Abstract
Mammalian H/ACA small nucleolar RNAs and telomerase RNA share common sequence and secondary structure motifs that form ribonucleoprotein particles (RNPs) with the same four core proteins, NAP57 (also dyskerin or in yeast Cbf5p), GAR1, NHP2, and NOP10. The assembly and molecular interactions of the components of H/ACA RNPs are unknown. Using in vitro transcription/translation in combination with immunoprecipitation of core proteins, UV-crosslinking, and electrophoretic mobility shift assays, we demonstrate the following. NOP10 associates with NAP57 as a prerequisite for NHP2 binding. Although NHP2 on its own binds RNA nonspecifically, this NAP57–NOP10–NHP2 core trimer specifically recognizes H/ACA RNAs. GAR1 associates independently with NAP57 near the pseudouridylase core of mature H/ACA RNPs. In contrast to other RNPs whose assembly is initiated by protein–RNA interactions, the four H/ACA core proteins form a protein-only particle that associates with H/ACA RNAs. Nonetheless, functional H/ACA snoRNPs assembled in cytosolic extracts are stable and do not exchange their RNA components, suggesting that new particle formation requires de novo synthesis.
Keywords: dyskeratosis congenita, H/ACA snoRNAs, pseudouridine synthase, ribonucleoprotein particle assembly, telomerase RNA
Introduction
Modification of ribosomal and spliceosomal small nuclear RNAs is essential for proper function of ribosomes in translation and of spliceosomes in pre-mRNA splicing, respectively (Yu et al, 1998; Bonnerot et al, 2003; King et al, 2003). In eucaryotes, such RNA modifications and, in the case of pre-ribosomal RNA, also some processing steps are mediated by hundreds of small nucleolar ribonucleoprotein particles (snoRNPs). Based on conserved secondary structure and sequence elements of their small nucleolar RNAs (snoRNAs), two major classes of snoRNPs can be distinguished, H/ACA and C/D. H/ACA snoRNPs primarily mediate site-specific pseudouridylation and C/D snoRNPs 2′-O-methylation of their target RNAs (for reviews, see Bachellerie and Cavaille, 1997; Smith and Steitz, 1997; Tollervey and Kiss, 1997; Decatur and Fournier, 2003). Each H/ACA snoRNP consists of the same four core proteins and a different snoRNA, which guides modification by site-directed base pairing (Ganot et al, 1997; Ni et al, 1997; Henras et al, 1998; Lafontaine et al, 1998; Watkins et al, 1998). In fact, the core particle of H/ACA snoRNPs, comprising the proteins NAP57, GAR1, NHP2, and NOP10 and an H/ACA snoRNA, is sufficient for site-specific pseudouridylation of ribosomal RNA (rRNA) in vitro (Wang et al, 2002).
In addition to H/ACA snoRNAs, mammalian telomerase RNA (hTR in humans), required for replicating telomeres, ends in an H/ACA motif and associates with the same four core proteins to form an H/ACA RNP (Mitchell et al, 1999a, 1999b; Dragon et al, 2000; Pogacic et al, 2000; Dez et al, 2001). This H/ACA domain of hTR appears solely required for stability and proper 3′-end processing of the RNA, as no substrate for pseudouridylation has been identified (Mitchell et al, 1999a) and as synthetic substrates complementary to the putative pseudouridylation pockets of the H/ACA domain failed to be modified in vitro (unpublished observation).
The four H/ACA core proteins are evolutionarily highly conserved, as documented by the existence of orthologs in yeast and archaea (Meier and Blobel, 1994; Henras et al, 1998; Watkins et al, 1998; Watanabe and Gray, 2000; Rozhdestvensky et al, 2003). All four proteins are concentrated in the nucleoli and Cajal bodies of every cell (Girard et al, 1992; Meier and Blobel, 1994; Henras et al, 1998; Pogacic et al, 2000). As judged from depletion studies in yeast, all proteins are essential for viability and, with the exception of GAR1, are required for the stability of H/ACA RNPs (Bousquet-Antonelli et al, 1997; Henras et al, 1998; Lafontaine et al, 1998; Watkins et al, 1998; Dez et al, 2001).
NAP57, originally identified in yeast as Cbf5p and also known as dyskerin in mammalian cells, is apparently the enzyme of H/ACA snoRNPs that catalyzes the isomerization of uridines to pseudouridines (Jiang et al, 1993; Meier and Blobel, 1994; Heiss et al, 1998; Lafontaine et al, 1998). This is based on its homology to proven eubacterial pseudouridine synthases and the severe defects in pseudouridylation caused by point mutations introduced in the catalytic domain of Cbf5p (Meier and Blobel, 1994; Nurse et al, 1995; Zebarjadian et al, 1999). The gene encoding human NAP57, DKC1, is mutated in the X-linked form of the bone marrow failure syndrome dyskeratosis congenita (DC) (Heiss et al, 1998). DC is a rare genetic disorder characterized by the triad of skin pigmentation, nail dystrophy, and mucosal leucoplakia, and by a predisposition to malignancy in rapidly dividing tissues (for recent reviews, see Dokal and Vulliamy, 2003; Marrone and Mason, 2003). Patient cells exhibit reduced levels of telomerase RNA and shortened telomeres, suggesting telomerase deficiency as the molecular basis for the disease (Mitchell et al, 1999b). However, a mouse model faithfully replicates the phenotype of the disease in the absence of any telomere defects, but with reduced levels of rRNA pseudouridylation (Ruggero et al, 2003). Altogether, DC mutations in NAP57 may, in general, affect the stability/function of H/ACA RNPs, of both small nucleolar and telomerase RNPs (Meier, 2003).
GAR1 (∼25 kDa), NHP2 (∼22 kDa), and NOP10 (∼10 kDa) are smaller proteins of unknown function. GAR1 consists of a central core domain flanked by two glycine-arginine-rich (GAR) domains (Girard et al, 1992). The core domain of yeast Gar1p is sufficient for viability and binds in vitro to two H/ACA snoRNAs (Girard et al, 1994; Bagni and Lapeyre, 1998). NHP2 is homologous to the ribosomal protein L30 and to the 15.5K/NHP2L1 protein (Snu13p in yeast) shared between C/D snoRNPs and U4 spliceosomal small nuclear RNPs (Henras et al, 1998; Watkins et al, 1998, 2000; Nottrott et al, 1999). Unlike its homologs, NHP2 does not bind to a specific RNA motif, but associates nonspecifically with RNA secondary structures (Henras et al, 2001). NOP10, although conserved like the other core proteins, contains no known motifs and measures only 64 amino acids (Henras et al, 1998).
Despite this detailed knowledge of the individual core components of H/ACA RNPs and the established importance of each component for the integrity of the particles, it remains completely elusive how these proteins interact with each other or the H/ACA RNA and how they are assembled. Here we report the in vitro assembly of H/ACA RNPs from individual components and delineate a protein–protein and protein–RNA interaction and assembly map. Furthermore, we investigate the impact of NAP57 mutations on H/ACA RNP assembly.
Results
Pseudouridylation target uridines UV-crosslink to NAP57 and GAR1
We previously showed that purified H/ACA snoRNP core particles consisting of NAP57, GAR1, NHP2, NOP10, and an H/ACA snoRNA were sufficient for faithful pseudouridylation of short site-specifically 32P-labeled rRNA substrates (Wang et al, 2002). To probe the catalytic core of these H/ACA snoRNPs by UV-crosslinking, we replaced by 4-thiouridine the site-specifically labeled uridine corresponding to nucleotide 4380 in human 28S rRNA. Incubation under assay conditions of this 4-thiouridine-substituted substrate with immunopurified H/ACA snoRNPs and exposure to 365 nm UV light resulted in two prominent 32P-labeled bands when analyzed by SDS–PAGE and autoradiography (Figure 1A, lane 1). After digestion of the crosslinked adducts by RNase A, they migrated faster, equivalent to positions of NAP57 and GAR1 (Figure 1A, lane 2). Identical incubation and UV irradiation of a substrate that was not 4-thiouridine substituted but otherwise identical yielded no 32P-labeled bands, confirming that crosslinking occurred through 4-thiouridine (not shown). When these experiments were repeated with the same substrate, but with the adjacent uridine (nucleotide 4379 in human 28S rRNA, which is not subject to pseudouridylation) substituted by 32P-labeled 4-thiouridine, phosphorimaging revealed 2.7- and 4.5-fold less crosslinking of the lower and upper bands, respectively. These data demonstrate that crosslinking is sensitive to the position of the 4-thiouridine in the substrate.
Figure 1.
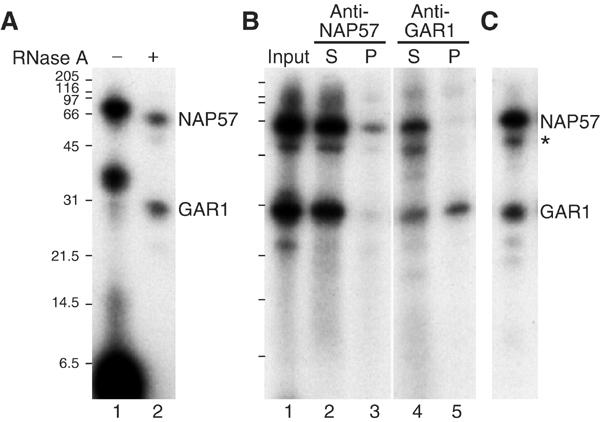
Pseudouridylation target uridine UV-crosslinks to NAP57 and GAR1. (A) Immunopurified box H/ACA snoRNPs were incubated with a site-specifically 32P-labeled and 4-thiouridine-substituted short rRNA substrate (corresponding to the site targeted by snoRNA E3) and UV irradiated. The proteins and crosslinked RNAs were eluted from protein A–sepharose beads and analyzed by 15% Tricine SDS–PAGE and autoradiography before (lane 1) and after RNase A treatment (lane 2). (B) After elution with SDS (lane 1), the proteins were re-precipitated in the presence of Triton X-100 with NAP57 (lanes 2 and 3) and GAR1 antibodies (lanes 4 and 5). The supernatants (lanes 2 and 4) and pellets (lanes 3 and 5) were analyzed as in (A). (C) The same as lane 1 in (B), but with a substrate corresponding to the site targeted by snoRNA U65. The migration positions of NAP57, GAR1, an NAP57 breakdown product (asterisk), and the molecular weight markers are indicated.
The identity of the two crosslinked bands as NAP57 and GAR1 was confirmed by precipitation with antibodies to the respective protein (Figure 1B). After UV-crosslinking, the immunopurified H/ACA snoRNPs (lane 1) were eluted by SDS denaturation, the SDS quenched by Triton X-100, and the proteins precipitated from the supernatants (lanes 2 and 4) by antibodies against NAP57 (lane 3) and GAR1 (lane 5). The precipitation of exclusively the upper and lower bands in each case confirmed their identity. The identity of NAP57 was further supported by the presence of a minor crosslink to a band migrating slightly below NAP57 (Figure 1C, asterisk), at the position of a NAP57 breakdown product that was routinely observed in immunopurified H/ACA snoRNPs (Wang et al, 2002). Importantly, identical crosslinking to NAP57 and GAR1 was observed, with a different 4-thiouridine-substituted substrate representing position 4363 in human 28S rRNA and whose pseudouridylation was guided by H/ACA snoRNA U65 (Figure 1C). Altogether, these data provided direct support that NAP57 is the pseudouridine synthase of H/ACA snoRNPs and that GAR1 also forms part of the catalytic core.
A protein-only core complex
Although the minimal composition of active H/ACA snoRNPs consists of only five components, nothing is known about the interactions between them. Since expression in bacteria and baculovirus of NAP57 yielded only denatured protein (data not shown), we used in vitro transcription/translation in rabbit reticulocyte lysate and subsequent immunoprecipitation to delineate the interactions between the four core proteins. When programmed with the individual cDNAs of the four core proteins, transcription/translation in the presence of 35S-methionine yielded one major protein band each, which migrated at the expected position (Figure 2A). All four proteins were also efficiently synthesized when their cDNAs were added simultaneously (Figure 2B, lane 1). Immunoprecipitation with anti-NAP57 peptide antibodies from the mixture of the four labeled proteins precipitated NAP57 (Figure 2B, lane 2) and removed it from the supernatant (compare lanes 4 and 5). Note that, for unknown reasons, immunoprecipitated NAP57 migrated more slowly, as multiple bands or a smear in our gel system (e.g., lane 2, vertical bar). In addition to NAP57, NHP2 and NOP10 were co-precipitated (lane 2). GAR1 was not always detectable in NAP57 precipitates (see below). Precipitation was specific because it was competed for by excess free NAP57 peptide against which the antibodies were raised (lane 3).
Figure 2.
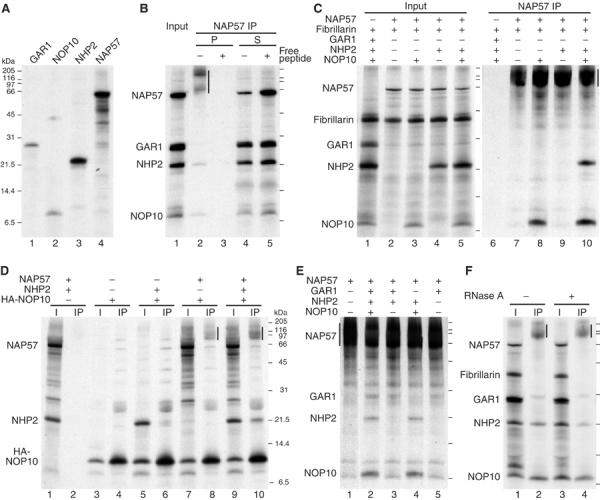
Immunoprecipitation of in vitro-translated snoRNP core proteins. (A) The individual cDNAs for the indicated H/ACA core proteins were transcribed and translated in rabbit reticulocyte lysate in the presence of 35S-methionine and analyzed by 15% Tricine SDS–PAGE and fluorography. (B) All four H/ACA core proteins were co-translated (lane 1) and precipitated with NAP57 peptide antibodies in the absence (lanes 2 and 4) and presence (lanes 3 and 5) of free competing peptide. Compared to the input (lane 1), twice the amount of pellets (lanes 2 and 3) and supernatants (lanes 4 and 5) were loaded. Note the anomalous migration of immunoprecipitated NAP57 (vertical bar). (C) H/ACA core proteins and fibrillarin as control were co-translated in various combinations and precipitated with NAP57 antibodies. The input (lanes 1–5) and the precipitates from 10-fold more (lanes 6–10) are shown. (D) Immunoprecipitations of various combinations of NAP57, NHP2, and HA-NOP10 with HA antibodies. The input (odd lanes) and precipitates (even) are shown, respectively. (E) NAP57 precipitates of different combinations of H/ACA core proteins including GAR1. (F) NAP57 precipitation in the presence of all core proteins was performed before (lanes 1 and 2) or after RNase A treatment (lanes 3 and 4). The input (odd lanes) and precipitates (even lanes) are shown. The migrating positions of molecular weight markers are indicated for each panel.
To dissect the association of the individual core proteins with NAP57, various combinations were expressed (Figure 2C, lanes 1–5) and precipitated with NAP57 antibodies (lanes 6–10). As a control, fibrillarin, a core protein of C/D snoRNPs, was included in all translations, but was absent from all precipitates (lanes 6–10). None of the other proteins was precipitated in the absence of NAP57 (lane 6), but NAP57 alone was (lane 7). NOP10 (lane 8), but not NHP2 (lane 9) alone, co-precipitated with NAP57. Precipitation of NHP2 required the presence of NOP10 (lane 10), suggesting that NOP10 mediated its association with NAP57, thereby forming a core trimer. In seven similar NAP57 co-precipitation experiments with at least two samples each, the molar ratio of NOP10/NHP2, as determined by phosphorimaging, was 2.0±0.7 (n=32). This further supported the NAP57 binding of NHP2 being NOP10 dependent and raised the possibility that there are two NOP10, and possibly NAP57, per one NHP2 in each core trimer.
For further analysis of this core trimeric complex, NOP10 was HA-tagged and precipitated with HA antibodies in the presence or absence of NAP57 and/or NHP2 (Figure 2D). HA-NOP10 itself (lane 3) was efficiently precipitated (lane 4), but none of the other proteins (lane 1) were present in the pellet in its absence (lane 2), documenting specificity. Trace amounts of NHP2 alone (lane 5) were co-precipitated with HA-NOP10 (lane 6), suggesting a weak interaction. However, NAP57 alone (lane 7) co-precipitated (lane 8, vertical bar), confirming the NAP57–NOP10 connection. When all three proteins were present (lane 9), HA-NOP10 efficiently precipitated NHP2 in addition to NAP57 (lane 10), confirming that both NAP57 and NOP10 are required for tight NHP2 binding.
For unknown reasons, GAR1 was not always detected in NAP57 precipitates, but was observed consistently within experiments where it occurred, for example, when excess NAP57 was used (Figure 2E). Note the many NAP57-derived background bands when NAP57 alone was precipitated (lane 1). All four proteins, including GAR1, were precipitated with NAP57 (lane 2), and GAR1, but not NHP2, co-precipitated with NAP57 when just the two proteins were added (lane 3). NHP2, as determined above, required NOP10 for co-precipitation (lane 4). Importantly, GAR1 precipitated with NAP57 alone (lane 5), indicating a GAR1–NAP57 interaction independent of NOP10 and NHP2.
We noted that the in vitro-translated GAR1 migrated as a doublet in the 15% Tricine SDS–PAGE system (e.g., Figure 2F, lane 3) and wondered if one of the two forms might preferentially associate with NAP57. GAR1 is known to be asymmetrically dimethylated on the arginine residues of its amino- and carboxy-terminal glycine/arginine-rich repeats (Frankel and Clarke, 1999; Xu et al, 2003), and the two bands could represent the methylated and unmethylated forms of GAR1. Indeed, addition of S-adenosyl methionine, the methyl donor for arginine methylation, and/or of recombinant protein arginine methyl transferase (PRMT1) converted the faster-migrating GAR1 band into the slower one (data not shown). These results indicated that there was some PRMT1 activity present in reticulocyte lysate, but insufficient for full methylation of GAR1. GAR1 association with NAP57, however, was unaffected or equally variable, whether its arginines were dimethylated or not (data not shown), consistent with the finding that a yeast GAR1 mutant without GAR domains functionally complemented a GAR1-depleted strain (Girard et al, 1994).
Since the four core proteins are usually associated with RNA, we tested if any of the RNAs in reticulocyte lysate might be involved in the observed complex formation (Figure 2F). For this purpose, the lysate was treated with RNase A after expression of the labeled proteins, but before precipitation with NAP57 antibodies. All four core proteins, but not fibrillarin, were precipitated regardless of prior incubation with RNase A (Figure 2F, compare lanes 2 and 4). In summary therefore, these co-precipitation data argue for a protein-only complex of the four H/ACA core proteins, consisting of a NAP57–NOP10–NHP2 core trimer and GAR1, associated with NAP57 (see Figure 6).
Figure 6.
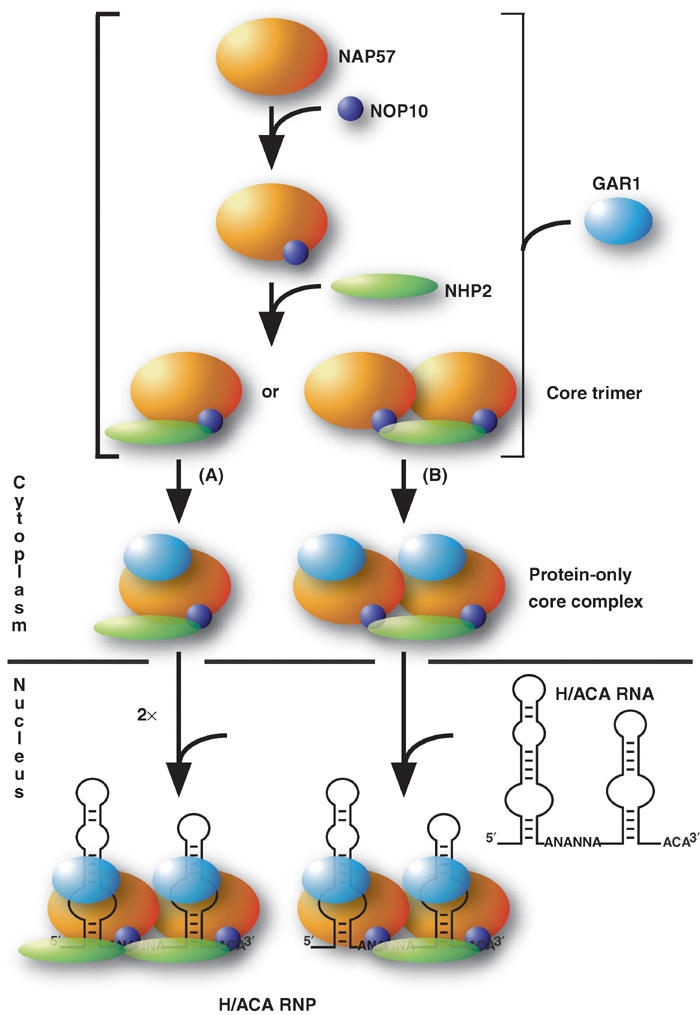
Schematic of H/ACA RNP assembly pathway and intra-RNP molecular interactions. The schematic is based on results from this study as explained in detail in the text. The sizes of proteins are depicted in approximate scale to their molecular weight.
Effect of NAP57 mutations on core protein complex formation
NAP57 is an evolutionary highly conserved protein containing a pseudouridylase domain (TruB) in its amino terminus and a pseudouridine synthase/archaeosine transglycosylase (PUA) domain in its carboxy-terminal half (Figure 3A; Aravind and Koonin, 1999). An aspartate residue in the TruB domain, corresponding to D126 in rat NAP57, apparently catalyzes the reaction by forming a covalent enzyme–substrate intermediate (Huang et al, 1998; Gu et al, 1999; Hoang and Ferre-D'Amare, 2001). The carboxy terminus of NAP57 stands out by its high charge density, mostly due to a large number of lysine residues (Jiang et al, 1993; Meier and Blobel, 1994). One of the hot spots of missense mutations in the DKC1 gene encoding human NAP57 is exon 3 (for reviews, see Dokal and Vulliamy, 2003; Marrone and Mason, 2003). In this exon, we selected the evolutionarily most highly conserved phenylalanine F37 in rat NAP57 and changed it to valine, which apparently causes DC in family DCR026 (Heiss et al, 1998).
Figure 3.
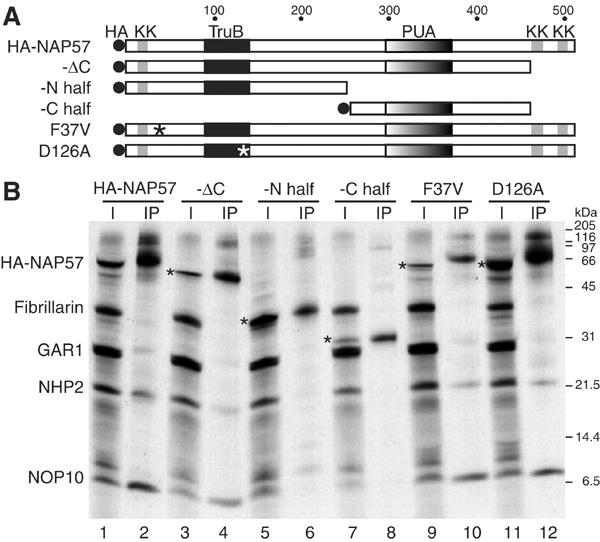
Co-immunoprecipitation of H/ACA core proteins with HA-NAP57 and derivatives. (A) Schematic representation of HA-tagged (HA) NAP57 and derived constructs with the highly conserved pseudouridylase (TruB) and pseudouridine synthase/archaeosine transglycosylase domains (PUA), the lysine clusters (KK), and point mutations (asterisks) highlighted. (B) HA-NAP57 and derivatives were co-translated with the other H/ACA core proteins and fibrillarin as described in the legend to Figure 2 and precipitated with anti-HA antibodies. Odd lanes correspond to input and even lanes to precipitates. The HA-tagged NAP57 derivatives are (amino acids in parentheses): HA-NAP57 (full-length, 1–509); -ΔC (1–466); -N half (1–259); C half (252–466); F37V, point mutation detected in a family with X-linked DC; D126A, point mutation of the aspartate required for pseudouridylase activity. The positions of the HA-NAP57 constructs are marked (asterisks).
We generated HA-tagged full-length, partial, and mutated NAP57 constructs following the above characteristics, and tested them for complex formation with the other core proteins in our in vitro translation/immunoprecipitation assay (Figure 3B). Incubation of the in vitro-translated proteins (Figure 3B, lane 1) with antibodies directed against the HA tag precipitated full-length HA-NAP57, NOP10, NHP2, and some GAR1, but not fibrillarin (lane 2), as was the case for untagged NAP57 (see Figure 2). While no difference in NAP57-associated proteins was observed when its lysine-rich C-terminus was missing (lane 4), none of the core proteins were present in the precipitates of the NAP57 amino- or carboxy-terminal halves alone (lanes 6 and 8). Furthermore, generation of the DC mutation F37V or catalytic inactivation of NAP57 by mutating aspartate 126 to alanine (D126A) had no apparent effect on core complex formation (lanes 10 and 12). Therefore, major structural alterations in NAP57, but not point mutations that were catalytically inactivating or associated with DC, interfered with its association with the other H/ACA core proteins.
Specific interaction of H/ACA sno- and telomerase RNAs with core proteins
Having established a protein-only complex of the core proteins, we investigated their association with H/ACA snoRNAs. First, we tested the RNA binding of recombinant NHP2 by electrophoretic mobility shift analysis, because its relative 15.5K/NHP2L1 of C/D snoRNPs binds to a conserved motif in C/D snoRNAs with high specificity (Watkins et al, 2000). Indeed, increasing amounts of GST–NHP2 fusion protein yielded an increased mobility shift of 32P-labeled H/ACA snoRNA E3 (Figure 4A). While excess unlabeled E3 snoRNA competed for the E3 gel shift (Figure 4B, lanes 1 and 2), the C/D snoRNA U3 competed even better (lanes 3 and 4), suggesting nonspecific binding of GST–NHP2 to E3. This interaction was not due to the GST moiety of our construct because GST–NOP10 did not bind E3 (Figure 4C, compare lanes 2 and 3). In addition, GST–NHP2 bound even more efficiently to the stem–loop structure formed by the 3′ untranslated region (3′ UTR) of a yeast mRNA (Chartrand et al, 1999), confirming its nonspecific RNA interaction (Figure 4C, lanes 4 and 5). These data with mammalian NHP2 are entirely consistent with the RNA-binding ability of yeast Nhp2p, as detailed elegantly by Henras et al (2001).
Figure 4.
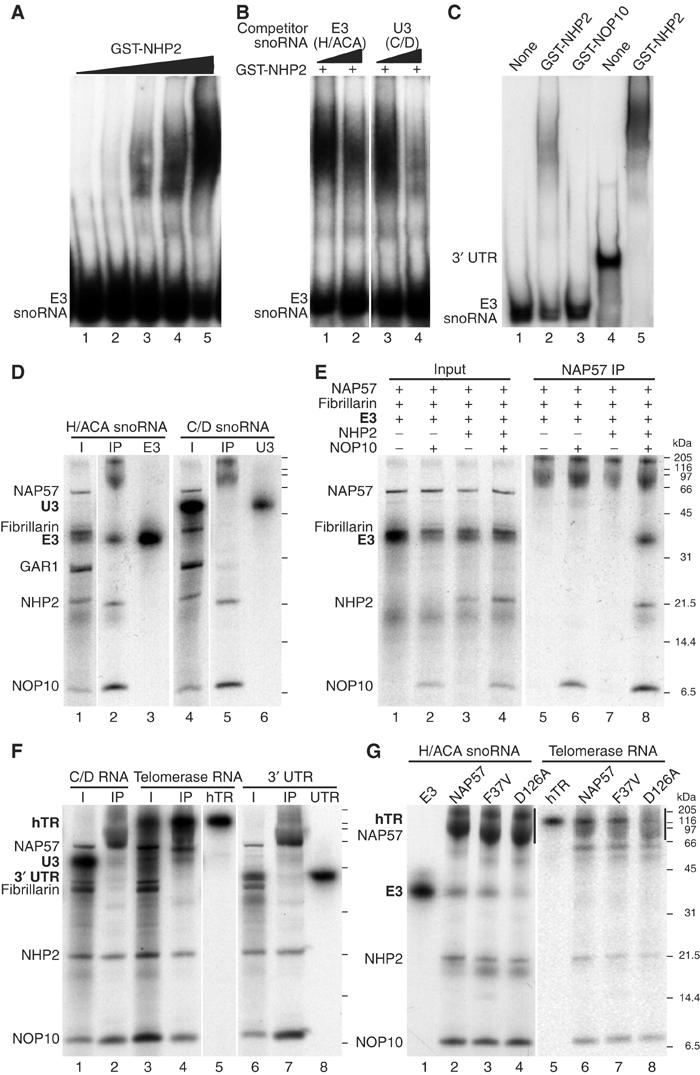
Electrophoretic mobility shift assay of 32P-labeled RNA with recombinant NHP2 (A–C) and co-immunoprecipitation of 32P-labeled RNA with in vitro-translated H/ACA core proteins (D–G). (A) Radiolabeled H/ACA snoRNA E3 was incubated with increasing amounts of purified, bacterially expressed GST-tagged NHP2 and analyzed by 4% native polyacrylamide gel electrophoresis and autoradiography. (B) Same as in (A) but in the presence of either unlabeled H/ACA snoRNA E3 (lanes 1 and 2) or C/D snoRNA U3 (lanes 3 and 4) at a 200- and 1000-fold molar excess (odd and even lanes, respectively). (C) Gel shift analysis of H/ACA snoRNA E3 (lanes 1–3) and of the stem–loop forming 3′ UTR of the Ash1 mRNA (lanes 4 and 5) in the absence of protein (lanes 1 and 4), the presence of recombinant GST–NHP2 (lanes 2 and 5), or of GST–NOP10 (lane 3). (D) The H/ACA core proteins and fibrillarin were co-translated in the presence of 32P-labeled H/ACA snoRNA E3 (lanes 1–3) and C/D snoRNA U3 (lanes 4–6) and precipitated with NAP57 antibodies as described in the legend to Figure 2. The input (lanes 1 and 4), the precipitates (lanes 2 and 5), and the labeled snoRNAs alone (lanes 3 and 6) are shown. (E) Same as in (D), except that only various combinations of the core trimer proteins and fibrillarin were used in the presence of 32P-labeled E3. The input (lanes 1–4) and the respective NAP57 precipitates (lanes 5–8) are depicted. (F) Co-precipitation of C/D snoRNA U3 (lanes 1 and 2), the H/ACA telomerase RNA hTR (lanes 3–5), and the 3′ UTR (see C; lanes 6–8) with the H/ACA core trimer. The input (lanes 1, 3, and 6), the NAP57 precipitates (lanes 2, 4, and 7), and the RNAs alone (lanes 5 and 8) are shown. (G) Co-precipitation of the H/ACA RNAs E3 (lanes 1–4) and hTR (lanes 5–8) with HA-NAP57 and derivatives (see Figure 3A) in the context of the core trimer. The RNAs alone (lanes 1 and 5) and the HA-antibody precipitates (lanes 2–4 and 5–8) are depicted. The position of the HA-NAP57 constructs is indicated (vertical lines).
Using NAP57 antibodies in our in vitro translation/immunoprecipitation assay, we tested if the context of the other H/ACA core proteins would alter the specificity of NHP2–RNA-binding (Figure 4D). Indeed, 32P-labeled H/ACA snoRNA E3, but not C/D snoRNA U3, precipitated with the 35S-methionine-labeled core complex (Figure 4D, lanes 2 and 5). To assay if H/ACA snoRNAs aided in NAP57–NHP2 interaction and what the minimal protein complex for their association was, E3 was incubated with a combination of core proteins (Figure 4E, lanes 1–4). E3 only co-precipitated with the NAP57–NOP10–NHP2 trimer (Figure 4E, lane 8), but not with NAP57 alone or in combination with either protein (lanes 5–7). Furthermore, GAR1 was not required for RNA binding (compare lanes 2 and 8 in Figure 4D and E, respectively). To test if the NAP57–NOP10–NHP2 trimer was responsible for the specificity of H/ACA snoRNA binding, the three proteins were precipitated in the presence of other 32P-labeled RNAs (Figure 4F). Neither C/D snoRNA U3 that competed for GST–NHP2 binding nor the mRNA 3′ UTR that bound efficiently to GST–NHP2 alone co-precipitated with the trimer (Figure 4F, lanes 1 and 2, and 6 and 7, respectively). However, human telomerase RNA (hTR), which ends in an H/ACA element, precipitated efficiently with the trimer (Figure 4F, lanes 3–5). Therefore, the core protein trimer NAP57–NOP10–NHP2 is responsible for the specific association of H/ACA RNAs.
Finally, we investigated if the above-described NAP57 point mutations (see Figure 3) would affect the association of H/ACA RNAs with the trimer (Figure 4G). The DC mutation F37V showed no apparent reduction in binding of E3 (Figure 4G, lanes 2 and 3) or hTR (lanes 6 and 7). However, in the context of the trimer, the catalytically inactivated mutant (D126A) precipitated both RNAs less efficiently than wild type (Figure 4G, lanes 4 and 8). When quantitated by phosphorimaging relative to the amount of precipitated NHP2, the D126A-containing trimer bound only 37% of E3 snoRNA and 49% of hTR compared to that with wild-type HA-NAP57. Note, as previously reported (Mitchell et al, 1999a), transcription of hTR yielded two bands, and despite gel purification of the upper (Figure 4F, lane 5, and 4G, lane 5) the lower band reappeared during incubation and also associated with the trimer, suggesting a folding variant (Figure 4F, lane 4, and 4G, lanes 6–8). In control experiments, the C/D snoRNA U3 did not bind to core complexes containing wild type or mutant HA-NAP57 constructs (data not shown). In summary, the D126A point mutation of NAP57 affected H/ACA RNA association but not core trimer formation.
Assembly of active and stable H/ACA snoRNPs
Although we apparently assembled intact H/ACA snoRNPs with all four in vitro-translated core proteins and an H/ACA snoRNA, no or only minimal pseudouridylase activity could be observed towards a cognate rRNA substrate (data not shown). This was most likely caused by the low amount of reconstituted snoRNPs and the variable association of GAR1 with them. Nevertheless, we had previously noticed that addition of exogenous H/ACA snoRNA to nuclear lysates boosted the pseudouridylase activity of these extracts above endogenous levels, suggesting the presence of assembly competent snoRNP core proteins or core complexes (Wang et al, 2002). Similarly, addition of H/ACA snoRNAs to nuclear extracts yielded snoRNP assembly, as analyzed by mobility shift assay (Dragon et al, 2000). We reasoned that cytosolic extracts might contain even more RNA-free core proteins. Indeed, addition of the H/ACA snoRNA E3 to inactive cytosolic S-100 extracts (Figure 5A, lane 1) yielded high pseudouridylase activity towards the E3 cognate rRNA substrate (lane 2), while addition of a different H/ACA snoRNA, E2, failed to reconstitute this activity (lane 3). As E3 itself is inactive (Wang et al, 2002), it apparently assembled into an active snoRNP together with the core proteins in the extract.
Figure 5.
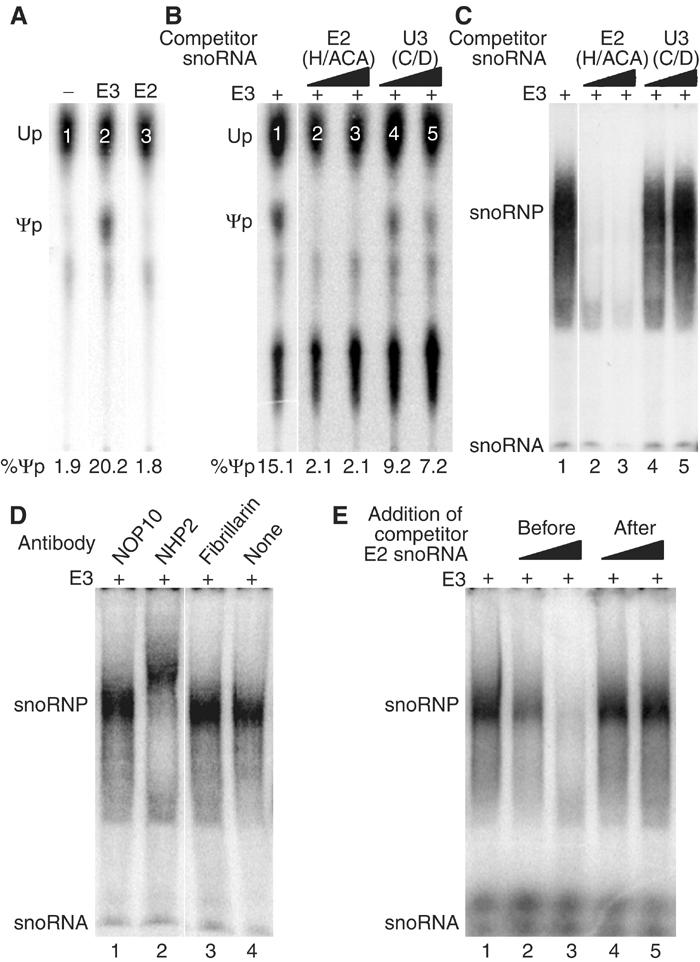
Assembly of functional E3 H/ACA snoRNPs in cytosolic extracts. (A) In vitro pseudouridylase assay. Autoradiograph of a thin-layer chromatogram of uridine (Up) and pseudouridine (Ψp) liberated from a short site-specifically 32P-labeled rRNA substrate (corresponding to the sequence modified by E3) after incubation with cytosolic S-100 extracts that had been incubated prior with unlabeled H/ACA snoRNAs E3 (lane 2) and E2 (lane 3) or no RNA (lane 1). The amount of pseudouridine produced is expressed as percent of uridine and pseudouridine combined. (B) Same as in (A), except that all extracts were first incubated with E3 in the presence of H/ACA snoRNA E2 (lanes 2 and 3) and C/D snoRNA U3 (lanes 4 and 5) in 10- and 100-fold molar excess (lanes 2 and 4, and 3 and 5, respectively). (C) Electrophoretic mobility shift analysis of 32P-labeled H/ACA snoRNA E3 after incubation with cytosolic S-100 extracts (lane 1) in the presence of unlabeled H/ACA snoRNA E2 (lanes 2 and 3) and C/D snoRNA U3 (lanes 4 and 5) in 10- and 1000-fold molar excess (lanes 2 and 4, and 3 and 5, respectively). The migrating position on the native gel of the labeled E3 snoRNA and the newly formed snoRNP is indicated on the autoradiograph. Note that most of E3 is degraded in the extracts unless incorporated into a snoRNP. (D) Same as lane 1 in (C), except that antibodies against the proteins indicated on top were added during snoRNP formation. Note the supershift with antibodies to NHP2 (lane 2). (E) Same as (C), except that unlabeled H/ACA snoRNA E2 was added before (lanes 2 and 3) or after (lanes 4 and 5) snoRNP formation in 10- and 1000-fold molar excess (lanes 2 and 4, and 3 and 5, respectively).
To test for the specificity of this assembly, we added together with E3 an excess of a different H/ACA (E2) and C/D (U3) snoRNA and assayed for pseudouridylase activity towards the E3 cognate rRNA substrate (Figure 5B). While E2 competed for the assembly and therefore activity (Figure 5B, lanes 2 and 3), U3 had only marginal effects (lanes 4 and 5). To monitor directly snoRNP assembly under these conditions, E3 snoRNA was uniformly 32P-labeled and snoRNP incorporation assayed by electrophoretic mobility shift analysis, as described previously (Dragon et al, 2000). One broad major shift was observed under these conditions (Figure 5C, lane 1), which was competed for by excess unlabeled E2 (lanes 2 and 3) but not U3 (lanes 4 and 5), as observed in the activity assay (Figure 5B). That at least NHP2, but not fibrillarin, was part of the RNP identified by mobility shift was confirmed by supershift analysis (Figure 5D, lanes 2 and 3). Antibodies to NOP10 failed to shift the band (Figure 5D, lane 1). This was possibly caused by masking of the epitope by NHP2 and NAP57 or by a lack of binding of these antibodies under the assay conditions.
To investigate if the core proteins associated with one snoRNA can switch to a different snoRNA, for example, to modify another site, we tested if unlabeled H/ACA snoRNA competed for snoRNP assembly when added after incubation with labeled E3 in our gel shift assembly assay (Figure 5E). When added before or simultaneously with labeled E3, unlabeled E2 competed for assembly (Figure 5E, lanes 2 and 3) but not when added after (lanes 4 and 5). Thus, functional H/ACA snoRNPs, once formed, are stable and unable to exchange their snoRNAs, suggesting that new particle formation requires de novo synthesis.
Discussion
Using in vitro assays, we delineate the molecular interactions of the five core components of H/ACA sno- and telomerase RNPs, NAP57, NOP10, NHP2, GAR1, and the RNA. The identified protein–protein and protein–RNA interactions suggest the following assembly path for these particles (Figure 6). NOP10 associates with NAP57 as a prerequisite for binding of NHP2 and consequent core trimer formation. GAR1 independently associates with NAP57 at a yet to be determined point during or after trimer formation. This assembly of a protein-only particle consisting of all four H/ACA core proteins likely occurs in the cytoplasm because nuclear import of GAR1, NHP2, and NOP10 appears to depend on their association with NAP57, which is the only core protein containing functional classic nuclear localization signals (Girard et al, 1994; Meier and Blobel, 1994; Heiss et al, 1999; Youssoufian et al, 1999; Pogacic et al, 2000). The protein-only core complex then enters the nucleus, where it associates with H/ACA RNAs to form functional and stable core particles with NAP57 and GAR1 situated at the pseudouridylation pockets of the H/ACA RNA hairpins. It will be interesting to determine where in the nucleus RNA binding occurs, for example, in the nucleoli or Cajal bodies, the major sites of H/ACA RNP accumulation, or at the sites of H/ACA RNA transcription and processing.
X-linked DC is mainly caused by missense mutations in the NAP57 gene. As NAP57 is part of an RNP, these mutations can be expected to affect the structure and function of the entire particle. This is best illustrated by depletion studies with its yeast ortholog Cbf5p, which leads to the depletion of all H/ACA snoRNAs and of Gar1p, presumably by destabilization of the particle, and to a loss of rRNA pseudouridylation (Lafontaine et al, 1998). Indeed, cells from X-linked DC patients show reduced levels of the H/ACA RNA hTR and shortened telomeres (Mitchell et al, 1999b). Our in vitro assembly system allowed us to differentiate between effects of NAP57 missense mutations on protein–protein or on protein–RNA interactions. We investigated two mutations, one observed in a family with DC (F37V) and the other that inactivates the pseudouridylase activity by replacing an aspartate in the catalytic center (D126A). The latter was shown in yeast to abolish rRNA pseudouridylation and to lead to a reduced level of H/ACA snoRNAs (Zebarjadian et al, 1999). While neither of the mutations appeared to interfere with assembly of the protein-only core complex (Figure 3B), the D126A mutation significantly reduced H/ACA RNA association (Figure 4G). Although only a single DC mutation has been identified in the highly conserved pseudouridylation domain of NAP57 (Knight et al, 1999), this result documents that NAP57 missense mutations can affect protein–RNA interactions in the RNP and could thereby cause reduced levels of H/ACA RNAs. Furthermore, these observations indicate that, in addition to NHP2, NAP57 participates in binding to H/ACA RNAs. While the effect of DC mutations in NAP57 may be subtler than the D126A exchange, for example, in the case of F37V, our assay will now allow testing the effects on RNP integrity of all other DC mutations identified in NAP57.
The established interactions in H/ACA RNPs could be direct or indirect because our studies in reticulocyte lysate cannot rule out participation of an intrinsic factor. However, several observations suggest that the interactions are direct. Based on Western blots with NAP57 antibodies (not shown) and on the fact that the observed interactions entirely depend on the exogenously translated proteins, for example, there is no intrinsic NOP10 to mediate NHP2–NAP57 interaction (Figure 4E, lane 7), reticulocyte lysate does not contain H/ACA snoRNPs or its precursors. As documented by purification of functional snoRNPs, the majority of H/ACA RNPs consist of exclusively the five core components and no additional factors (Wang et al, 2002). The same defined composition of core particles was previously determined in purified yeast H/ACA snoRNPs (Lübben et al, 1995; Henras et al, 1998; Watkins et al, 1998). A direct interaction between NAP57 and GAR1 is also consistent with the identification of Cbf5p, the yeast NAP57 ortholog, in a yeast two-hybrid screen with Gar1p as bait (Henras et al, 1998). Furthermore, genetic depletion studies of the evolutionaily conserved yeast H/ACA snoRNP core proteins support the association map defined in this work (Figure 6). Thus, depletion of any of the three trimer proteins, but not of GAR1, results in a specific loss of H/ACA snoRNAs and GAR1 (Henras et al, 1998; Lafontaine et al, 1998; Watkins et al, 1998). In yeast cells therefore, the trimer also appears to provide the platform for RNA and GAR1 binding. In summary, although additional factors may function during biogenesis of H/ACA snoRNPs, for example, Shq1p, Naf1p, Rnt1p, and/or Rvb2p in yeast (King et al, 2001; Dez et al, 2002; Fatica et al, 2002; Tremblay et al, 2002; Yang et al, 2002), the molecular interactions defined in this study for mature H/ACA particles appear to be direct.
The stoichiometry of the individual components of H/ACA core particles remains to be established. Several arguments suggest that each H/ACA RNA assembles with a complement of two of each of the core proteins (Figure 6, pathway A). Eucaryotic H/ACA RNAs are characterized by two hairpins, both of which can contain pseudouridylation pockets and consequently both require the pseudouridylase NAP57 and GAR1, which we located near the catalytic core by UV-crosslinking. Purified yeast H/ACA snoRNPs exhibit a two-domain structure by electron microscopy and their derived molecular weight accommodates two copies of each core protein and one snoRNA (Lübben et al, 1995; Watkins et al, 1998). Moreover, many trypanosomal and archaeal H/ACA snoRNAs possess only a single hairpin, indicating that the full complement of core proteins can bind to a single hairpin (Liang et al, 2001; Tang et al, 2002). However, other evidence suggests an asymmetrical distribution of core proteins (Figure 6, pathway B). For example, the molar ratio of NOP10/NHP2 is close to two in all NAP57 precipitates, indicating either the precipitation of twice as many NAP57–NOP10 as NAP57–NOP10–NHP2 complexes or, pertinent to the present argument, that the precipitated complex contains only one NHP2 for every two NOP10 and NAP57, that is, (NAP57–NOP10)2–NHP2. Although eucaryotic H/ACA snoRNAs are structurally and functionally symmetrical with two equivalent hairpins followed each by a conserved H (ANANNA) or ACA (ANA) motif, mutation of either element abolishes pseudouridylation guided by both hairpins (Bortolin et al, 1999). This suggests the presence of a common factor, for example, NHP2, that anchors two pseudouridylases, one for each hairpin. Asymmetrical binding of one NHP2 per H/ACA RNA despite two symmetrical structural and sequence elements would mirror the asymmetric binding of its homolog 15.5K/NHP2L1 to symmetrical C/D snoRNAs (Cahill et al, 2002; Weinstein Szewczak et al, 2002). Finally, it is possible that both pathways (Figure 6A and B) coexist in eucaryotic cells as not all H/ACA RNAs and for that matter RNPs may be created equally. For example, not all snoRNAs are affected equally by the depletion of individual core proteins in yeast (Henras et al, 1998, 2001). Additionally, U17 and hTR differ from other H/ACA RNAs in that their 3′ hairpins alone, unlike those of E3 and E2, are sufficient to compete for RNP assembly in the nuclear extract (Dragon et al, 2000). These data are consistent with our finding that the 3′ hairpin of E3, with or without the H box, failed to be precipitated by the core trimer (data not shown).
Although the core trimer is required for the specificity of H/ACA RNA recognition, mostly NHP2 appears responsible for binding. For example, NHP2 alone binds RNA nonspecifically and a protein of its mobility UV crosslinks most prominently to H/ACA RNAs in in vitro-assembled snoRNPs (this study; Dragon et al, 2000; Henras et al, 2001). Moreover, NHP2 apparently evolved from the archaeal L7Ae, which is also the common ancestor of the 15.5K/NHP2L1 and the ribosomal L30 proteins and which binds specifically to K-turn motifs of individual hairpins in archaeal H/ACA RNAs (Rozhdestvensky et al, 2003). Since typical K-turn motifs are apparently absent from eucaryotic H/ACA RNAs (Rozhdestvensky et al, 2003), NHP2 may have lost its RNA-binding specificity during evolution but regained it by teaming up with NAP57 and NOP10. This RNA binding of NHP2 appears to occur at a distance from the target uridine, which was efficiently crosslinked to NAP57 and GAR1 but not to NHP2.
While crosslinking of the target uridine to NAP57 could be anticipated because NAP57 had been identified as the putative pseudouridylase of H/ACA snoRNPs, crosslinking to GAR1 was not. It is not clear if the crosslink to GAR1 occurred in the same substrate conformation as that to NAP57, or if there are two conformations, one that brings the target uridine closer to the enzyme and one that has it situated closer to GAR1. Either way, GAR1 may serve as a clamp to hold the substrate RNA in place. Such a model is supported by the requirement of GAR1 for snoRNP association with higher order structures in yeast nucleoli (Bousquet-Antonelli et al, 1997). Our system now allows further dissecting if and how mutations in NAP57 or the other core proteins and H/ACA RNAs affect their molecular interactions, which apparently lie at the heart of the molecular mechanism of DC.
Materials and methods
In vitro transcription
All RNAs were transcribed using a T7-MEGA shortscript transcription kit (Ambion, Austin, TX) and gel purified as described previously (Wang et al, 2002). For labeling purposes, [α-32P]-UTP was included in the reaction. The DNA templates for the respective RNAs were constructed as follows: rat E3 and E2 snoRNAs were previously described (Wang et al, 2002); hTR under the T7 promoter was amplified from pGRN33 (Feng et al, 1995; from Geron, Menlo Park, CA) using primers U250 and U251 (for primer sequences, see Supplementary Table I); rat U3 snoRNA and the 3′ UTR of yeast Ash1 mRNA were generated by linearizing U3pSP64 (a kind gift from Susan Baserga) with RsaI and pRS168 (Long et al, 2000) with HindIII, respectively.
For all other methods, see Supplementary data.
Supplementary Material
Supplementary Material
Acknowledgments
We are indebted to John Aris, Susan Baserga, Pascal Chartrand, Witek Filipowicz, Michael Henry, Angus Lamond, Vanda Pogacic, Charles Query, and Rob Singer for the generous gift of reagents. We thank Xavier Darzacq, Charles Query, and Susan Smith for critical comments on the manuscript and throughout this work. This study was supported by a grant from the American Cancer Society.
References
- Aravind L, Koonin EV (1999) Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol 48: 291–302 [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaille J (1997) Guiding ribose methylation of rRNA. Trends Biochem Sci 22: 257–261 [DOI] [PubMed] [Google Scholar]
- Bagni C, Lapeyre B (1998) Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem 273: 10868–10873 [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Pintard L, Lutfalla G (2003) Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol Cell 12: 1309–1315 [DOI] [PubMed] [Google Scholar]
- Bortolin ML, Ganot P, Kiss T (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Henry Y, Gélugne J-P, Caizergues-Ferrer M, Kiss T (1997) A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J 16: 4770–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill NM, Friend K, Speckmann W, Li ZH, Terns RM, Terns MP, Steitz JA (2002) Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J 21: 3816–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–336 [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ (2003) RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 278: 695–698 [DOI] [PubMed] [Google Scholar]
- Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y (2001) Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res 29: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y (2002) Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol Cell Biol 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I, Vulliamy T (2003) Dyskeratosis congenita: its link to telomerase and aplastic anaemia. Blood Rev 17: 217–225 [DOI] [PubMed] [Google Scholar]
- Dragon F, Pogacic V, Filipowicz W (2000) In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol 20: 3037–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Dlakic M, Tollervey D (2002) Naf1 p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514 [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B (1995) The RNA component of human telomerase. Science 269: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Frankel A, Clarke S (1999) RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem Biophys Res Commun 259: 391–400 [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Girard JP, Bagni C, Caizergues-Ferrer M, Amalric F, Lapeyre B (1994) Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J Biol Chem 269: 18499–18506 [PubMed] [Google Scholar]
- Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B (1992) GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J 11: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Liu Y, Santi DV (1999) The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc Natl Acad Sci USA 96: 14270–14275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS, Girod A, Salowsky R, Wiemann S, Pepperkok R, Poustka A (1999) Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum Mol Genet 8: 2515–2524 [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I (1998) X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M (2001) Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res 29: 2733–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR (2001) Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939 [DOI] [PubMed] [Google Scholar]
- Huang L, Pookanjanatavip M, Gu X, Santi DV (1998) A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 37: 344–351 [DOI] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon H-J, Fouquet C, Carbon J (1993) An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol Cell Biol 13: 4884–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ (2001) A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol 21: 7731–7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ (2003) Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425–435 [DOI] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Aalfs CM, McMahon C, Richmond P, Jones A, Hennekam RC, Poustka A, Mason PJ, Dokal I (1999) Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal–Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br J Haematol 107: 335–339 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D (1998) The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 12: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu L, Michaeli S (2001) Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J Biol Chem 276: 40313–40318 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P (2000) She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübben B, Fabrizio P, Kastner B, Lührmann R (1995) Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J Biol Chem 270: 11549–11554 [DOI] [PubMed] [Google Scholar]
- Marrone A, Mason PJ (2003) Dyskeratosis congenita. Cell Mol Life Sci 60: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT (2003) Dissecting dyskeratosis. Nat Genet 33: 116–117 [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G (1994) NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol (correction appeared in 140: 447) 127: 1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K (1999a) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K (1999b) A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R (1999) Functional interaction of a novel 15.5 kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J 18: 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J (1995) Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA 1: 102–112 [PMC free article] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol 20: 9028–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A (2003) Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP (2003) Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262 [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89: 669–672 [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A (2002) Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Kiss T (1997) Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol 9: 337–342 [DOI] [PubMed] [Google Scholar]
- Tremblay A, Lamontagne B, Catala M, Yam Y, Larose S, Good L, Elela SA (2002) A physical interaction between Gar1p and Rnt1pi is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol Cell Biol 22: 4792–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Query CC, Meier UT (2002) Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate RRNA independently of their association with phosphorylated Nopp140. Mol Cell Biol 22: 8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW (2000) Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R (1998) Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all box H/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4: 1549–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R (2000) A common core RNP structure shared between the small nuclear box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Weinstein Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA (2002) Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem Biol 9: 1095–1107 [DOI] [PubMed] [Google Scholar]
- Xu C, Henry PA, Setya A, Henry MF (2003) In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA 9: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G (2002) The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J Biol Chem 277: 45235–45242 [DOI] [PubMed] [Google Scholar]
- Youssoufian H, Gharibyan V, Qatanani M (1999) Analysis of epitope-tagged forms of the dyskeratosis congenital protein (dyskerin): identification of a nuclear localization signal. Blood Cells Mol Dis 25: 305–309 [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J (1999) Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol 19: 7461–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


