Abstract
Histone acetyltransferases (HATs) and ATP-dependent chromatin remodeling factors (ADCRs) are involved in selective gene regulation via modulation of local chromatin configuration. Activation of the recombination hotspot ade6-M26 of Schizosaccharomyces pombe is mediated by a cAMP responsive element (CRE)-like sequence, M26, and a heterodimeric ATF/CREB transcription factor, Atf1·Pcr1. Chromatin remodeling occurs meiotically around M26. We examined the roles of HATs and ADCRs in chromatin remodeling around M26. Histones H3 and H4 around M26 were hyperacetylated in an M26- and Atf1-dependent manner early in meiosis. SpGcn5, the S. pombe homolog of Gcn5p, was required for the majority of histone H3 acetylation around M26 in vivo. Deletion of gcn5+ caused a significant delay in chromatin remodeling but only partial reduction of M26 meiotic recombination frequency. The snf22+ (a Swi2/Snf2-ADCR homologue) deletion and snf22+gcn5+ double deletion abolished chromatin remodeling and significant reduction of meiotic recombination around M26. These results suggest that HATs and ADCRs cooperatively alter local chromatin structure, as in selective transcription activation, to activate meiotic recombination at M26 in a site-specific manner.
Keywords: ATP-dependent chromatin remodeling factor, chromatin remodeling, CRE, histone acetylation, meiotic recombination hotspot
Introduction
Eucaryotic chromosomal DNA is packaged into a highly condensed chromatin structure, which inhibits various DNA-associated processes, such as replication, transcription, and recombination, presumably by preventing the loading of trans-acting factors onto target DNA sites. Thus, the local chromatin structure around cis-acting DNA sites should be converted into an open configuration before the initiation of DNA-associated processes (Wolffe, 1997).
Chromatin-modifying machineries, such as histone acetyltransferase (HAT) complexes and ATP-dependent chromatin remodeling factors (ADCRs), are often recruited to promoters via sequence-specific DNA binding proteins, bound to their target sites, to activate transcription (Cosma et al, 1999; Krebs et al, 1999; Agalioti et al, 2000). Chromatin-modifying machineries may also activate other DNA-associated reactions. For example, histone acetylation is involved in DNA repair and site-specific V(D)J recombination (McBlane et al, 1995; McMurry and Krangel, 2000; Bird et al, 2002).
Homologous recombination is elevated markedly in meiosis and contributes to the genetic diversity of the next generation and the proper segregation of meiotic chromosomes. Most meiotic recombination in Saccharomyces cerevisiae is initiated by transient, meiosis-specific DNA-double strand breaks (DSBs) that map to hotspots for meiotic gene conversion. Such DSB sites are often found in transcription promoters that show hypersensitivity to nucleases (Ohta et al, 1994; Wu and Lichten, 1994). Meiotic DSBs also initiate recombination in fission yeast (Cervantes et al, 2000; Zenvirth and Simchen 2000). These observations suggest that the chromatin structure and its modification, possibly mediated by some sequence-specific transcriptional activators, are important to regulate the initiation of meiotic homologous recombination. In support of this idea, certain transcription factors influence DSB formation at ‘α-hotspots' in S. cerevisiae (Petes, 2001).
The ade6-M26 (M26) locus in the fission yeast Schizosaccharomyces pombe is a well-characterized meiotic recombination hotspot that provides a good model system for studies of recombination regulation. The ade6-M26 allele is a single G/T transversion in the 5′ end of the ade6 coding region (Ponticelli et al, 1988; Szankasi et al, 1988). This mutation creates a nonsense codon and also confers an up to 15-fold, meiosis-specific elevation of recombination as compared to control alleles such as ade6-M375 (M375) (Gutz, 1971; Ponticelli et al, 1988; Schuchert et al, 1991). The ade6-M375 allele is also a G/T base substitution that creates a nonsense mutation in the codon adjacent to that altered by the M26 mutation, but M375 does not show the hotspot activity. Thus, the ade-M375 allele provides an excellent negative control for studies of hotspot recombination at ade6-M26.
Recent studies have revealed a molecular basis for hotspot recombination at ade6-M26. Atf1 and Pcr1, which both belong to the ATF/CREB transcription factor family, bind as a heterodimer specifically to a heptameric DNA sequence, ATGACGT, which is created by the M26 mutation and is similar to the CRE (cAMP Response Element, TGACGT) sequence (Wahls and Smith, 1994; Kon et al, 1997). Binding of the Atf1·Pcr1 complex to this ‘M26 DNA site' is required for the recombination hotspot activity (Wahls and Smith, 1994; Kon et al, 1997). Interestingly, the Atf1·Pcr1 heterodimer is a transcription factor involved in meiotic induction, but it is dispensable for the steady-state levels of ade6 transcription (Wahls and Smith, 1994; Kon et al, 1997). Activation of meiotic recombination at M26 is at least partly due to meiosis-specific formation of DSBs around M26, which are dependent upon Rec12 (the S. pombe homologue of Spo11) and Pcr1 (Steiner et al, 2002).
We previously reported that the chromatin structure around ade6-M26 is remodeled meiotically (Mizuno et al, 1997). The nucleosome phasing along the ade6 open reading frame is lost, and in turn a micrococcal nuclease (MNase)-sensitive site appears de novo. This process, referred to as M26 chromatin remodeling, is regulated by meiosis-inducing signaling pathways (Mizuno et al, 2001). In addition, similar chromatin remodeling occurs around naturally occurring CRE-related sequences, such as ctt1+ and fbp1+ promoter sequences (Mizuno et al, 2001; Hirota et al, 2003), controlled by cAMP-dependent kinase (PKA) and stress-activated kinase (SAPK) pathways and fission yeast Tup1-like corepressors (Hirota et al, 2003). This suggests that M26 chromatin remodeling might reflect an intrinsic physiological response occurring at S. pombe natural CRE-related sequences.
These findings, together with the dual roles of the Atf1·Pcr1 heterodimer in meiotic development and hotspot activation (Kon et al, 1998), suggest that the induction of recombination in meiosis may be partly regulated at the chromatin or DNA accessibility level, and may be coupled in some way to transcriptional regulation in a developmental stage-specific manner. To further understand the mechanisms underlying the induction of meiotic recombination, we have studied the roles of histone acetylation in M26 chromatin remodeling. Here we report roles of the Atf1·Pcr1-, SpGcn5 HAT-mediated histone acetylation, and SpSnf22 ADCR-like factor in chromatin remodeling and meiotic recombination at the M26 hotspot. These results provide important insights into molecular mechanisms on site-specific chromatin regulation at CRE-like sequences in the activation of meiotic recombination.
Results
Atf1 and Pcr1 are required for chromatin remodeling at ade6-M26
Activation of the M26 recombination hotspot requires Atf1·Pcr1 (Kon et al, 1997), which is constitutively expressed and binds to the M26 DNA site, as revealed by a methylation interference assay (Kon et al, 1998) and a chromatin immunoprecipitation (Ch-IP) assay (W. Wahls, unpublished results). To study the molecular basis of M26 chromatin remodeling, we first analyzed the effects of atf1Δ and pcr1Δ (null) mutations on chromatin structure. Wild-type (atf1+ pcr1+), atf1Δ, pcr1Δ, and atf1Δ pcr1Δ mutant cells were cultured in a pre-sporulation medium and were then cultured in a sporulation medium prior to analysis of chromatin structure. In atf1+ pcr1+ cells, the chromatin structure around ade6-M26 changed by 3 h of meiosis, while little or no change was observed around M375 (Figure 1A). However, in atf1Δ or pcr1Δ cells, the M26 DNA site-dependent chromatin remodeling was not observed at 3 h and even at 6 h (data not shown) after meiotic induction. Similarly, atf1Δ pcr1Δ double mutant cells did not exhibit chromatin remodeling at M26 (Figure 1A). We conclude that both Atf1 and Pcr1 are strictly required for the meiotic chromatin remodeling around ade6-M26.
Figure 1.
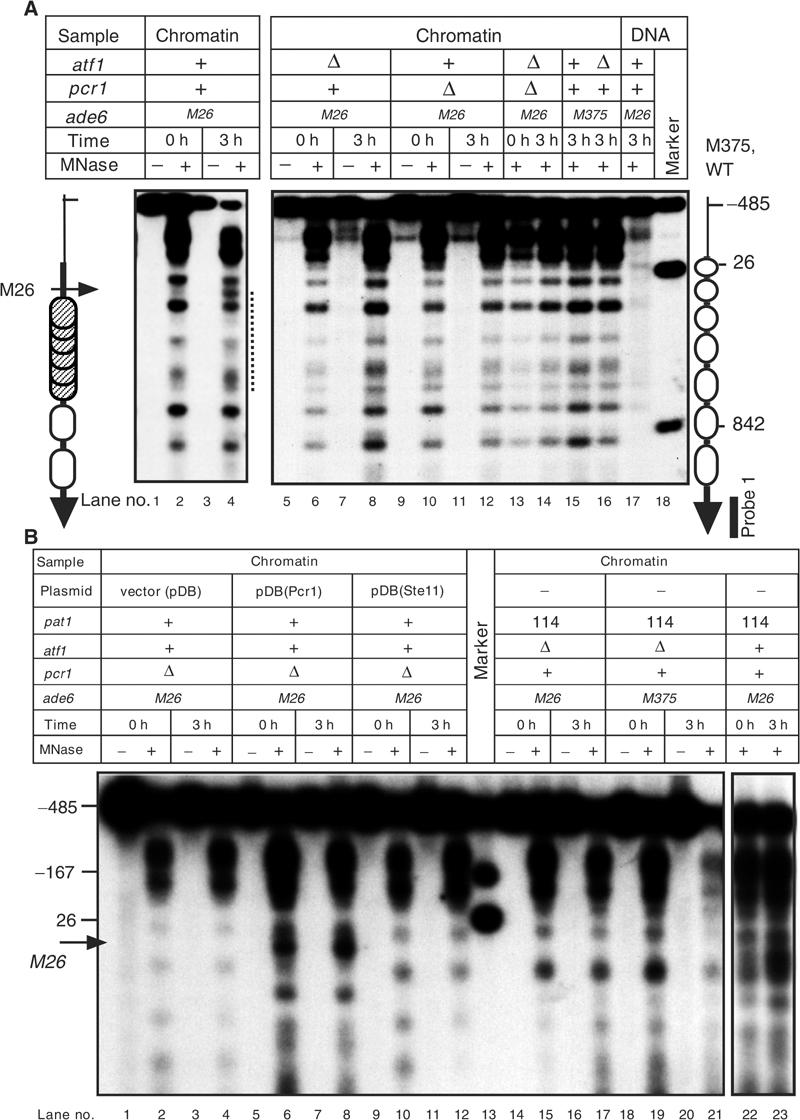
Atf1·Pcr1 is required for meiotic chromatin remodeling around ade6-M26. (A) Disruption of atf1+ or pcr1+ abolishes meiotic chromatin remodeling around ade6-M26. Diploid strains ELD205 (ade6-M26), WSP779 (ade6-M26, atf1Δ), WSP857 (ade6-M26, pcr1Δ), WSP859 (ade6-M26, atf1Δ, pcr1Δ), ELD203 (ade6-M375), and WSP780 (ade6-M375, atf1Δ) were cultured in presporulation medium (lanes 0 h). Cells were then transferred to sporulation medium and cultured further for 3 h (lanes 3 h). Chromatin isolated from cells was digested with 0 (lanes −) or 20 (lanes +) units/ml of MNase and analyzed as described (Mizuno et al, 1997). Probe 1 was used for indirect end labeling. The vertical and the horizontal arrows indicate the ade6 ORF and the position of the M26 mutation, respectively. Numbers by the right vertical arrow show the positions in nucleotides of the Xho I (−485), Bam HI (26), and Hind III (842) sites relative to the first A of the ade6 coding region. Open and hatched ovals represent phased and randomly positioned nucleosomes, respectively. The broken line by lane 4 indicates the region where the chromatin structure is remodeled. (B) Ectopically induced meiosis does not induce chromatin remodeling in atf1Δ and pcr1Δ cells. (Lanes 1–12) Chromatin structures of WSP857 (ade6-M26, atf1+, pcr1Δ) diploids harboring the empty vector (lanes vector(pDB)), the Pcr1 expressing plasmid (lanes pDB(Pcr1)), and the Ste11 expressing plasmid (lanes pDB(Ste11)) were analyzed. Experiments were performed as in Figure 1A. (Lanes 14–23) The K213 (ade6-M26, atf1Δ, pat1-114), K214 (ade6-M375, atf1Δ, pat1- 114), and GP1725x (ade6-M26, pat1-114) cells were cultured for 24 h in presporulation medium (lanes 0 h). Cells were transferred to sporulation medium and cultured at 34°C for 3 h (lanes 3 h). Chromatin analyses were performed as in Figure 1A. Overexpression of Pcr1 caused chromatin remodeling in premeiotic cells (lane 6). This may be due to artificial activation of chromatin remodeling by enhanced binding of Atf1·Pcr1 to the M26 site.
As Atf1 and Pcr1 are transcription factors that induce some genes during meiotic differentiation (Takeda et al, 1995; Kanoh et al, 1996; Watanabe and Yamamoto, 1996), it was possible that the deletion of atf1+ or pcr1+ indirectly affected the chromatin remodeling around ade6-M26. Kon et al. previously ruled out the possibility that the loss of atf1+ or pcr1+ decreased ade6 transcription and consequently abolished the chromatin remodeling (Kon et al, 1997). The entire loss of the M26 chromatin remodeling in the atf1Δ and the pcr1Δ strains is also unlikely to be due to their deficiencies in the meiotic proficiency, since nearly half of the mutant cells underwent sporulation. However, it remained possible that Atf1·Pcr1 regulated the expression of other meiotic factors that mediated the chromatin remodeling.
To test this hypothesis more rigorously, we used two approaches to bypass the requirement for the Atf1·Pcr1 heterodimer in meiotic induction. First, we induced meiosis ectopically by expressing Ste11 (Sugimoto et al, 1991) in pcr1Δ cells. Second, we induced meiosis ectopically in atf1Δ cells by inactivation of the Pat1 kinase (Iino and Yamamoto, 1985). Each approach alleviated the early meiotic defects of pcr1Δ and atf1Δ cells (Watanabe and Yamamoto, 1996), but did not compensate for the loss of the M26 chromatin remodeling, even though meiosis was almost fully induced in these mutant cells (Figure 1B). These results, together with the fact that Atf1·Pcr1 occupies the M26 site in cells, led us to conclude that the M26 chromatin remodeling is mediated directly in cis by the Atf1·Pcr protein bound to the M26 DNA site.
Histones around ade6-M26 are hyperacetylated
HATs are often recruited by sequence-specific transcriptional activators, and thereafter convert the local chromatin configuration into a state suitable for active transcription (Brown et al, 2000). This led us to speculate that the Atf1·Pcr1 heterodimer might recruit HATs to the M26 site and mediate the chromatin remodeling through histone acetylation. To test this idea, we used Ch-IP for the analysis on the state of histone acetylation around M26. Cells were induced to enter meiosis and cell extracts were subjected to Ch-IP using antibodies specific for acetylated isoforms of histones. The DNA from the immunoprecipitated chromatin and the input material was placed on slot blots and was hybridized with either total genomic DNA (serving as a reference for global histone acetylation levels) or with DNA sequences around the M26 hotspot.
We reproducibly detected stronger signals on the blots of immunoprecipitated chromatin from the M26 cells as compared to those from the M375 cells (Figure 2A). Quantitative analysis demonstrated that the histones H3 and H4 around ade6-M26 were highly acetylated at 0.5 h after the meiotic induction. The histone H3 and H4 acetylation levels at M26 (about 20% of the total input fraction) were 2- and 1.5-fold higher, respectively, than those at ade6-M375, (Figure 2B and C), and were ∼20-fold higher than the global (genome average) levels (Figure 2B and C, about 1% of the total input fraction). The relatively high levels of histone acetylation around ade6-M375 (5–10% of the total input fraction), compared to the global levels, may be due to the active transcription of the ade6 gene, which is an essential housekeeping gene. The histone acetylation levels gradually decreased to 50% of their maximal values by 2 h after the meiotic induction, a time point when premeiotic DNA synthesis was almost completed and the M26 chromatin remodeling was initiated (Mizuno et al, 1997). Separate experiments revealed that the global levels of histone acetylation in cells harboring ade6-M26 were similar to those in ade6-M375 cells (Figure 2B-C, broken lines). These results demonstrate that histones at the ade6 locus are hyperacetylated in an M26 DNA site-dependent manner.
Figure 2.
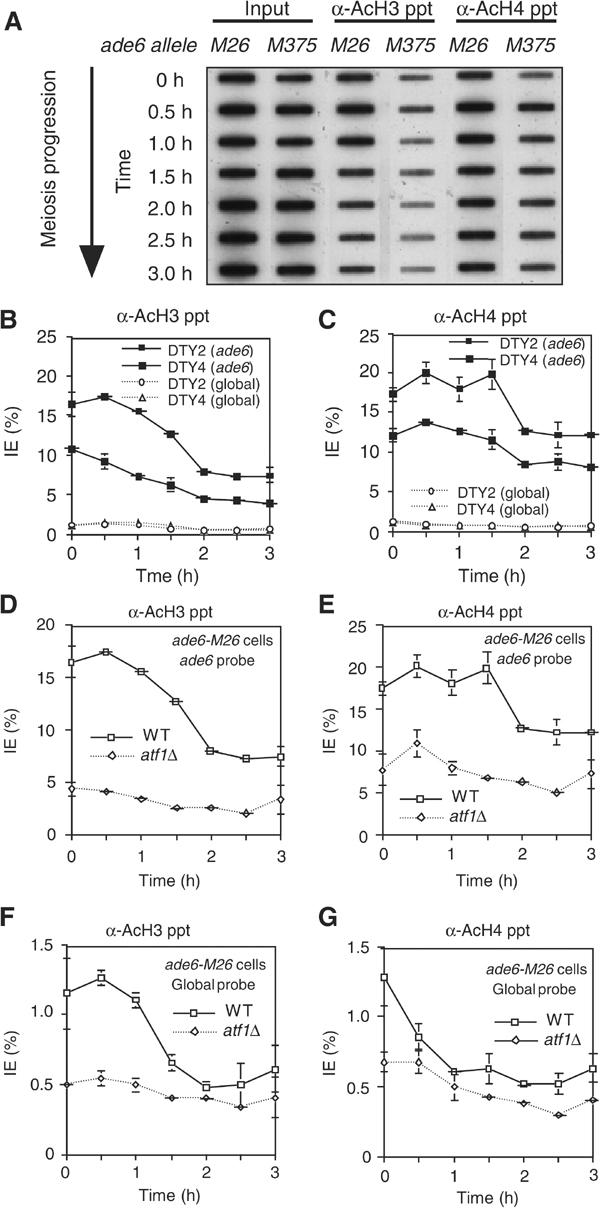
Histones H3 and H4 around ade6-M26 are highly acetylated in an Atf1-dependent manner during meiosis. Chromatin obtained from DTY2 (ade6-M26), DTY4 (ade6-M375), and WSP779 (atf1Δ, ade6-M26) diploid cells was immunoprecipitated with antiacetylated histone H3 and H4 antibodies. DNA obtained from input and immunoprecipitated material was applied to slot blots. (A) Example of primary data showing autoradiograms of slot blots probed for the ade6-M26 or ade6-M375 region. (B–G) Quantitative data are expressed as the ratio of the bound to the input material (immunoprecipitation efficiency: IE). All data were averages of at least three independent experiments. Labels inset into each figure panel indicate the relevant cell genotype and the probe used for each experiment. The data for WT in panels D–G are the same as those in panels B, C (solid line) and B, C (broken line). (B, C) Effects of ade6 alleles on acetylation of histone H3 and H4, respectively. (D, E) Requirement for Atf1 in histone H3 and H4 acetylation at ade6-M26, respectively. (F, G) Role of Atf1 in global (genome average) acetylation of histone H3 and H4, respectively.
We next applied the Ch-IP analysis to atf1Δ cells, in which no meiotic chromatin remodeling was observed around M26 (see Figure 1). As shown in Figure 2D and E, the acetylation of histones H3 and H4 around ade6-M26 was greatly reduced in the atf1Δ cells, indicating that histone acetylation around M26 requires Atf1. Interestingly, the global levels of histone acetylation were also reduced significantly in atf1Δ cells (Figure 2F and G).
S. pombe Gcn5p is a HAT
We next sought to identify the HAT that may be involved in histone acetylation around M26. Referring to the amino-acid sequences of the conserved HAT domains and bromodomains of budding yeast and human Gcn5p, we searched for a fission yeast homologue of Gcn5p by a BLAST search. One gene, SPAC1952.05, showed the highest similarity to yeast and human Gcn5p (Figure 3A). SPAC1952.05 was previously identified as a gene encoding a putative S. pombe homologue of Gcn5p, and the gene product was found in a SAGA-like complex (Mitsuzawa et al, 2001), although it was unknown whether the protein had HAT activity.
Figure 3.
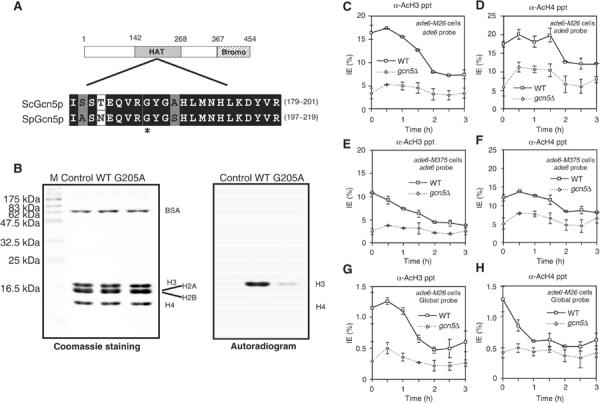
HAT activity of SpGcn5 in vitro and in vivo around M26 (A) Sequence alignments of ScGcn5p and SpGcn5 showing identical (dark gray) and conserved (light gray) amino acids in motif ‘A' of the HAT domain. The asterisk indicates the glycine residue replaced with alanine in SpGcn5(G205A). (B) SpGcn5 has histone (H3>H4) acetyltransferase activity in vitro. Recombinant SpGcn5 and SpGcn5(G205A) proteins were purified and assayed for HAT activity by incubation with human histone octamers and [14C] acetyl-CoA. Labeled proteins were separated by SDS–PAGE, stained with Coomassie brilliant blue (left panel), and subjected to autoradiography (right panel) to reveal acetylated histones (control, no enzyme). (C–H) Chromatin obtained from DTY9 (gcn5Δ, ade6-M26) and DTY11 (gcn5Δ, ade6-M375) cells was analyzed as in Figure 2. All data are averages of at least three independent experiments. Labels inset into each figure panel indicate the relevant cell genotype and the probe used for each experiment. The data for WT in panels C–H are same as those in Figures 2B, C, F and G. Requirement for Gcn5 in histone H3 (C, E) and H4 (D, F) acetylation at ade6-M26 (C, D) and at ade6-M375 (E, F). (G, H) Role of Gcn5 in global (genome average) acetylation of histone H3 and H4, respectively.
To test the hypothesis that SPAC1952.05 encoded a HAT enzyme, we produced and purified bacterially expressed, wild-type protein and a protein with an alanine substitution at the glycine205. The Gly205 residue is in the putative acetyl CoA-binding site and corresponds to an amino acid required for the HAT activity of budding yeast Gcn5p (Kuo et al, 1998) (Figure 3A). We then assayed the potential HAT activities of these proteins, using human histone octamers as substrates (Figure 3B).
The wild-type protein had HAT activity, with preference toward histone H3 compared to H4 (∼30-fold, Figure 3B, lane WT), which is similar to the specificity of the budding yeast Gcn5p (Kuo et al, 1996). The protein with the G205A substitution exhibited very low HAT activity (Figure 3B, lane G205A). We concluded that SPAC1952.05 encodes a functional HAT enzyme homologous to human and budding yeast Gcn5, and therefore use the names ‘gcn5+' and ‘SpGcn5' to refer to the S. pombe gene and protein, respectively.
To determine whether SpGcn5 was responsible for the acetylation of histones at ade6 in vivo, we deleted the gcn5+ gene. Haploid gcn5Δ (null) mutants were viable and grew slightly slower than the gcn5+ cells. Diploid gcn5Δ mutants underwent meiosis I normally, except that the meiosis I process seems slightly quick and a small fraction of the cells (less than 10%) entered meiosis prematurely when cultured in a presporulation medium (Supplemental data 1). In addition, the gcn5Δ cells produced asci with four viable spores (78% spore viability). Moreover, northern analyses revealed that loss of gcn5+ did not affect the transcriptional induction of the meiosis-specific recombination genes, rec6+ and rec12+ (data not shown). These phenotypes differ markedly from those of budding yeast, where the deletion of GCN5 completely inhibits meiotic events (Burgess et al, 1999).
The Ch-IP experiments revealed that histone H3 acetylation levels around ade6-M26 were much lower in gcn5Δ cells than in gcn5+ cells (∼75% reduction, Figure 3C). A less severe decrease in histone H4 acetylation was observed, consistent with the substrate preference of SpGcn5 toward histone H3 (Figure 3D), while such in vivo effects might include an indirect influence on acetylation by other HATs in the absence of SpGcn5. We also observed a significant reduction of histone H3 acetylation around ade6-M375 (Figure 3E) and in the entire genome (Figure 3G) in gcn5Δ cells. Notably, in gcn5Δ cells the absolute levels of histone H3 acetylation around ade6-M26 were very similar to those around ade6-M375. These results demonstrate that SpGcn5 is required for the acetylation of histone H3 around ade6 and in the whole genome (see discussion), and more importantly it is required for the M26-specific hyperacetylation.
Effects of gcn5+ deletion on genome-wide DSB formation and meiotic recombination
We next examined the effects of gcn5+ deletion on the meiotic DSB formation on whole chromosomes (Figure 4A) using haploid strains with the pat1-114 rad50S background, which enabled us to detect the accumulation of discrete meiotic DSB bands in highly synchronized meiosis. In gcn5+ cells, we detected the accumulation of smeared chromosomal bands, reflecting meiotic DSBs, after 4 h in sporulation culture (Figure 4A lanes 4 and 5). The abundance of these subchromosomal fragments was markedly reduced at 4 h in gcn5Δ cells (Figure 4A lane 9), while gcn5Δ cells undergo slightly quicker meiosis I (see above). However, the final extent of chromosomal breakage in the gcn5+ and gcn5Δ cells was similar at a later time point (Figure 4A, lane 10). These results indicate that SpGcn5 is required for the proper timing of the DSB formation, but has only a nominal effect on the final level of the genome-wide DSB formation.
Figure 4.
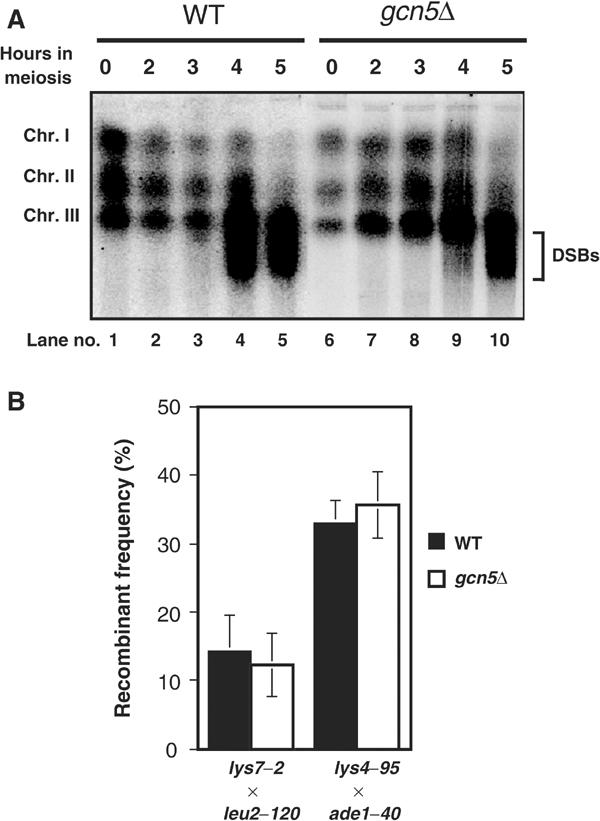
Effects of gcn5+ disruption on the genome-wide DSB formation and recombination. (A) Effects of gcn5Δ mutation upon meiotic DSBs in whole chromosomes. Genomic DNA was prepared from K341 (ade6-M26, rad50S, lanes WT) and TY24 (ade6-M26, rad50S, gcn5Δ, lanes gcn5Δ) cells cultured for the indicated times in sporulation medium, and was analyzed by pulsed-field gel electrophoresis. The electrophoresis image was obtained by an FM BIO II image analyzer (HITACHI) after Syber Green staining. The smeared DNA shows the broken DNA generated during meiosis. (B) Effects of the gcn5Δ mutation upon intergenic recombination. Standard genetic crosses between haploids containing appropriate markers were performed, and spores were plated on complete medium. Then the colonies arising from the spores were replicated onto appropriate test media to check the prototrophy and to measure the recombination frequencies. Each recombination frequency represents average of five independent crosses.
To determine whether the gcn5Δ mutation influences global recombination, we measured the frequency of intergenic recombination (crossing-over) between lys7 and leu2 on chromosome I and between lys4 and ade1 on chromosome II (Figure 4B). We found that the gcn5+ disruption did not significantly affect the recombinant frequencies. A possibility of overestimating recombinants in gcn5Δ mutants was eliminated, since there is no significant effect of the gcn5Δ mutation on viable or diploid spore formation. Thus, we concluded that although SpGcn5 is required for proper timing of meiotic DSB formation, it is dispensable for intergenic, nonhotspot recombination, at least for the two intervals tested.
Effects of gcn5+ disruption on chromatin remodeling, DSB formation, and recombination at the ade6-M26 hotspot
We studied the effects of the gcn5Δ mutation on chromatin remodeling, meiotic DSB formation, and recombination at the ade6-M26 hotspot. In wild-type diploid cells, the M26 chromatin remodeling occurred by 2–3 h after meiotic induction (Figures 1 and 5A). However, in gcn5Δ diploid cells, chromatin remodeling was not observed after 3 h, but partial remodeling occurred at 4.5 h (data not shown) and at 6 h (Figure 5A). Similar results were obtained with highly synchronized meioses in pat1-114 cells (data not shown). These results demonstrate that SpGcn5 is involved in the M26-dependent chromatin remodeling during meiosis.
Figure 5.
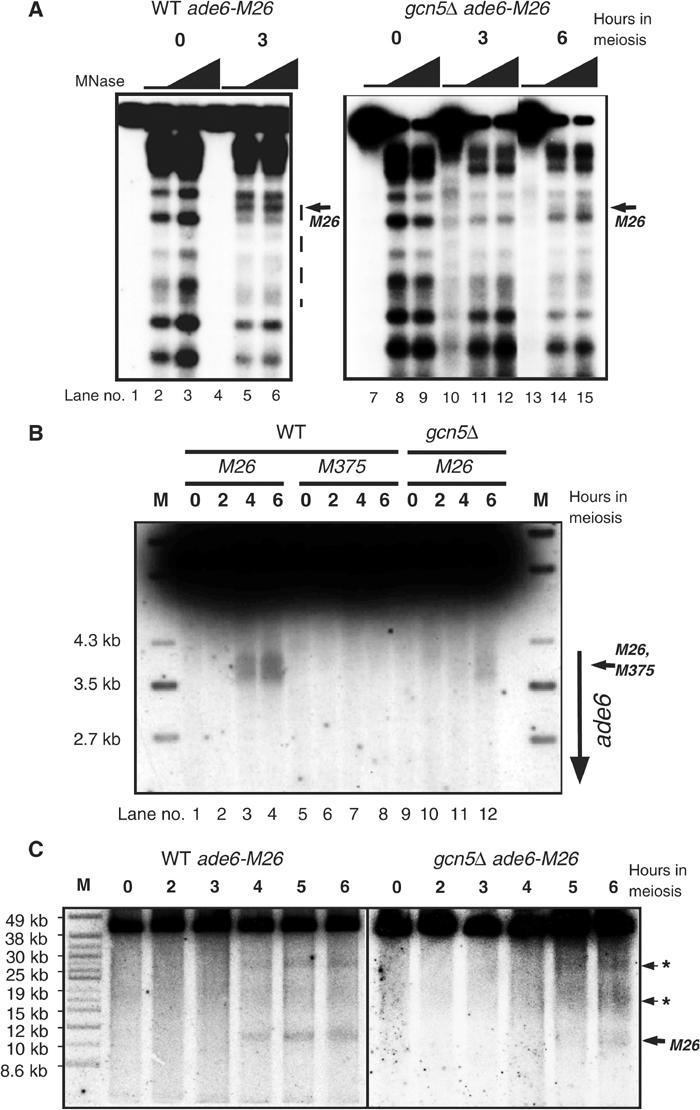
Effects of gcn5+ disruption on the chromatin remodeling, DSB formation, and recombination at M26. (A) Requirement for SpGcn5 in meiotic chromatin remodeling. Meiotic chromatin remodeling at the ade6 locus in DTY2 (ade6-M26, lanes WT) and DTY9 (ade6-M26, gcn5Δ, lanes gcn5Δ) diploids was analyzed as in Figure 1, using 0, 20, and 30 units/ml of MNase. Three independent experiments gave the same results. The broken line by lane 6 indicates the region where the chromatin structure is remodeled. (B) Requirement for SpGcn5 in meiotic DSB formation around M26. Genomic DNA was prepared as in Figure 4A, was digested with Afl II, and analyzed by Southern analysis. Horizontal and vertical arrows indicate the positions of the M26 (or M375) sites and the ade6 ORF, respectively. (C) Meiotic DSBs at the M26-independent sites adjacent to M26 in the gcn5Δ strain. Genomic DNA was prepared as in Figure 4A, was digested with Pac I, and analyzed by pulsed-field gel electrophoresis followed by Southern hybridization. The horizontal arrowhead and the asterisks indicate the position of M26 (or M375) and the adjacent breakages, respectively.
We subsequently analyzed meiotic DSB formation around ade6-M26 using genomic DNA prepared from the pat1-114 rad50S haploid cells (Figure 5B). In the gcn5+ cells, the meiotic DSB signals around ade6-M26 could be detected after 4 h in sporulation culture, and accumulated as meiosis proceeded. The DSB frequency at 6 h was 0.62±0.065% (n=3). On the other hand, no DSB signal could be detected in gcn5Δ cells after 4 h, and even after 6 h only reduced levels of meiotic DSBs were detected. The final level of the DSBs around M26 was 0.32±0.095% (n=3), almost half of that in gcn5+ strain. The delayed rate and the decreased amount of DSB formation in gcn5Δ cells paralleled the effects of gcn5Δ on chromatin remodeling at M26 (Figure 5A).
We also analyzed other M26 hotspot-independent DSB sites located upstream of the ade6 locus (Figure 5C). A slight delay in the DSB formation was observed at the M26-independent DSB sites adjacent to the ade6 locus, consistent with the delay in the genome-wide DSB formation. However, the final levels of DSB formation at those M26-independent sites were not significantly affected in gcn5Δ. On the other hand, DSBs around the M26 site occurred even later than those at the M26-independent sites, and were significantly impaired in the final DSB levels; the DSBs around M26 in gcn5+ cells (123% relative to one of the upstream DSB site) exhibited 42% reduction in gcn5Δ cells (71% relative to the upstream DSB site). Thus, the reduction in the final DSB level in gcn5Δ cells is specific to the M26 site.
We further tested the effects of the gcn5+ disruption on intragenic meiotic recombination at ade6. The recombination frequency (gene conversion) was measured by scoring the frequencies of Ade+ recombinants in random spores after zygotic meioses (Figure 6C). Recombination frequencies from crosses harboring M26 (hotspot) were reproducibly and significantly lower (60.3%) in gcn5Δ cells than in gcn5+ cells (Figure 6C). The reduction in the gene conversion frequencies was only partial but proportional to the decrease in the DSB frequencies (51.6%) in the gcn5Δ mutants. Importantly, no significant effects on recombination at M375 were detected in gcn5Δ cells (Figure 6C).
Figure 6.
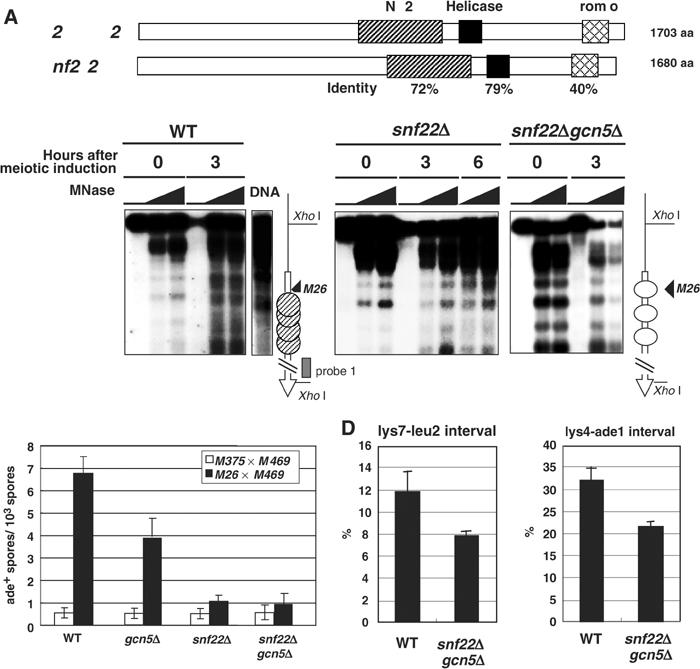
Effects of the snf22+ deletion on meiotic chromatin remodeling and recombination at M26. (A) Primary structures of S. cerevisiae Swi2/Snf2 and S. pombe SpSnf22. Hatched, filled, and cross-hatched boxes represent SNF2, helicase domains, and bromodomains, respectively. Identity of amino-acid sequences of each domain was indicated by percentages. (B) Requirement for SpSnf22 in meiotic chromatin remodeling at M26. Meiotic chromatin remodeling at the ade6 locus in DTY2 (ade6-M26, lanes WT), D13A1B1 (ade6-M26, snf22Δ, lanes snf22Δ), and D13G5A1B1 (ade6-M26, snf22Δ, gcn5Δ, lanes snf22Δgcn5Δ) diploids was analyzed as in Figure 1, using 0, 20, and 30 units/ml of MNase. Note that the relative intensity of each band varies in different chromatin preparations, although the patterns and positions of the MNase-sensitive sites are reproducible in each experiment. Schematic drawings of nucleosome mapping are the same as in Figure 1. (C) Requirement for SpSnf22 in meiotic recombination at ade6-M26. Standard genetic crosses between the ade6-M26 haploid strain and the ade6-469 haploid strain were performed, and the spores were plated on nonselective medium to determine the viable cell titer (T) and on selective medium lacking adenine to determine the ade6+ recombinant titer (R). The recombinant frequency in each experiment is R/T. Data are the recombinant frequencies from 2–3 independent experiments. (D, E) Effects of the snf22Δ mutation upon intergenic recombination. Standard genetic crosses and measurements of the recombination frequencies between lys7-leu2 (D) and lys4-ade1 (E) intervals were as described in Figure 4. Each recombination frequency represents average of two independent crosses.
Effects of snf22+ disruption on chromatin remodeling and meiotic recombination at the ade6-M26 hotspot
The gcn5+ deletion has only partial effects on meiotic recombination. This may be because other chromatin remodeling mechanisms can partially compensate the Gcn5 function. ADCRs are candidates for such alternative mechanisms. Hence we looked for ADCRs homologues and examined their mutant effects on the chromatin and recombination at M26. Using similarity to the budding yeast Swi2/Snf2 protein, we identified three possible ADCR genes. One of them, referred as snf22+ (SPACC 1620.14c), has a bromodomain and Swi2/Snf2-like conserved ATP-binding/helicase motifs (Figure 6A). The snf22+ deletion mutants exhibited inefficient mating, but once they formed zygotes, they produced spores almost normally. Using the snf22Δ and the gcn5Δ snf22Δ double mutants, the meiotic chromatin and recombination frequency around M26 were analyzed. In the snf22Δ and snf22Δgcn5Δ strains, we detected no significant chromatin changes even after 3–6 h of meiotic induction (Figure 6B). Importantly, meiotic recombination frequencies at M26 in these strains were greatly reduced (∼1/6 of the normal M26 levels) almost close to the negative control M375 levels (Figure 6C). On the other hand, the snf22Δgcn5Δ mutant only partially influenced genome-wide recombination of the lys2-leu2 and lys4-ade1 intervals (ca. 30% of the wild-type levels, Figure 6D and E). These results indicate an epistatic relationship between gcn5+ and snf22+ in meiotic chromatin regulation and recombination activation at M26.
Discussion
We studied the molecular mechanisms leading to activation of the M26 meiotic recombination hotspot, especially with regard to chromatin remodeling. We report that: (1) the Atf1·Pcr1 complex, essential for M26 hotspot activity, is indispensable for chromatin remodeling; (2) histones H3 and H4 are hyperacetylated around M26 in an M26- and Atf1-dependent manner; (3) SpGcn5p is a HAT in vitro, and is involved in the hotspot-specific histone acetylation around M26 and global histone acetylation; (4) Gcn5p is important for the chromatin remodeling and DSB formation around M26; and (4) Swi2/Snf2-like SpSnf22 is absolutely required for meiotic chromatin remodeling and recombination around M26. These results suggest a mechanism on activation of the M26 recombination hotspot.
Role of Atf1·Pcr1 in M26 activation
Chromatin remodeling at the M26 meiotic recombination hotspot fails to occur in mutants lacking any subunits of the Atf1·Pcr1 heterodimer and in strains harboring mutations in the DNA binding site for Atf1·Pcr1. In addition, the chromatin remodeling defects in atf1Δ or pcr1Δ mutants cannot be complemented by the inactivation of the Pat1 kinase or by the overproduction of Ste11, situations that alleviate the lack of transcription activation of the downstream genes in the absence of the Atf1·Pcr1 complex. These results provide compelling evidence that the Atf1·Pcr1 complex bound to the M26 DNA site directly mediates chromatin remodeling in cis, rather than by some indirect mechanism, such as transcriptional activation of the downstream genes.
Then, how does Atf1·Pcr1 induce chromatin remodeling? One can consider a possibility that Atf1·Pcr1 itself might mediate chromatin remodeling through histone acetylation, since S. pombe Atf1 is similar to human ATF2 with intrinsic HAT activity (Kawasaki et al, 2000). However, this may not be the case, because no HAT activity was detected in the purified recombinant Atf1 protein (data not shown). Furthermore, neither Atf1 nor Pcr1 share the conserved amino-acid residues that are important for HAT activity. An alternative possibility is that the binding of Atf1·Pcr1 to M26 may simply interfere with the local nucleosome positioning. This possibility is also ruled out, because Atf1·Pcr1 is expressed (Wahls and Smith, 1994; Kon et al, 1998) and binds to the M26 site under culture conditions in which M26 chromatin remodeling cannot be detected at all (Wahls and Smith, 1994; Kon et al, 1998).
From these, it is likely that the Atf1·Pcr1·M26 complex acts to recruit chromatin-modifying machinery such as HATs and ADCRs to the M26 site. Similar mechanisms are often found in transcription regulation sites. For example, the loading of the Drosophila NURF (ADCR) is achieved indirectly, via the binding to the GAGA-binding factor occupying its target sites on the DNA (Tsukiyama et al, 1994). In addition, the ATF/CREB family transcription factors in higher eucaryotes recruit coactivator proteins (e.g., CBP) with HAT activity (Ogryzko et al, 1996) to transcription promoters with CRE-like sequences.
Roles of SpGcn5 HATs and SpSnf22 in M26 activation
The present results indicate that the histones around the M26 site are hyperacetylated, and that the elements required for acetylation are identical to those required for chromatin remodeling at M26. The histone acetylation levels at ade6-M26 are significantly higher than those at ade6-M375 (Figure 2A–C), demonstrating a requirement for the M26 DNA site. This hotspot-specific histone hyperacetylation requires the Atf1·Pcr1 heterodimer (Figure 2D and E) and the SpGcn5 HAT (Figure 3C and D). These results suggest that the Atf1·Pcr1·M26 complex may recruit SpGcn5 to the M26 site (Figure 7). However, preliminary in vitro experiments failed to reveal direct interactions between purified Atf1 and purified SpGcn5 (data not shown). Thus, as proposed in Figure 7, it is likely that Atf1 recruits SpGcn5 via adaptor proteins, like the Tra1p subunit of the SAGA complex and the NuA4 complex in S. cerevisiae (Brown et al, 2001).
Figure 7.
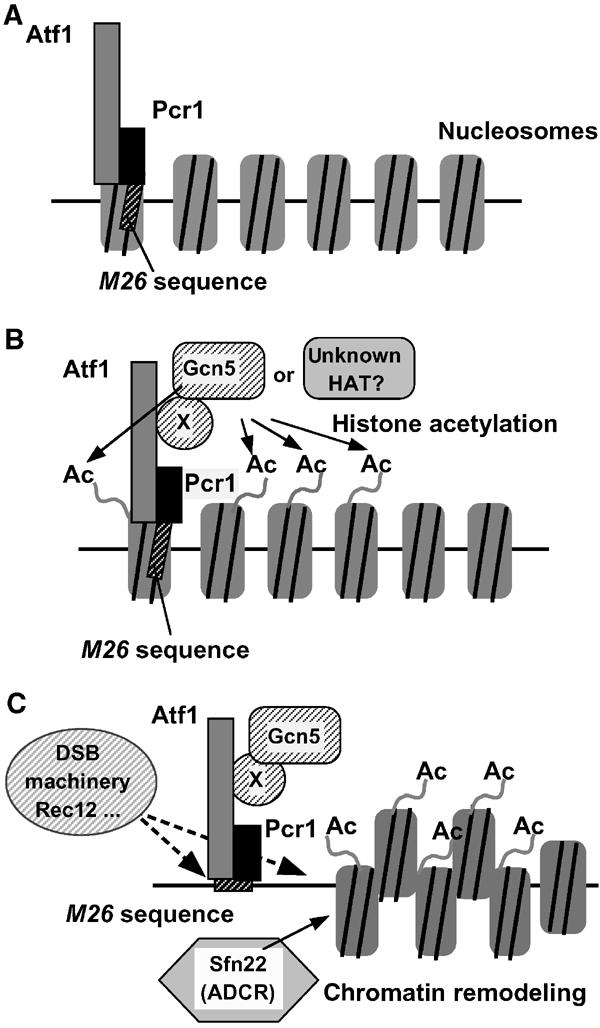
A regulatory model of meiotic recombination at M26 by HAT-mediated chromatin remodeling. (A) In cells lacking Gcn5 HAT, the histones around M26 are supposed to be hypoacetylated. In this situation, chromatin remodeling and subsequent DSB breaks occur very slowly. (B) Histones around M26 are hyperacetylated by Gcn5 HAT and possibly other HATs, which are introduced to the M26 site via the interaction with the Atf1·Pcr1 protein complex and a putative adaptor protein (shown as protein X). (C) Acetylated histones may be preferential targets of other ATP-dependent chromatin remodeling proteins (ADCR), such SpSnf22. DSB machinery, such as Rec12, is recruited to the open chromatin region created around M26, thereby activating meiotic recombination locally.
The loss of gcn5+ causes a delay and impairment of meiotic chromatin remodeling and DSB formation at the M26 site (Figure 5). This indicates that the SpGcn5-mediated histone acetylation facilitates chromatin remodeling and DSB formation at M26, but is not absolutely required for them. It is suggested that the function of SpGcn5 is to promote the rate of remodeling by marking histones with acetylation to be interacted with bromodomain-containing ADCRs, as suggested in the case of budding yeast (Hassan et al, 2001). This idea is also consistent with the previous observation that the deletion of budding yeast GCN5 caused a delay of chromatin remodeling at the PHO8 promoter (Barbaric et al, 2001). Finally, regions with chromatin structure altered by ADCRs may provide preferential loading sites of the meiotic DSB machinery such as Rec12; thereby DSB formation may be accelerated.
The gcn5+ deletion has only partial effects on M26-dependent meiotic recombination. It should be noticed that the deletion of the ADCR-like factor snf22+ causes a drastic reduction of meiotic recombination frequency at M26, and the snf22Δ gcn5Δ double mutant generally exhibits a deficiency very similar to the snf22Δ single mutant. Thus, snf22+ and gcn5+ have an epistatic relationship, possibly existing on the same pathway for chromatin remodeling. Therefore, the partial effects of the gcn5+ deletion are very likely explained by the presence of redundant HAT pathways prior to the action of ADCRs. For example, the acetylation of histone H4, which still occurs at significant levels in gcn5Δ mutants, may function redundantly with acetylation of histone H3 for the activation of recombination. In the budding yeast, histone H4 is preferentially acetylated by HAT called Esa1p (Clarke et al, 1999). Although Esa1p is supposed to be important in later DSB repair processes (Bird et al, 2002), it will be interesting to study the roles of Esa1p.
Interestingly, the genome-wide acetylation levels of histones H3 and H4 are decreased to similar, very low levels in atf1Δ as well as gcn5Δ cells (Figures 2F and G and 3G and H). One possibility is that Atf1 activates transcription of the gcn5+ gene. This seems unlikely, since there are no Atf1·Pcr1 binding sites upstream of gcn5+ and the atf1Δ mutation has no detectable effects on gcn5+ transcription (data not shown). It thus seems likely that Atf1 participates in the majority of the SpGcn5-mediated histone acetylation. However, the number of Atf1 binding sites is limited in the S. pombe genome to explain such global effects, implicating additional indirect functions of Atf1 in the SpGcn5-mediated histone acetylation.
In contrast to the partial effects of gcn5+ deletion, the deletion of the snf22+ confers very severe defects on meiotic chromatin remodeling and recombination. This indicates the vital role of the SpSnf22 in chromatin remodeling, and suggests that chromatin remodeling is a prerequisite to the M26 hotspot activation. It is most likely that SpSnf22 functions as an ADCR for chromatin remodeling at M26, considering conserved ADCR domains in SpSnf22. It is interesting to examine the biochemical activity of SpSnf22 using an in vitro reconstitution system in future experiments.
Possible molecular mechanisms of M26 activation and diverse roles of transcription factors in chromatin
The present results provide important insights into the molecular process of the M26 recombination activation. As reported previously, the Atf1·Pcr1 heterodimer occupies the M26 sites already in mitotic cells (Wahls and Smith, 1994; Kon et al, 1998). Maximal histone acetylation occurs subsequently, at the very beginning of the meiotic induction (0–1 h, see Figure 2). Chromatin remodeling appears by 2–3 h and meiotic DSBs accumulate at around 4 h, followed by completion of the later recombination processes prior to meiosis I. These differences in the timing of the events reveal a temporal sequence of the processes for the M26 hotspot activation (see Figure 7): (1) Atf1·Pcr1 binding to M26, (2) HAT recruitment and histone acetylation, (3) ADCR recruitment and chromatin remodeling, and (4) loading of recombination initiating proteins.
In this notion, Atf1·Pcr1 may be better thought as a ‘chromatin landmark' for the entry of HATs and ADCRs, rather than a canonical ‘transcription factor'. Atf1·Pcr1 functions to activate recombination at M26, but might act to stimulate transcription at other loci, depending on the local patterns of bound factors and cellular signals. This idea raises a question of unexpected diverse roles of transcription factors in various DNA-related reactions. Analyses on the versatility of transcription factors in chromatin would be an interesting aspect of a future study.
Materials and methods
Yeast strains, media, sporulation, and genetic analyses
S. pombe strains used in this study are listed in Supplementary data 2. Media and sporulation conditions were as described (Mizuno et al, 1997). Strain constructions, meiotic crosses, and analyses of recombination frequencies were as described (Ponticelli et al, 1988).
Identification and disruption of the gcn5+ and snf22+ gene
After BLAST searches using the amino-acid sequences of S. cerevisiae Swi2/Snf2 helicase domain, and HAT domain and bromodomain of the S. cerevisiae and human Gcn5p, directed against all translated open reading frames (ORFs) in the S. pombe genome, we identified several ORFs encoding proteins with significant homology. Among these, we focused on SPAC1952.05 and SPACC 1620.14c genes, which exhibit the highest homology to GCN5 and Swi2/Snf2, respectively. SPAC1952.05 is identical to the recently reported S. pombe gcn5+ (Mitsuzawa et al, 2001). Gene disruptions of gcn5+ and snf22+ were performed by a one-step gene disruption, which inserted ura4+ and kanMX into the open reading frame of gcn5+ and snf22+, and was confirmed by Southern hybridization.
HAT assay of SpGcn5
To prepare the recombinant 6 × -histidine tagged version of wild-type SpGcn5 (His-SpGcn5), a DNA fragment (1366 bp) encoding the gcn5+ gene was cloned into pET-15b and expressed in Escherichia coli BL21. The His-SpGcn5 protein was purified with TALON metal affinity resin (Clontech) according to the manufacturer's instructions. A mutated version of His-SpGcn5 (His-SpGcn5-G205A) was prepared as follows. A plasmid encoding SpGcn5-G205A was constructed by mutation of the gcn5+ cDNA using the QuikChange Site-Directed Mutagenesis Kit (Stratagene), followed by subcloning into the pET-15b vector. The His-SpGcn5-G205A protein was expressed and purified as described above. The HAT activities of His-SpGcn5 and His-SpGcn5-G205A were assayed using purified human core histones, as described elsewhere (Ogryzko et al, 1996).
Analyses of chromatin structure and mRNA
Analysis of chromatin structure was as described (Mizuno et al, 1997). Total RNA from S. pombe cells was prepared and analyzed by Northern blotting, as described (Kon et al, 1998).
Ch-IP using antiacetylated histones
Ch-IP was performed as described elsewhere (Rundlett et al, 1998) with slight modifications. Briefly, cell extracts prepared from crosslinked cells were incubated with antiacetylated histone H3 (AcLys(9/14)) and antiacetylated histone H4 (AcLys(5/8/12/16)) (Upstate). The immune complexes were collected by Dynabeads-ProteinA (Dynal) and washed thoroughly. The DNAs present in the cell extracts and in the immunoprecipitates were purified and applied to a nylon membrane by using a slot blot with a vacuum manifold. The membranes were hybridized to radioactively labeled probes for total genomic DNA or for the ade6-M26 region (probe M26: corresponds to nucleotides126–692 relative to the first A of the ade6 coding region). Hybridization signals were quantified using a Fuji BAS2500 Image Analyzer. All Ch-IP data presented were averages of three independent experiments.
Detection of meiotic DSB
Meiosis-specific DSBs around ade6-M26 were detected by the following two methods, as described (Steiner et al, 2002; Young et al, 2002). Genomic DNA embedded in agarose plugs was digested with the restriction endonuclease Pac I. The DNA was subjected to pulse field gel electrophoresis followed by Southern blotting. Hybridization probes were amplified by PCR using S. pombe genomic DNA as a template with the primer pairs (FW 5′-AAACGACTCACACTTTATAGGAGCAAC-3′; RV 5′-AGGCAGCCTCAAAAGCCC-3′), gel purified, and labeled with 32P. Alternatively, plugs digested with Afl II were analyzed by standard agarose gel electrophoresis followed by Southern blotting. Hybridization probes were amplified by PCR using S. pombe genomic DNA as a template with primer pairs (FW 5′-GCTCCAAGGCAAAATATGTC-3′; RV 5′-AACATACAGGTTGGATCTTAAG-3′), gel purified, and labeled with 32P. To monitor meiotic breaks of the entire chromosome, the DNA embedded in plugs was analyzed by pulsed-field gel electrophoresis, as described (Cervantes et al, 2000).
Note added in proof
Accession numbers for gcn5+ is AB162439 and Snf22+ is AB162438.
Supplementary Material
Supplemental data 1
Supplemental data 2
Acknowledgments
We thank Drs M Grunstein, N Suka, M Yanagida, T Tomonaga for the Ch-IP methods, Drs G Smith, W Steiner, T Kamakura, K Ogino, and H Masai for DSB detection protocols, and Dr J Kohli for providing strains. We also thank Ms Y Ichikawa and Ms R Nakazawa for DNA sequencing, and Ms Y Sakuma and K Kobayashi for technical assistance. We are grateful to Dr T Yasuda for histone octamers. We thank Drs MK Davidson and MJ Lichten for critical reading of the manuscript and Dr Y Watanabe for technical comments. This work was supported by grants from the HFSP; the ‘Bioarchitect Research Program' of RIKEN; the CREST program of JST; the Ministry of Education, Science, Culture, & Sports, Japan; and the National Institutes of Health, USA (GM62244 and GM62801).
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 [DOI] [PubMed] [Google Scholar]
- Barbaric S, Walker J, Schmid A, Svejstrup JQ, Hörz W (2001) Increasing the rate of chromatin remodeling and gene activation-a novel role for the histone acetyltransferase Gcn5. EMBO J 20: 4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant G, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337 [DOI] [PubMed] [Google Scholar]
- Brown CE, Lechner T, Howe L, Workman JL (2000) The many HATs of transcription coactivators. Trends Biochem Sci 25: 15–19 [DOI] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N (1999) GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci USA 96: 6835–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR (2000) Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell 5: 883–888 [DOI] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L (1999) Esa1p is an essential acetyltransferase required for cell cycle progression. Mol Cell Biol 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311 [DOI] [PubMed] [Google Scholar]
- Gutz H (1971) Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics 69: 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize Swi/Snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hirota K, Hoffman CS, Shibata T, Ohta K (2003) Fission yeast Tup1-like repressors repress chromatin remodeling at the fbp1+ promoter and the ade6-M26 recombination hotspot. Genetics 165: 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Yamamoto M (1985) Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet 198: 416–421 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M (1996) Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1: 391–408 [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405: 195–200 [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP (1997) Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA 94: 13765–13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Schroeder SC, Krawchuk MD, Wahls WP (1998) Regulation of the Mts1-Mts2-dependent ade6-M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol Cell Biol 18: 7575–7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev 13: 1412–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272 [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev 12: 627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83: 387–395 [DOI] [PubMed] [Google Scholar]
- McMurry MT, Krangel MS (2000) A role for histone acetylation in the developmental regulation of V(D)J recombination. Science 287: 495–49810642553 [Google Scholar]
- Mitsuzawa H, Seino H, Yamao F, Ishihama A (2001) Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J Biol Chem 276: 17117–17124 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T (1997) The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev 11: 876–886 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Hasemi T, Ubukata T, Yamada T, Lehmann E, Kohli J, Watanabe Y, Iino Y, Yamamoto M, Fox ME, Smith GR, Murofushi H, Shibata T, Ohta K (2001) Counteracting regulation of chromatin remodeling at a fission yeast CRE-related recombination hotspot by SAPK, cAMP-dependent kinase, and meiosis regulators. Genetics 159: 1467–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959 [DOI] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A (1994) Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J 13: 5754–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2: 360–369 [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Sena EP, Smith GR (1988) Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics 119: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835 [DOI] [PubMed] [Google Scholar]
- Schuchert P, Langsford M, Kaslin E, Kohli J (1991) A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J 10: 2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR (2002) Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol Cell 9: 847–855 [DOI] [PubMed] [Google Scholar]
- Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev 5: 1990–1999 [DOI] [PubMed] [Google Scholar]
- Szankasi P, Heyer WD, Schuchert P, Kohli J (1988) DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination hot spot allele ade6-M26. J Mol Biol 204: 917–925 [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N (1995) Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J 14: 6193–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C (1994) ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367: 525–532 [DOI] [PubMed] [Google Scholar]
- Wahls WP, Smith GR (1994) A heteromeric protein that binds to a meiotic homologous recombination hot spot: correlation of binding and hot spot activity. Genes Dev 8: 1693–1702 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M (1996) Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol 16: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP (1997) Chromatin: Structure and function, 3rd edn San Diego, USA: Academic Press [Google Scholar]
- Wu TC, Lichten M (1994) Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518 [DOI] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR (2002) Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell 9: 253–263 [DOI] [PubMed] [Google Scholar]
- Zenvirth D, Simchen G (2000) Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr Genet 38: 33–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data 1
Supplemental data 2


