare variant was associated with a low fenofibrate response [OR = 6.46 (95% CI: 1.4, 30.8); P = 0.02] (120). Among the rare SNPs identified, 1 variant in intron 1 lies 35 base pairs upstream of exon 2 near 2 putatively conserved transcription factor binding sites and located in a region where chromatin modification marks have been identified: H3K4me1 and H3K4me3 (120).
In addition, in 861 subjects, Frazier-Wood et al. (121) identified PPARα variants associated with fenofibrate response with respect to reductions in LDL cholesterol (rs135550 and rs135549) and TGs (rs4253701) and changes in the concentration of plasma inflammation markers. Furthermore, the PPARα gene variant rs9626730 showed an association with fasting IL-2 [(P = 0.0002; q = 0.018); q value represents P value corrected for the number of tests run], and 4 other SNPs showed suggestive associations with TNF-α. This highlights the importance of genetic variation in PPAR-α in protecting against the effects on inflammatory markers.
In contrast, studies have not shown a significant association between the postfenofibrate treatment level of TGs and the L162V polymorphism in hypertriglyceridemic subjects (122), nondiabetic hyperlipidemic subjects (123), and subjects with type 2 diabetes (119).
PPAR-α in transgenerational environmental reprogramming of metabolism
In eukaryotic cells, DNA is packaged into chromatin, and covalent modifications on the histone proteins of the chromatin or on the nucleotide bases of the DNA can influence the expression of genes. When these modifications are heritable from 1 cell generation to the next, such modifications are referred to as epigenetic modifications and can bring about lasting changes in gene expression (124–126). Epigenetic alterations that arise around the time of conception or during early embryogenesis are amplified during development by cell division and somatic maintenance and thus affect a high proportion of cells in the fully grown organism (124). One implication of epigenetic inheritance systems is that they provide a potential mechanism by which parents could transfer information to their offspring about the environmental conditions that they experience (127).
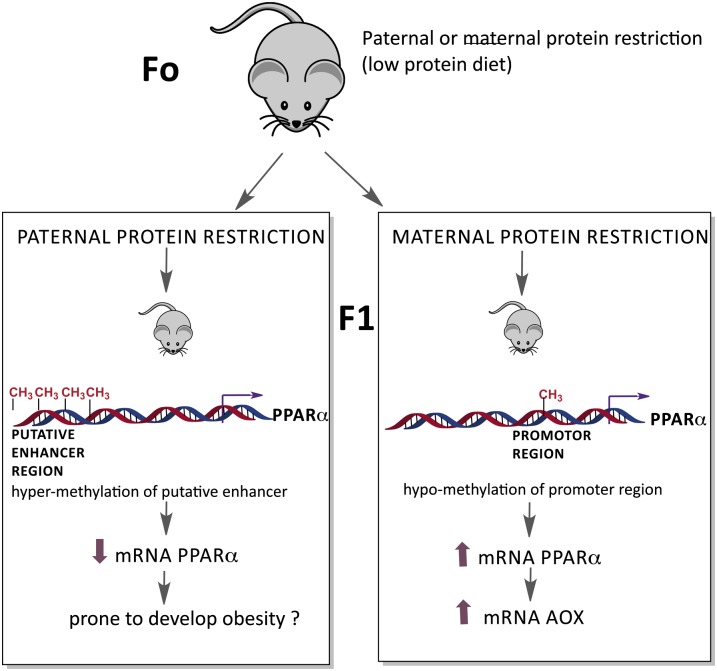
These epigenetic modifications can alter the expression of specific genes, such as PPAR-α, depending on the type of diet consumed. For instance, in rats fed a protein-restricted diet throughout pregnancy, PPAR-α gene methylation was 20.6% lower (P < 0.001), resulting in an 10.5-fold higher expression in the liver of the offspring after weaning, compared with dams fed a control diet (128). In a genomic screen for transgenerational effects of paternal diet, the expression of hundreds of genes changed in the offspring of males fed a low-protein diet with upregulation of genes coding for lipid and cholesterol biosynthetic pathways. Epigenomic profiling in the livers of offspring identified changes in cytosine methylation at a putative enhancer for PPARα, and these changes correlated with the downregulation of this gene (127). This transgenerational effect on liver metabolism was not linked to changes in DNA methylation in the sperm of the males under various dietary regimes.
In addition, hypomethylation of the PPARα promoter induced in the F1 offspring generation by maternal protein restriction during pregnancy was transmitted to the F2 offspring generation. As a result, hypomethylation tended (P < 0.1) to increase the expression of PPARα and its target gene acyl-CoA oxidase in the F1 and F2 males (129) (Fig. 3). Thus, environmental factors such as nutritional condition can generate long-lasting effects on gene expression associated with a specific chromatin state; however, despite considerable progress during recent years, many questions remain regarding the mechanisms that are involved in how the environment triggers these alterations in the epigenome.
Figure 3.
The effect of diet on PPAR-α expression mediated by changes in DNA methylation. Paternal protein restriction (low-protein diet) results in changes in cytosine methylation at a putative enhancer for PPARα, and these changes correlate with the downregulation of this gene in the offspring (127). In addition, hypomethylation of the PPAR-α promoter induced by maternal protein restriction during pregnancy is correlated with an increase in the expression of PPARα and its target gene acyl-CoA oxidase (129).
Conclusions
Recent studies demonstrate that the role of PPAR-α is not limited only to metabolism; however, its expression in primarily metabolic tissues (brown adipose tissue, liver, and kidney) and its role in the regulation of energetic homeostasis enable its function as a controller of metabolic adaptation under different nutritional states. For instance, PPAR-α is involved in the homeostasis of lipids in the fasted state as well as during the acute postprandial response to dietary fat.
Lifestyle, drugs, and dietary modifications, including consumption of foods that can activate PPAR-α, can affect its metabolic responses; however, the specific response will depend on the interactions between these factors and the genetic variants. Therefore, environmental factors and genetic background should be considered to determine the effect of different polymorphisms on disease risk. In addition, it is important to note that diet can have long-lasting effects on PPAR-α gene expression, and parental exposures may induce other epigenetic effects in addition to the current genetic and environmental factors underlying complex diseases.
Currently, the PPAR-α genetic variants studied and associated with clinical and biological effects are few; however, the expanding use of next-generation DNA sequencing technologies, including chromatin immunoprecipitation followed by DNA sequencing and global DNA methylation analysis, will allow the identification of additional variants and epigenetic modifications that may interact with some dietary nutrients leading to particular metabolic responses that are associated with the development of some diseases.
Acknowledgments
All authors have read and approved the final manuscript.
Footnotes
Abbreviations used: 16:0/18:1-GPC, phospholipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; PPRE, peroxisome proliferator responsive element; SNP, single nucleotide polymorphism; TC, total cholesterol; UTR, untranslated region.
Literature Cited
- 1.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–4 [DOI] [PubMed] [Google Scholar]
- 2.Jay MA, Ren J. Peroxisome proliferator-activated receptor (PPAR) in metabolic syndrome and type 2 diabetes mellitus. Curr Diabetes Rev. 2007;3:33–9 [DOI] [PubMed] [Google Scholar]
- 3.Vamecq J, Latruffe N. Medical significance of peroxisome proliferator-activated receptors. Lancet. 1999;354:141–8 [DOI] [PubMed] [Google Scholar]
- 4.Fruchart JC. Peroxisome proliferator-activated receptor-alpha (PPARalpha): at the crossroads of obesity, diabetes and cardiovascular disease. Atherosclerosis. 2009;205:1–8 [DOI] [PubMed] [Google Scholar]
- 5.Semple RK, Chatterjee VK, O'Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest. 2006;116:581–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson-Jurica MA, Schrader WT, O'Malley BW. Steroid receptor family: structure and functions. Endocr Rev. 1990;11:201–20 [DOI] [PubMed] [Google Scholar]
- 7.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–91 [DOI] [PubMed] [Google Scholar]
- 8.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–87 [DOI] [PubMed] [Google Scholar]
- 9.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704 [DOI] [PubMed] [Google Scholar]
- 10.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–50 [DOI] [PubMed] [Google Scholar]
- 11.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–25 [PubMed] [Google Scholar]
- 12.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyer C, Keller H, Mahfoudi A, Laudet V, Krey G, Wahli W. Positive regulation of the peroxisomal beta-oxidation pathway by fatty acids through activation of peroxisome proliferator-activated receptors (PPAR). Biol Cell. 1993;77:67–76 [DOI] [PubMed] [Google Scholar]
- 14.Osada S, Tsukamoto T, Takiguchi M, Mori M, Osumi T. Identification of an extended half-site motif required for the function of peroxisome proliferator-activated receptor alpha. Genes Cells. 1997;2:315–27 [DOI] [PubMed] [Google Scholar]
- 15.Krey G, Keller H, Mahfoudi A, Medin J, Ozato K, Dreyer C, Wahli W. Xenopus peroxisome proliferator activated receptors: genomic organization, response element recognition, heterodimer formation with retinoid X receptor and activation by fatty acids. J Steroid Biochem Mol Biol. 1993;47:65–73 [DOI] [PubMed] [Google Scholar]
- 16.Viswakarma N, Jia Y, Bai L, Vluggens A, Borensztajn J, Xu J, Reddy JK. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010; 2010. [DOI] [PMC free article] [PubMed]
- 17.Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, Jr, McKee DD, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin PG, Guillou H, Lasserre F, Dejean S, Lan A, Pascussi JM, Sancristobal M, Legrand P, Besse P, Pineau T. Novel aspects of PPARalpha-mediated regulation of lipid and xenobiotic metabolism revealed through a nutrigenomic study. Hepatology. 2007;45:767–77 [DOI] [PubMed] [Google Scholar]
- 21.Jump DB, Botolin D, Wang Y, Xu J, Christian B, Demeure O. Fatty acid regulation of hepatic gene transcription. J Nutr. 2005;135:2503–6 [DOI] [PubMed] [Google Scholar]
- 22.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–82 [DOI] [PubMed] [Google Scholar]
- 23.Lin Q, Ruuska SE, Shaw NS, Dong D, Noy N. Ligand selectivity of the peroxisome proliferator-activated receptor alpha. Biochemistry. 1999;38:185–90 [DOI] [PubMed] [Google Scholar]
- 24.Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–22 [DOI] [PubMed] [Google Scholar]
- 25.Sanderson LM, Degenhardt T, Koppen A, Kalkhoven E, Desvergne B, Muller M, Kersten S. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol Cell Biol. 2009;29:6257–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanderson LM, de Groot PJ, Hooiveld GJ, Koppen A, Kalkhoven E, Muller M, Kersten S. Effect of synthetic dietary triglycerides: a novel research paradigm for nutrigenomics. PLoS One. 2008; 3:e1681. [DOI] [PMC free article] [PubMed]
- 27.Gloerich J, van den Brink DM, Ruiter JP, van Vlies N, Vaz FM, Wanders RJ, Ferdinandusse S. Metabolism of phytol to phytanic acid in the mouse, and the role of PPARalpha in its regulation. J Lipid Res. 2007;48:77–85 [DOI] [PubMed] [Google Scholar]
- 28.Goto T, Takahashi N, Kato S, Egawa K, Ebisu S, Moriyama T, Fushiki T, Kawada T. Phytol directly activates peroxisome proliferator-activated receptor alpha (PPARalpha) and regulates gene expression involved in lipid metabolism in PPARalpha-expressing HepG2 hepatocytes. Biochem Biophys Res Commun. 2005;337:440–5 [DOI] [PubMed] [Google Scholar]
- 29.Radler U, Stangl H, Lechner S, Lienbacher G, Krepp R, Zeller E, Brachinger M, Eller-Berndl D, Fischer A, Anzur C, et al. A combination of (omega-3) polyunsaturated fatty acids, polyphenols and L-carnitine reduces the plasma lipid levels and increases the expression of genes involved in fatty acid oxidation in human peripheral blood mononuclear cells and HepG2 cells. Ann Nutr Metab. 2011;58:133–40 [DOI] [PubMed] [Google Scholar]
- 30.Esposito E, Mazzon E, Paterniti I, Dal Toso R, Pressi G, Caminiti R, Cuzzocrea S. PPAR-alpha contributes to the anti-inflammatory activity of verbascoside in a model of inflammatory bowel disease in mice. PPAR Res. 2010; 2010:917312. [DOI] [PMC free article] [PubMed]
- 31.Tsukamoto T, Nakata R, Tamura E, Kosuge Y, Kariya A, Katsukawa M, Mishima S, Ito T, Iinuma M, Akao Y, et al. Vaticanol C, a resveratrol tetramer, activates PPARalpha and PPARbeta/delta in vitro and in vivo. Nutr Metab (Lond). 2010;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue H, Jiang XF, Katayama T, Osada S, Umesono K, Namura S. Brain protection by resveratrol and fenofibrate against stroke requires peroxisome proliferator-activated receptor alpha in mice. Neurosci Lett. 2003;352:203–6 [DOI] [PubMed] [Google Scholar]
- 33.Mizuno CS, Ma G, Khan S, Patny A, Avery MA, Rimando AM. Design, synthesis, biological evaluation and docking studies of pterostilbene analogs inside PPARalpha. Bioorg Med Chem. 2008;16:3800–8 [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Shin HJ, Kim SY, Kim JH, Lee YS, Kim DH, Lee MO. Genistein enhances expression of genes involved in fatty acid catabolism through activation of PPARalpha. Mol Cell Endocrinol. 2004;220:51–8 [DOI] [PubMed] [Google Scholar]
- 35.Kim YI, Hirai S, Takahashi H, Goto T, Ohyane C, Tsugane T, Konishi C, Fujii T, Inai S, Iijima Y, et al. 9-oxo-10(E),12(E)-Octadecadienoic acid derived from tomato is a potent PPAR alpha agonist to decrease triglyceride accumulation in mouse primary hepatocytes. Mol Nutr Food Res. 2011;55:585–93 [DOI] [PubMed] [Google Scholar]
- 36.Jia Y, Kim JY, Jun HJ, Kim SJ, Lee JH, Hoang MH, Hwang KY, Um SJ, Chang HI, Lee SJ. The natural carotenoid astaxanthin, a PPAR-alpha agonist and PPAR-gamma antagonist, reduces hepatic lipid accumulation by rewiring the transcriptome in lipid-loaded hepatocytes. Mol Nutr Food Res. 2012;56:878–88 [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki K, Kuromitsu J, Tanaka I. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator- activated receptor alpha agonists. Biochem Biophys Res Commun. 2002;290:1114–22 [DOI] [PubMed] [Google Scholar]
- 38.Reifel-Miller A, Otto K, Hawkins E, Barr R, Bensch WR, Bull C, Dana S, Klausing K, Martin JA, Rafaeloff-Phail R, et al. A peroxisome proliferator-activated receptor alpha/gamma dual agonist with a unique in vitro profile and potent glucose and lipid effects in rodent models of type 2 diabetes and dyslipidemia. Mol Endocrinol. 2005;19:1593–605 [DOI] [PubMed] [Google Scholar]
- 39.Velkov T, Rimmer KA, Headey SJ. Ligand-enhanced expression and in-cell assay of human peroxisome proliferator-activated receptor alpha ligand binding domain. Protein Expr Purif. 2010;70:260–9 [DOI] [PubMed] [Google Scholar]
- 40.Kane CD, Stevens KA, Fischer JE, Haghpassand M, Royer LJ, Aldinger C, Landschulz KT, Zagouras P, Bagley SW, Hada W, et al. Molecular characterization of novel and selective peroxisome proliferator-activated receptor alpha agonists with robust hypolipidemic activity in vivo. Mol Pharmacol. 2009;75:296–306 [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Phillips DI, Wang C, Byrne CD. Human skeletal muscle PPARalpha expression correlates with fat metabolism gene expression but not BMI or insulin sensitivity. Am J Physiol Endocrinol Metab. 2004;286:E168–75 [DOI] [PubMed] [Google Scholar]
- 42.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–66 [DOI] [PubMed] [Google Scholar]
- 43.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–8 [DOI] [PubMed] [Google Scholar]
- 45.Motojima K, Passilly P, Peters JM, Gonzalez FJ, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J Biol Chem. 1998;273:16710–4 [DOI] [PubMed] [Google Scholar]
- 46.Aoyama T, Peters JM, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez FJ. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J Biol Chem. 1998;273:5678–84 [DOI] [PubMed] [Google Scholar]
- 47.Peters JM, Aoyama T, Cattley RC, Nobumitsu U, Hashimoto T, Gonzalez FJ. Role of peroxisome proliferator-activated receptor alpha in altered cell cycle regulation in mouse liver. Carcinogenesis. 1998;19:1989–94 [DOI] [PubMed] [Google Scholar]
- 48.Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotman N, Wahli W. Fatty acid synthesis and PPARalpha hand in hand. Chem Biol. 2009;16:801–2 [DOI] [PubMed] [Google Scholar]
- 51.Mandard S, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol Life Sci. 2004;61:393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah A, Rader DJ, Millar JS. The effect of PPAR-alpha agonism on apolipoprotein metabolism in humans. Atherosclerosis. 2010;210:35–40 [DOI] [PubMed] [Google Scholar]
- 53.Prieur X, Coste H, Rodriguez JC. The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-alpha and contains a novel farnesoid X-activated receptor response element. J Biol Chem. 2003;278:25468–80 [DOI] [PubMed] [Google Scholar]
- 54.Erol E, Kumar LS, Cline GW, Shulman GI, Kelly DP, Binas B. Liver fatty acid binding protein is required for high rates of hepatic fatty acid oxidation but not for the action of PPARalpha in fasting mice. FASEB J. 2004;18:347–9 [DOI] [PubMed] [Google Scholar]
- 55.Fernandez-Alvarez A, Alvarez MS, Gonzalez R, Cucarella C, Muntane J, Casado M. Human SREBP1c expression in liver is directly regulated by peroxisome proliferator-activated receptor alpha (PPARalpha). J Biol Chem. 2011;286:21466–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–25 [DOI] [PubMed] [Google Scholar]
- 57.Reitman ML. FGF21: a missing link in the biology of fasting. Cell Metab. 2007;5:405–7 [DOI] [PubMed] [Google Scholar]
- 58.Kersten S, Mandard S, Escher P, Gonzalez FJ, Tafuri S, Desvergne B, Wahli W. The peroxisome proliferator-activated receptor alpha regulates amino acid metabolism. FASEB J. 2001;15:1971–8 [DOI] [PubMed] [Google Scholar]
- 59.Makowski L, Noland RC, Koves TR, Xing W, Ilkayeva OR, Muehlbauer MJ, Stevens RD, Muoio DM. Metabolic profiling of PPARalpha−/− mice reveals defects in carnitine and amino acid homeostasis that are partially reversed by oral carnitine supplementation. FASEB J. 2009;23:586–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheikh K, Camejo G, Lanne B, Halvarsson T, Landergren MR, Oakes ND. Beyond lipids, pharmacological PPARalpha activation has important effects on amino acid metabolism as studied in the rat. Am J Physiol Endocrinol Metab. 2007;292:E1157–65 [DOI] [PubMed] [Google Scholar]
- 61.Hiukka A, Maranghi M, Matikainen N, Taskinen MR. PPARalpha: an emerging therapeutic target in diabetic microvascular damage. Nat Rev Endocrinol. 2010;6:454–63 [DOI] [PubMed] [Google Scholar]
- 62.Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, Fruchart JC, Tedgui A, Haegeman G, Staels B. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem. 1999;274:32048–54 [DOI] [PubMed] [Google Scholar]
- 63.Ramanan S, Kooshki M, Zhao W, Hsu FC, Robbins ME. PPARalpha ligands inhibit radiation-induced microglial inflammatory responses by negatively regulating NF-kappaB and AP-1 pathways. Free Radic Biol Med. 2008;45:1695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23:351–63 [DOI] [PubMed] [Google Scholar]
- 65.Zhou J, Zhang S, Xue J, Avery J, Wu J, Lind SE, Ding WQ. Activation of peroxisome proliferator-activated receptor alpha (PPARalpha) suppresses hypoxia-inducible factor-1alpha (HIF-1alpha) signaling in cancer cells. J Biol Chem. 2012;287:35161–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas M, Burk O, Klumpp B, Kandel BA, Damm G, Weiss TS, Klein K, Schwab M, Zanger UM. Direct transcriptional regulation of human hepatic cytochrome p450 3a4 (cyp3a4) by peroxisome proliferator-activated receptor alpha (PPARalpha). Mol Pharmacol. 2013;83:709–18 [DOI] [PubMed] [Google Scholar]
- 67.Sapone A, Peters JM, Sakai S, Tomita S, Papiha SS, Dai R, Friedman FK, Gonzalez FJ. The human peroxisome proliferator-activated receptor alpha gene: identification and functional characterization of two natural allelic variants. Pharmacogenetics. 2000;10:321–33 [DOI] [PubMed] [Google Scholar]
- 68.Myers KA, Lambe KG, Aldridge TC, Macdonald N, Tugwood JD. Amino acid residues in both the DNA-binding and ligand-binding domains influence transcriptional activity of the human peroxisome proliferator-activated receptor alpha. Biochem Biophys Res Commun. 1997;239:522–6 [DOI] [PubMed] [Google Scholar]
- 69.Flavell DM, Pineda Torra I, Jamshidi Y, Evans D, Diamond JR, Elkeles RS, Bujac SR, Miller G, Talmud PJ, Staels B, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43:673–80 [DOI] [PubMed] [Google Scholar]
- 70.Naito H, Yamanoshita O, Kamijima M, Katoh T, Matsunaga T, Lee CH, Kim H, Aoyama T, Gonzalez FJ, Nakajima T. Association of V227A PPARalpha polymorphism with altered serum biochemistry and alcohol drinking in Japanese men. Pharmacogenet Genomics. 2006;16:569–77 [DOI] [PubMed] [Google Scholar]
- 71.Burns KA, Vanden Heuvel JP. Modulation of PPAR activity via phosphorylation. Biochim Biophys Acta. 2007; 1771:952–960. [DOI] [PMC free article] [PubMed]
- 72.Yong EL, Li J, Liu MH. Single gene contributions: genetic variants of peroxisome proliferator-activated receptor (isoforms alpha, beta/delta and gamma) and mechanisms of dyslipidemias. Curr Opin Lipidol. 2008;19:106–12 [DOI] [PubMed] [Google Scholar]
- 73.Tanaka T, Ordovas JM, Delgado-Lista J, Perez-Jimenez F, Marin C, Perez-Martinez P, Gomez P, Lopez-Miranda J. Peroxisome proliferator-activated receptor alpha polymorphisms and postprandial lipemia in healthy men. J Lipid Res. 2007;48:1402–8 [DOI] [PubMed] [Google Scholar]
- 74.Andrulionyte L, Kuulasmaa T, Chiasson JL, Laakso M. Single nucleotide polymorphisms of the peroxisome proliferator-activated receptor-alpha gene (PPARA) influence the conversion from impaired glucose tolerance to type 2 diabetes: the STOP-NIDDM trial. Diabetes. 2007;56:1181–6 [DOI] [PubMed] [Google Scholar]
- 75.Flavell DM, Ireland H, Stephens JW, Hawe E, Acharya J, Mather H, Hurel SJ, Humphries SE. Peroxisome proliferator-activated receptor alpha gene variation influences age of onset and progression of type 2 diabetes. Diabetes. 2005;54:582–6 [DOI] [PubMed] [Google Scholar]
- 76.Vohl MC, Lepage P, Gaudet D, Brewer CG, Betard C, Perron P, Houde G, Cellier C, Faith JM, Despres JP, et al. Molecular scanning of the human PPARa gene: association of the L162v mutation with hyperapobetalipoproteinemia. J Lipid Res. 2000;41:945–52 [PubMed] [Google Scholar]
- 77.Lacquemant C, Lepretre F, Pineda Torra I, Manraj M, Charpentier G, Ruiz J, Staels B, Froguel P. Mutation screening of the PPARalpha gene in type 2 diabetes associated with coronary heart disease. Diabetes Metab. 2000;26:393–401 [PubMed] [Google Scholar]
- 78.Robitaille J, Brouillette C, Houde A, Lemieux S, Perusse L, Tchernof A, Gaudet D, Vohl MC. Association between the PPARalpha-L162V polymorphism and components of the metabolic syndrome. J Hum Genet. 2004;49:482–9 [DOI] [PubMed] [Google Scholar]
- 79.Tai ES, Demissie S, Cupples LA, Corella D, Wilson PW, Schaefer EJ, Ordovas JM. Association between the PPARA L162V polymorphism and plasma lipid levels: the Framingham Offspring Study. Arterioscler Thromb Vasc Biol. 2002;22:805–10 [DOI] [PubMed] [Google Scholar]
- 80.Uthurralt J, Gordish-Dressman H, Bradbury M, Tesi-Rocha C, Devaney J, Harmon B, Reeves EK, Brandoli C, Hansen BC, Seip RL, et al. PPARalpha L162V underlies variation in serum triglycerides and subcutaneous fat volume in young males. BMC Med Genet. 2007;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sparso T, Hussain MS, Andersen G, Hainerova I, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O. Relationships between the functional PPARalpha Leu162Val polymorphism and obesity, type 2 diabetes, dyslipidaemia, and related quantitative traits in studies of 5799 middle-aged white people. Mol Genet Metab. 2007;90:205–9 [DOI] [PubMed] [Google Scholar]
- 82.Shin MJ, Kanaya AM, Krauss RM. Polymorphisms in the peroxisome proliferator activated receptor alpha gene are associated with levels of apolipoprotein CIII and triglyceride in African-Americans but not Caucasians. Atherosclerosis. 2008;198:313–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doney AS, Fischer B, Lee SP, Morris AD, Leese G, Palmer CN. Association of common variation in the PPARA gene with incident myocardial infarction in individuals with type 2 diabetes: a Go-DARTS study. Nucl Recept. 2005;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tai ES, Collins D, Robins SJ, O'Connor JJ, Jr, Bloomfield HE, Ordovas JM, Schaefer EJ, Brousseau ME. The L162V polymorphism at the peroxisome proliferator activated receptor alpha locus modulates the risk of cardiovascular events associated with insulin resistance and diabetes mellitus: the Veterans Affairs HDL Intervention Trial (VA-HIT). Atherosclerosis. 2006;187:153–60 [DOI] [PubMed] [Google Scholar]
- 85.Mazzotti DR, Singulane CC, Ota VK, Rodrigues TP, Furuya TK, de Souza FJ, Cordeiro BG, Magalhaes C, Chen ES, Jacomini A, et al. PPARalpha polymorphisms as risk factors for dyslipidemia in a Brazilian population. Mol Genet Metab. 2011;102:189–93 [DOI] [PubMed] [Google Scholar]
- 86.Yamakawa-Kobayashi K, Ishiguro H, Arinami T, Miyazaki R, Hamaguchi HA. Val227Ala polymorphism in the peroxisome proliferator activated receptor alpha (PPARalpha) gene is associated with variations in serum lipid levels. J Med Genet. 2002;39:189–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hara M, Wang X, Paz VP, Iwasaki N, Honda M, Iwamoto Y, Bell GI. Identification of three missense mutations in the peroxisome proliferator-activated receptor alpha gene in Japanese subjects with maturity-onset diabetes of the young. J Hum Genet. 2001;46:285–8 [DOI] [PubMed] [Google Scholar]
- 88.Chan E, Tan CS, Deurenberg-Yap M, Chia KS, Chew SK, Tai ES. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis. 2006;187:309–15 [DOI] [PubMed] [Google Scholar]
- 89.Liu MH, Li J, Shen P, Husna B, Tai ES, Yong EL. A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin. Mol Endocrinol. 2008;22:1078–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Naito H, Kamijima M, Yamanoshita O, Nakahara A, Katoh T, Tanaka N, Aoyama T, Gonzalez FJ, Nakajima T. Differential effects of aging, drinking and exercise on serum cholesterol levels dependent on the PPARA-V227A polymorphism. J Occup Health. 2007;49:353–62 [DOI] [PubMed] [Google Scholar]
- 91.Jamshidi Y, Montgomery HE, Hense HW, Myerson SG, Torra IP, Staels B, World MJ, Doering A, Erdmann J, Hengstenberg C, et al. Peroxisome proliferator–activated receptor alpha gene regulates left ventricular growth in response to exercise and hypertension. Circulation. 2002;105:950–5 [DOI] [PubMed] [Google Scholar]
- 92.Flavell DM, Jamshidi Y, Hawe E, Pineda Torra I, Taskinen MR, Frick MH, Nieminen MS, Kesaniemi YA, Pasternack A, Staels B, et al. Peroxisome proliferator-activated receptor alpha gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation. 2002;105:1440–5 [DOI] [PubMed] [Google Scholar]
- 93.Balcerzyk A, Zak I, Krauze J. Synergistic effect between polymorphisms of PPARA and ABCA1 genes on the premature coronary artery disease. Acta Cardiol. 2007;62:233–8 [DOI] [PubMed] [Google Scholar]
- 94.Purushothaman S, Ajitkumar VK, Renuka Nair R. Association of PPARalpha intron 7 polymorphism with coronary artery disease: a cross-sectional study. ISRN Cardiol. 2011;2011:816025. [DOI] [PMC free article] [PubMed]
- 95.Chen ES, Mazzotti DR, Furuya TK, Cendoroglo MS, Ramos LR, Araujo LQ, Burbano RR, Smith Mde A. Association of PPARalpha gene polymorphisms and lipid serum levels in a Brazilian elderly population. Exp Mol Pathol. 2010;88:197–201 [DOI] [PubMed] [Google Scholar]
- 96.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 2007;104: Suppl 1:8655–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aragones J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40:170–80 [DOI] [PubMed] [Google Scholar]
- 98.Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–5 [DOI] [PubMed] [Google Scholar]
- 99.Ge RL, Simonson TS, Cooksey RC, Tanna U, Qin G, Huff CD, Witherspoon DJ, Xing J, Zhengzhong B, Prchal JT, et al. Metabolic insight into mechanisms of high-altitude adaptation in Tibetans. Mol Genet Metab. 2012;106:244–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ordovas JM. Genotype-phenotype associations: modulation by diet and obesity. Obesity (Silver Spring). 2008;16: Suppl 3:S40–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Price PT, Nelson CM, Clarke SD. Omega-3 polyunsaturated fatty acid regulation of gene expression. Curr Opin Lipidol. 2000;11:3–7 [DOI] [PubMed] [Google Scholar]
- 102.Tai ES, Corella D, Demissie S, Cupples LA, Coltell O, Schaefer EJ, Tucker KL, Ordovas JM. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C–III concentrations in the Framingham Heart Study. J Nutr. 2005;135:397–403 [DOI] [PubMed] [Google Scholar]
- 103.Bouchard-Mercier A, Godin G, Lamarche B, Perusse L, Vohl MC. Effects of peroxisome proliferator-activated receptors, dietary fat intakes and gene-diet interactions on peak particle diameters of low-density lipoproteins. J Nutrigenet Nutrigenomics. 2011;4:36–48 [DOI] [PubMed] [Google Scholar]
- 104.Paradis AM, Fontaine-Bisson B, Bosse Y, Robitaille J, Lemieux S, Jacques H, Lamarche B, Tchernof A, Couture P, Vohl MC. The peroxisome proliferator-activated receptor alpha Leu162Val polymorphism influences the metabolic response to a dietary intervention altering fatty acid proportions in healthy men. Am J Clin Nutr. 2005;81:523–30 [DOI] [PubMed] [Google Scholar]
- 105.Caron-Dorval D, Paquet P, Paradis AM, Rudkowska I, Lemieux S, Couture P, Vohl MC. Effect of the PPAR-Alpha L162V polymorphism on the cardiovascular disease risk factor in response to n-3 polyunsaturated fatty acids. J Nutrigenet Nutrigenomics. 2008;1:205–12 [DOI] [PubMed] [Google Scholar]
- 106.Volcik KA, Nettleton JA, Ballantyne CM, Boerwinkle E. Peroxisome proliferator-activated receptor [alpha] genetic variation interacts with n-6 and long-chain n-3 fatty acid intake to affect total cholesterol and LDL-cholesterol concentrations in the Atherosclerosis Risk in Communities Study. Am J Clin Nutr. 2008;87:1926–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alsaleh A, Frost GS, Griffin BA, Lovegrove JA, Jebb SA, Sanders TA, O'Dell SD. PPARgamma2 gene Pro12Ala and PPARalpha gene Leu162Val single nucleotide polymorphisms interact with dietary intake of fat in determination of plasma lipid concentrations. J Nutrigenet Nutrigenomics. 2011;4:354–66 [DOI] [PubMed] [Google Scholar]
- 108.AlSaleh A, Sanders TA, O'Dell SD. Effect of interaction between PPARG, PPARA and ADIPOQ gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc Nutr Soc. 2012;71:141–53 [DOI] [PubMed] [Google Scholar]
- 109.Tonkin A, Hunt D, Voysey M, Kesaniemi A, Hamer A, Waites J, Mahar L, Mann S, Glasziou P, Forder P, et al. Effects of fenofibrate on cardiovascular events in patients with diabetes, with and without prior cardiovascular disease: The Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. Am Heart J. 2012;163:508–14 [DOI] [PubMed] [Google Scholar]
- 110.Tenenbaum A, Fisman EZ, Motro M, Adler Y. Atherogenic dyslipidemia in metabolic syndrome and type 2 diabetes: therapeutic options beyond statins. Cardiovasc Diabetol. 2006;5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keating GM. Fenofibrate: a review of its lipid-modifying effects in dyslipidemia and its vascular effects in type 2 diabetes mellitus. Am J Cardiovasc Drugs. 2011;11:227–47 [DOI] [PubMed] [Google Scholar]
- 112.Keech A, Simes RJ, Barter P, Best J, Scott R, Taskinen MR, Forder P, Pillai A, Davis T, Glasziou P, et al. Effects of long-term fenofibrate therapy on cardiovascular events in 9795 people with type 2 diabetes mellitus (the FIELD study): randomised controlled trial. Lancet. 2005;366:1849–61 [DOI] [PubMed] [Google Scholar]
- 113.Taskinen MR. Diabetic dyslipidaemia: from basic research to clinical practice. Diabetologia. 2003;46:733–49 [DOI] [PubMed] [Google Scholar]
- 114.Scott R, O'Brien R, Fulcher G, Pardy C, D'Emden M, Tse D, Taskinen MR, Ehnholm C, Keech A. Effects of fenofibrate treatment on cardiovascular disease risk in 9,795 individuals with type 2 diabetes and various components of the metabolic syndrome: the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. Diabetes Care. 2009;32:493–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Belfort R, Berria R, Cornell J, Cusi K. Fenofibrate reduces systemic inflammation markers independent of its effects on lipid and glucose metabolism in patients with the metabolic syndrome. J Clin Endocrinol Metab. 2010;95:829–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Madej A, Okopien B, Kowalski J, Zielinski M, Wysocki J, Szygula B, Kalina Z, Herman ZS. Effects of fenofibrate on plasma cytokine concentrations in patients with atherosclerosis and hyperlipoproteinemia IIb. Int J Clin Pharmacol Ther. 1998;36:345–9 [PubMed] [Google Scholar]
- 118.Keating GM, Ormrod D. Micronised fenofibrate: an updated review of its clinical efficacy in the management of dyslipidaemia. Drugs. 2002;62:1909–44 [DOI] [PubMed] [Google Scholar]
- 119.Foucher C, Rattier S, Flavell DM, Talmud PJ, Humphries SE, Kastelein JJ, Ayyobi A, Pimstone S, Frohlich J, Ansquer JC, et al. Response to micronized fenofibrate treatment is associated with the peroxisome-proliferator-activated receptors alpha G/C intron7 polymorphism in subjects with type 2 diabetes. Pharmacogenetics. 2004;14:823–9 [DOI] [PubMed] [Google Scholar]
- 120.Irvin MR, Zhang Q, Kabagambe EK, Perry RT, Straka RJ, Tiwari HK, Borecki IB, Shimmin LC, Stuart C, Zhong Y, et al. Rare PPARA variants and extreme response to fenofibrate in the Genetics of Lipid-Lowering Drugs and Diet Network Study. Pharmacogenet Genomics. 2012;22:367–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frazier-Wood AC, Ordovas JM, Straka RJ, Hixson JE, Borecki IB, Tiwari HK, Arnett DK. The PPAR alpha gene is associated with triglyceride, low-density cholesterol and inflammation marker response to fenofibrate intervention: the GOLDN study. Pharmacogenomics J. Epub 2012 May 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brisson D, Ledoux K, Bosse Y, St-Pierre J, Julien P, Perron P, Hudson TJ, Vohl MC, Gaudet D. Effect of apolipoprotein E, peroxisome proliferator-activated receptor alpha and lipoprotein lipase gene mutations on the ability of fenofibrate to improve lipid profiles and reach clinical guideline targets among hypertriglyceridemic patients. Pharmacogenetics. 2002;12:313–20 [DOI] [PubMed] [Google Scholar]
- 123.Puckey LH, Knight BL. Variation at position 162 of peroxisome proliferator-activated receptor alpha does not influence the effect of fibrates on cholesterol or triacylglycerol concentrations in hyperlipidaemic subjects. Pharmacogenetics. 2001;11:619–24 [DOI] [PubMed] [Google Scholar]
- 124.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109 [DOI] [PubMed] [Google Scholar]
- 125.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–96 [DOI] [PubMed] [Google Scholar]
- 126.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–6 [DOI] [PubMed] [Google Scholar]
- 129.Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA. Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr. 2007;97:435–9 [DOI] [PMC free article] [PubMed] [Google Scholar]