Abstract
The yeast Snf1 protein kinase and its animal homologue, the AMP-activated protein kinase, play important roles in metabolic regulation, by serving as energy gauges that turn off energy-consuming processes and mobilize energy reserves during low-energy conditions. The closest homologue of these kinases in plants is Snf1-related protein kinase 1 (SnRK1). We have cloned two SnRK1-encoding genes, PpSNF1a and PpSNF1b, in the moss Physcomitrella patens, where gene function can be studied directly by gene targeting in the haploid gametophyte. A snf1a snf1b double knockout mutant is viable, but lacks all Snf1-like protein kinase activity. The mutant has a complex phenotype that includes developmental abnormalities, premature senescence and altered sensitivities to plant hormones. Remarkably, the double knockout mutant also requires continuous light, and is unable to grow in a normal day–night light cycle. This suggests that SnRK1 is needed for metabolic changes that help the plant cope with the dark hours of the night.
Keywords: AMPK, metabolic regulation, Physcomitrella , Snf1, SnRK1
Introduction
The ability to monitor the energy status of cells and tissues, and to adjust the metabolism of available nutrients accordingly, is important to all eukaryotes. One example of such regulation is found in the yeast Saccharomyces cerevisiae, which preferentially ferments the favoured carbon source glucose. In order to save energy, enzymatic reactions that are needed to metabolize other carbon sources are inhibited in the presence of glucose, mainly by transcriptional repression. This mechanism is known as glucose repression (Ronne, 1995; Johnston, 1999) and affects a large number of genes that are required for the uptake and metabolism of alternative carbon sources as well as genes required for oxidative growth. A key component in glucose repression is the Snf1 protein kinase (Celenza and Carlson, 1986), which regulates the nucleocytoplasmic localization of the Mig1 repressor (Nehlin and Ronne, 1990) by differential phosphorylation in a glucose-dependent manner (De Vit et al, 1997; Smith et al, 1999).
Genes encoding Snf1-related kinases have been isolated from all eukaryotic kingdoms and it is thought that these kinases share an evolutionary conserved function as metabolic sensors or energy gauges (Hardie et al, 1998). In animals, the kinase is known as the AMP-activated protein kinase, AMPK, since it is regulated by the ATP:AMP ratio. In plants, there are several Snf1-related kinases, which fall into three major subfamilies referred to as SnRK1, SnRK2 and SnRK3 (SnRK: Snf1-related protein kinase) (Halford and Hardie, 1998; Halford et al, 2003, 2004). The SnRK2 and SnRK3 subfamilies are rather divergent in sequence and appear to be unique to plants, while the SnRK1 kinases, based on sequence similarity, are the closest homologues of the yeast Snf1 kinase and the mammalian AMPK.
The biochemical properties of SnRK1 kinases have been extensively studied in several plant species (Halford et al, 2003), and the identification of substrates like HMG-CoA reductase, sucrose phosphate synthase and nitrate reductase (Dale et al, 1995; Sugden et al, 1999) is consistent with a function in metabolic control. This conclusion is also supported by antisense experiments. Thus, Purcell et al (1998) found that potato SnRK1 is needed for sucrose induction of sucrose synthase expression and Zhang et al (2001) showed that barley SnRK1 is required for the formation of functional pollen, possibly because it is needed for starch accumulation. Furthermore, experiments in wheat embryos suggested a role for SnRK1 in regulating the expression of α-amylase (Laurie et al, 2003). Although many details remain to be elucidated, these findings suggest a role for plant SnRK1 kinases in regulating the carbon and energy metabolism, which is similar to the proposed roles of the yeast Snf1 kinase and the mammalian AMPK (Hardie et al, 1998).
The moss Physcomitrella patens has recently emerged as a powerful model system in plant functional genomics following the discovery that gene targeting by one-step gene disruption works in this plant, with frequencies comparable to those in yeast (reviewed by Schaefer, 2002). Furthermore, the main vegetative growth phase of Physcomitrella is the haploid gametophyte, which makes it possible to study the phenotype of recessive loss-of-function mutations such as gene knockouts without further crosses. In order to learn more about the function of the Snf1-related kinases in plants, we have cloned two Physcomitrella genes, PpSNF1a and PpSNF1b, encoding SnRK1 kinases and characterized their in vivo functions by targeted gene knockouts. Single knockout mutants had no obvious phenotypes except a reduced Snf1-like kinase activity in protonemal tissue extracts. The double knockout mutant is devoid of Snf1-like kinase activity, and has a severe pleiotropic phenotype that includes an inability to grow in a normal day–night light cycle.
Results
Two closely related genes encode type 1 Snf1-related kinases in Physcomitrella
We used degenerative PCR combined with 5′- and 3′-RACE to clone sequences encoding Snf1-related kinases from the moss P. patens. As described in Materials and methods, we obtained full-length cDNAs and the corresponding genomic sequences of two closely related genes that we named PpSNF1a and PpSNF1b. Both genes have 10 exons that are spliced to form cDNAs encoding highly similar proteins of 542 and 545 amino-acid residues, respectively. The kinase domains (amino acids 19–273) have 96.5% identical residues, while the rest of the two proteins show 67.4% identity to each other. In order to determine if there are any other closely related genes in Physcomitrella, we performed low-stringency Southern blots with probes from the kinase domain-encoding regions. These probes are specific for PpSNF1a and PpSNF1b, respectively, but crosshybridization between the two genes could easily be detected (Supplementary Figure 1). No other hybridizing bands were seen, suggesting that there are no other genes in Physcomitrella that are closely related to PpSNF1a and PpSNF1b.
A phylogenetic analysis based on amino-acid sequences clearly shows that PpSnf1a and PpSnf1b belong to the type 1 (SnRK1) subfamily of the Snf1-related kinases (Figure 1 and Supplementary Figure 2). As expected, they also have an exon–intron organization resembling that of other plant SnRK1-encoding genes, such as Arabidopsis AKIN10 (Le Guen et al, 1992). We proceeded to examine the expression of PpSNF1a and PpSNF1b. Since mRNA levels were too low to be easily detected in Northern blots, we used quantitative RT–PCR analysis with internal standards as described by Siebert and Larrick (1992). Total RNA was prepared from young protonemal tissue that had been exposed to different treatments, including growth in the dark or in intense light, and growth in the presence of 0.2 M glucose or mannitol. We found that none of the treatments tested had any significant effect on the expression of either PpSNF1a or PpSNFb (Supplementary Figure 3). We conclude that both genes seem to be constitutively expressed in young protonemal tissue.
Figure 1.
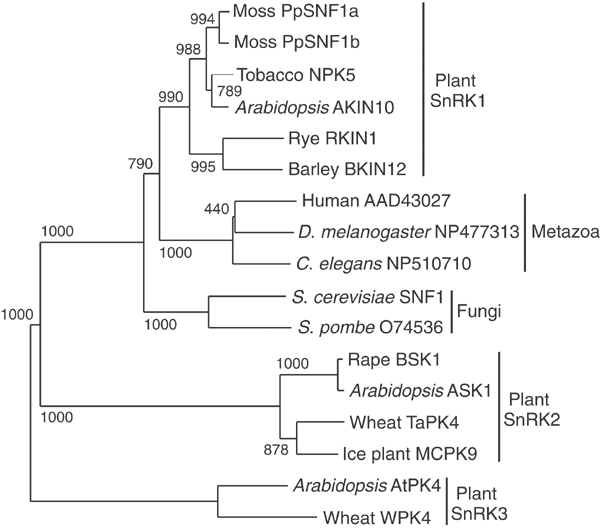
Phylogenetic tree of Snf1-related kinases showing the positions of PpSnf1a and PpSnf1b. The conserved kinase domain amino-acid sequences corresponding to residues 20–272 in PpSnf1a and PpSnf1b were aligned using ClustalX (Thompson et al, 1997) with default settings. The tree was computed from the aligned sequences using the neighbour-joining method with correction for multiple substitutions and exclusion of gaps in the alignment. Numbers shown are bootstrap values for branch points based on 1000 samples. The three subfamilies of Snf1-related kinases that have been identified in plants (Halford and Hardie, 1998) are also shown.
PpSnf1a and PpSnf1b can complement a yeast snf1 mutant and are regulated by the Snf4 protein in yeast
Several plant SnRK1 kinases, first exemplified by RKIN1 (Alderson et al, 1991), can complement the glucose derepression defect of a yeast snf1 mutant, thereby enabling it to grow on other carbon sources than glucose. We therefore proceeded to test if the PpSNF1a and PpSNF1b cDNAs can complement the glucose derepression defects of snf1 when expressed from the methionine-regulated MET3 promoter in yeast. The yeast SNF1 gene expressed from the same promoter was included as a control. As shown in Figure 2, we found that both PpSNF1a and PpSNF1b can support growth of the snf1 strain on the non-repressing carbon source raffinose, indicating functional complementation of the snf1 defect. However, neither cDNA could support growth of the snf1 strain on lactate, which is a more stringent test for glucose derepression. We conclude that PpSNF1a and PpSNF1b can partially complement a snf1 mutant in yeast.
Figure 2.
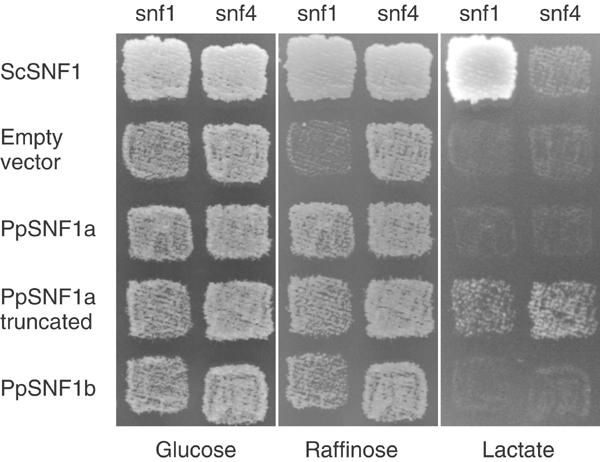
Complementation of yeast snf1 and snf4 mutants by the PpSNF1a and PpSNF1b cDNAs. Yeast strains H316 (snf1) and H1307 (snf4) were transformed with plasmids expressing the yeast SNF1 gene, the PpSNF1a cDNA, the PpSNF1b cDNA, or a truncated PpSNF1a cDNA encoding only the kinase domain. The empty vector pJO177 was also included as a control. Growth was tested on synthetic media containing either glucose, raffinose or lactate, but lacking uracil and methionine in order to select for maintenance of the plasmids and derepression of the MET3 promoter.
The yeast Snf1 kinase is a heterotrimeric complex made up of the catalytic α subunit encoded by SNF1, a β subunit encoded by the partially redundant SIP1, SIP2 and GAL83 genes, and a γ subunit encoded by SNF4 (Jiang and Carlson, 1997). A model has been proposed for how Snf1 is regulated in which a C-terminal regulatory domain binds to the kinase domain and blocks the active site in the presence of glucose. During glucose depletion, Snf4 binds to the Snf1 regulatory domain, thus relieving this autoinhibition (Johnston, 1999). Consistent with this model, a deletion of the C-terminal regulatory domain makes the yeast Snf1 kinase partly independent of Snf4 (Celenza and Carlson, 1989). We proceeded to test PpSNF1a and PpSNF1b for their ability to suppress a yeast snf4 mutant. Since snf4 mutants can grow on raffinose, growth on lactate was used to score the phenotype (Figure 2). We found that neither PpSNF1a nor PpSNF1b can suppress the snf4 phenotype, indicating that they are dependent on Snf4 for their ability to function in yeast. Significantly, we further found that a deletion of the C-terminal domain in PpSnf1a made it independent of Snf4, enabling the snf4 mutant to grow on lactate (Figure 2). We conclude that PpSnf1a and PpSnf1b not only can complement the kinase activity of Snf1 but are also regulated by Snf4.
Targeted disruptions show that PpSNF1a and PpSNF1b have similar functions in vivo
We proceeded to make knockout constructs for both PpSNF1a and PpSNF1b (Figure 3). To facilitate the generation of double mutants, different selection markers were used in the two constructs: neomycin resistance for PpSNF1a and hygromycin resistance for PpSNF1b. Moss protoplasts were transformed as described by Schaefer et al (1991). Three independent clones from each gene disruption were selected for verification. For each clone, the presence of the disruption in the chromosomal DNA was checked by PCR and the disappearance of the transcript was confirmed by RT–PCR (Figures 3C and D). Initial phenotypic analyses failed to reveal any phenotypes associated with either knockout mutant, except for a partial effect on Snf1-related kinase activity (see below). This suggested that the two genes are functionally redundant, and that a double knockout mutant would be required. Such mutants were generated by transforming a verified snf1a mutant with the PpSNF1b knockout construct. As with the single knockout mutants, three independent clones were verified (Figures 3C and D) and then further analysed. Unlike the single disruptions, the double knockouts had a number of distinct phenotypes, which are further described below. We conclude that the proteins encoded by PpSNF1a and PpSNF1b have similar or identical functions in vivo. We further note that all clones sharing the same genotype behaved identically under all circumstances tested (for an example, see Figure 5A). Therefore, one representative clone of each knockout, referred to as the snf1a, snf1b and snf1a snf1b mutant, is shown in most of the figures below.
Figure 3.
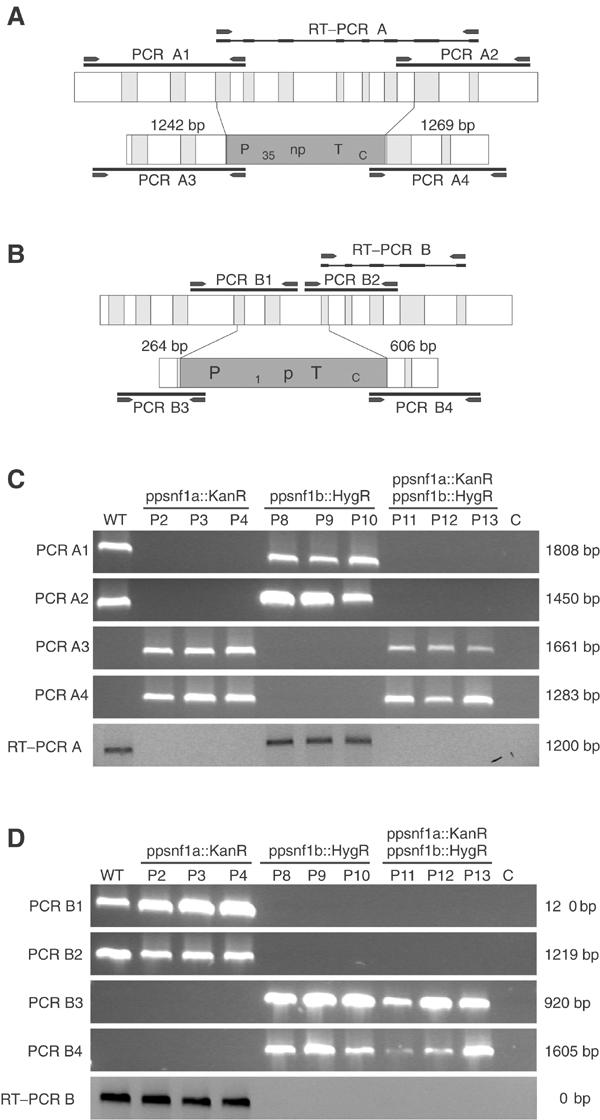
Gene targeting of PpSNF1a and PpSNF1b. (A) Overview of the PpSNF1a gene and the PpSNF1a targeting construct, in which exons 3–8 were replaced by the G418 resistance cassette. The PCR and RT–PCR products used to verify the deletions in (C) are shown as black bars. The primers used to amplify these products are shown as arrowheads. (B) Overview of the PpSNF1b gene and the PpSNF1b targeting construct, in which exons 4–6 were replaced by the hygromycin resistance cassette. The PCR and RT–PCR products used to verify deletions in (D) are shown as black bars. The primers used to amplify these products are shown as arrowheads. (C) Verification of PpSNF1a knockouts. Genomic DNA was prepared from the wild type (WT) and three independent knockouts of each type (P2–P13) grown on BCD medium under standard conditions. The PCR products A1–A4 were obtained using this DNA as template and the PpSNF1a-specific primers shown in (A). RT–PCR products obtained with total RNA prepared from the same strains and PpSNF1a-specific primers are shown below. (D) Verification of PpSNF1b knockouts. The same strains were used as in (C). The PCR products B1–B4 were obtained using this DNA as template and the PpSNF1b-specific primers shown in (B). RT–PCR products obtained with total RNA prepared from the same strains and PpSNF1b-specific primers are shown below.
Figure 5.
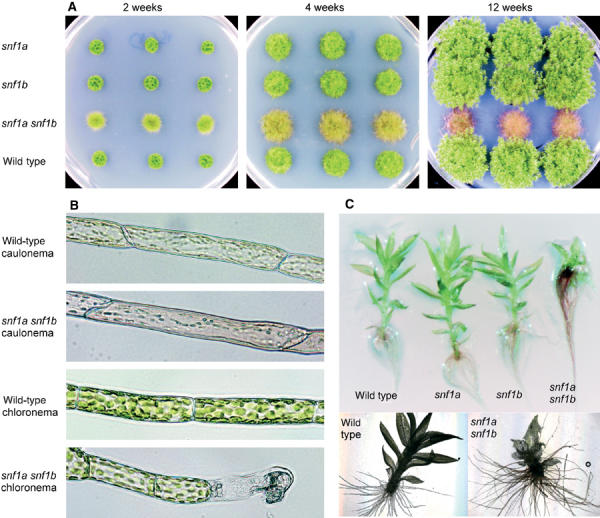
Developmental phenotypes of the snf1a snf1b double mutant. (A) Colony morphology of the wild type, the snf1a and snf1b single mutants, and the snf1a snf1b double mutant after 2, 4 and 12 weeks under standard growth conditions (BCD media, 2000 lux, continuous light). Three independent knockout mutants are shown in each case. (B) Morphology of caulonemal air filaments and chloronemal filaments from the wild type and the snf1a snf1b double mutant. (C) Leafy shoots from 4-week-old colonies of the wild type, the snf1a and snf1b single mutants, and the snf1a snf1b double mutant.
PpSnf1a and PpSnf1b together account for all Snf1-like kinase activity in protonemal tissue
Snf1-related kinases from plants, animals and fungi all share similar substrate specificities (Hardie et al, 1998) and can therefore be assayed with the same synthetic peptide substrates. A widely used substrate is the SAMS peptide (Davies et al, 1989), which is a modified version of the sequence around the AMPK target site in rat acetyl-CoA carboxylase. SAMS-peptide phosphorylating activity in plants was first described by Mackintosh et al (1992), and the SAMS peptide has subsequently been used to characterize several Snf1-related enzymes in plants (Halford et al, 2003). We proceeded to assay SAMS-peptide phosphorylating activity in crude extracts from Physcomitrella young protonemal tissue (Figure 4A). The assay was linear within the range shown both for total protein added and for time (data not shown). We found that protein extracts from wild-type Physcomitrella grown under standard conditions have a SAMS phosphorylating activity of 26.5±0.9 pmol mg−1 min−1 (Figure 4A). Growth for 20 h in the dark with or without 0.15 M glucose, or treatment with 1 μM auxin (IAA) or 1 μM cytokinin (BAP) for 20 h did not significantly affect the SAMS phosphorylating activity (Supplementary Figure 4). We conclude that the Snf1-like kinase activity in Physcomitrella does not seem to be regulated by these environmental conditions, although it is possible that some in vivo regulation exists that is not retained in the extracts.
Figure 4.
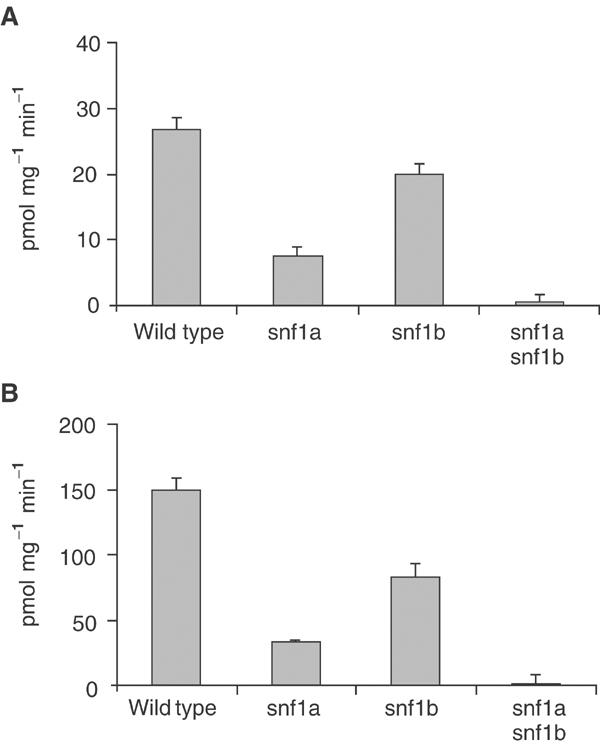
PpSNF1a and PpSNF1b together account for all SAMS phosphorylating activity in Physcomitrella. The enzyme activity is expressed as pmol phosphate incorporated into SAMS peptide per minute and mg of protein. The numbers shown are averages±standard deviations for three samples. For each sample, the activity without added peptide was subtracted from the activity with added peptide. (A) Activities in crude extracts of the wild type, the snf1a mutant, the snf1b mutant and the snf1a snf1b double mutant. (B) Activities in ammonium sulphate-precipitated fractions (15–30% saturation) of the wild type, the snf1a mutant, the snf1b mutant and the snf1a snf1b double mutant.
We next assayed the kinase activity in our knockout mutants. As shown in Figure 4A, both single mutants have significantly reduced activities as compared to the wild type. This shows that both proteins possess Snf1-like kinase activity, with PpSnf1a being the most active enzyme under the conditions tested. Significantly, no activity could be detected in the snf1a snf1b double knockout mutant (Figure 4A). This suggests that PpSnf1a and PpSnf1b are the only two Snf1-like kinases in protonemal tissue. This is consistent with the fact that the enzyme activities in the two single knockout mutants add up to the activity in the wild type, indicating that PpSnf1a contributes 72% and PpSnf1b 28% of the total activity. To verify these findings, we proceeded to assay SAMS-peptide phosphorylating activity after partial purification of the kinase by ammonium sulphate precipitation (see Materials and methods). Most of the activity was recovered in the 15–30% ammonium sulphate fraction, where the specific activity had been increased 5.6-fold to 150 pmol mg−1 min−1 in the wild type (Figure 4B). This is consistent with results from other plants where Snf1-like kinase activity was recovered in a 0–35% ammonium sulphate fraction resulting in a 3.5-fold purification (Ball et al, 1994). The results obtained with the knockout mutants were similar to those described above, with a clearly reduced kinase activity in either single mutant and no detectable activity in the double mutant. Furthermore, the contributions of each kinase as estimated from the single knockout data were identical to those in crude extracts (72% for PpSnf1a and 28% for PpSnf1b). We conclude that PpSnf1a and PpSnf1b together account for all Snf1-like kinase activity in young protonemal tissue.
The snf1a snf1b double mutant has severe pleiotropic phenotypes affecting growth, development and senescence
The formation of the Physcomitrella colony from regenerating spores or protoplasts starts with a filamentous tissue type called protonemata that grow by apical cell divisions (Reski, 1998). There are two types of filaments: chloronemata and caulonemata. The regenerating cells first develop into photosynthetically active chloronemal filaments, which occasionally differentiate into caulonemal cells characterized by oblique cell walls and smaller and fewer chloroplasts. Caulonemal side branch initials can give rise to buds, which subsequently produce gametophores, also known as leafy shoots. Each leafy shoot resembles a small plant with leaves, and has root-like rhizoids at its base.
We found that the snf1a snf1b double knockout mutant has several pronounced phenotypes that affect the development of both protonemata and leafy shoots. First, there are striking effects on protonemal filament formation. Thus, while young wild-type colonies consist mainly of a dense network of chloronemal filaments, the snf1a snf1b double mutant has a large excess of caulonemal filaments, which grow up into the air (Figures 5A and B). Such caulonemal air filaments are formed sporadically also in the wild type, but in the double mutant they comprise a major part of the colony. Furthermore, those few chloronemal filaments that are formed in the snf1a snf1b double mutant are aberrant, with shorter cells and rapid death of the apical cell, particularly under low light conditions (Figure 5B).
Second, there are severe effects on gametophore formation and development in the snf1a snf1b double mutant. Very few leafy shoots are formed in the double mutant, as compared to the wild type. Furthermore, those leafy shoots that are formed have shorter stems and smaller leaves with developmental abnormalities ranging from subtle to severe. Interestingly, leafy shoots from the double mutant instead have a large excess of rhizoids (Figure 5C). This suggests that the balance between leaf and rhizoid formation has been disturbed in the double mutant.
A third pronounced phenotype of the snf1a snf1b double mutant is its premature senescence. Initially, the double mutant colonies grow faster than wild-type colonies, due to the excess of caulonemal filaments, which promote radial growth. However, the double mutant colonies essentially cease to grow after 4 weeks (Figure 5A). The cells become increasingly pigmented and the colony assumes a wilty appearance. In contrast, the wild-type and the single mutant colonies continue to grow for several weeks under the same conditions with no signs of senescence (Figure 5A).
The snf1a snf1b double mutant is hypersensitive to auxin and hyposensitive to cytokinin
Growth and development in mosses, as in higher plants, is regulated by plant hormones. Specifically, auxins promote caulonema and rhizoid formation in Physcomitrella and inhibit the formation of secondary chloronema and leafy shoots, while cytokinins induce bud formation (Ashton et al, 1979). We therefore proceeded to examine how our mutants respond to these plant hormones. Wild-type Physcomitrella produces colonies with a low-density morphology in the presence of 1 μM auxin (Figure 6A), and even higher concentrations inhibit growth (not shown). Interestingly, the snf1a snf1b double mutant shows a pronounced hypersensitivity to auxin, with significant growth inhibition at 0.1 μM auxin and no growth at all at 1 μM auxin (Figure 6A).
Figure 6.
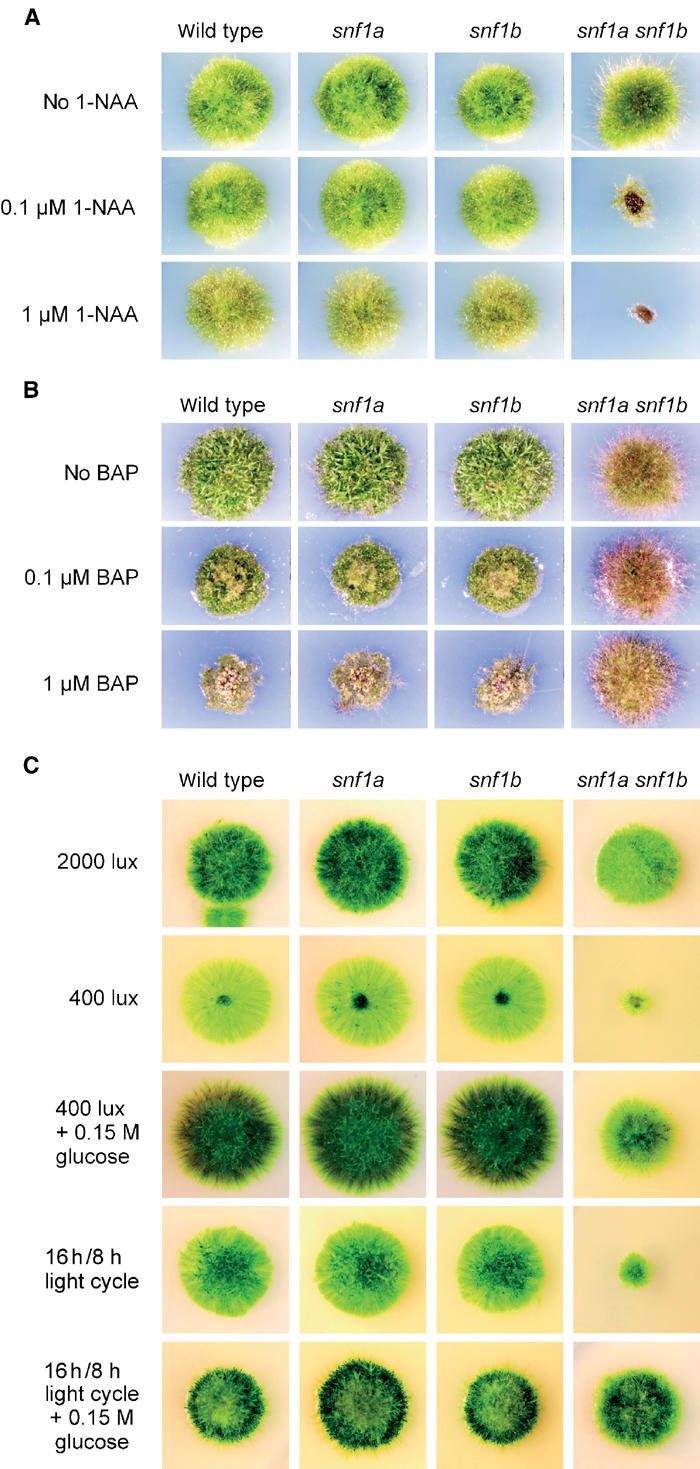
Effects of the snf1a snf1b double mutant on growth under different conditions. Small pieces of fresh protonemal tissue were transferred to tester plates on day one. The plates were then incubated under different conditions for 3–4 weeks. In each panel, the wild type and the snf1a and snf1b single mutants are also shown as controls. (A) Growth in the presence of the auxin 1-naphthaleneacetic acid (1-NAA). (B) Growth in the presence of the cytokinin 6-benzylaminopurine (BAP). (C) Growth in either high or low continuous light, or in a 16 h/8 h day–night light cycle.
Cytokinins at low concentrations induce bud formation in Physcomitrella, whereas higher concentrations inhibit growth and promote the formation of calluses (Ashton et al, 1979). Consistent with this, we could detect callus formation, accompanied by a reduction in colony size, when the wild-type strain was grown in the presence 0.1 μM cytokinin (Figure 6B). In contrast, the snf1a snf1b double mutant is hardly affected at all by concentrations up to 1 μM cytokinin (Figure 6B). We conclude that the snf1a snf1b double mutant is hyposensitive to cytokinin.
The snf1a snf1b double mutant is unable to grow in low light or in a normal day–night light cycle
Since Snf1-related protein kinases have been proposed to share a common function as gauges of cellular energy levels (Hardie et al, 1998), we proceeded to test how the snf1a snf1b double mutant responds to conditions of limited or variable energy supply. As shown in Figure 6C, we found that while the double mutant grows well in high light (2000 lux), it fails to grow at low light intensities (400 lux). Light microscopy analysis of the double mutant tissue grown at 400 lux revealed short chloronemal filaments with a very high incidence of dead apical cells, as previously described in Figure 5B. Significantly, the ability of the double mutant to grow in low light can be partially restored by adding an external carbon source (Figure 6C). This suggests that the failure to grow in low light is not due to a light signalling defect, but instead reflects a failure to adapt to a reduced energy supply.
The most obvious example of reduced light conditions that all plants must be able to cope with is the night. We therefore proceeded to examine how the double mutant is affected by growth in a normal (16 h/8 h) day–night light cycle instead of the continuous light used in all preceding experiments. Remarkably, we found that the double mutant is unable to grow in a day–night light cycle, even though high light (2000 lux) is provided for 16 h each day (Figure 6C). Also in this case, growth of the double mutant could be partially restored by the addition of external glucose (Figure 6C). This result shows that the Snf1-related kinase is essential not only for growth in the presence of a fixed but limited energy supply, but also for growth when the energy supply is plentiful but varies during the day. We conclude that the type 1 Snf1-related protein kinases encoded by PpSNF1a and PpSNF1b are essential for growth in low or variable light, which is consistent with the notion that these kinases are important for handling low-energy conditions.
The snf1a snf1b double mutant is deficient in starch accumulation
Plants accumulate starch within the chloroplasts during the day, which is subsequently mobilized to serve as an energy source during the night. Experiments in Arabidopsis have shown that this transient accumulation and breakdown of starch is important for growth in a day–night light cycle (Caspar et al, 1985, 1991; Lin et al, 1988), and antisense experiments in wheat have shown that the SnRK1 kinase is important for expression of α-amylase, a key enzyme in starch mobilization (Laurie et al, 2003). The inability of the snf1a snf1b double mutant to grow either in the dark or in a 16 h/8 h day–night light cycle therefore suggested that starch metabolism might be affected in the mutant. In order to test this possibility, we assayed the starch content in protonemal tissue from the wild type and the double mutant at different points following a transfer from continuous light to darkness. As shown in Figure 7, we found that the starch content in light-grown double mutant is reduced to 41% of the wild-type value, which suggests that starch accumulation is deficient in the mutant. In contrast, both the wild type and the mutant were able to break down accumulated starch following transfer into the dark, and the difference in starch content disappeared after 48 h in the dark.
Figure 7.
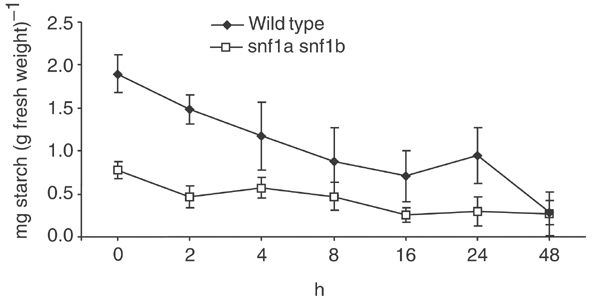
Reduced starch content in the snf1a snf1b double mutant. Protonemal tissue of the wild type and the snf1a snf1b double mutant was grown for 9 days in constant high light (2000 lux) and then transferred to darkness. The starch content was quantified after different times in the dark. The error bars show standard errors of mean for six independent samples.
Discussion
The yeast Snf1 kinase and its animal homologue AMPK play key roles in metabolic regulation. Thus, it is thought that these kinases function as energy gauges that help the cell to adapt to low-energy conditions by turning off energy-consuming processes and mobilizing energy reserves (Hardie et al, 1998). In plants, there are several Snf1-related kinases (Halford and Hardie, 1998; Halford et al, 2003), but the closest homologue of Snf1 is SnRK1. To learn more about the function of SnRK1 in plants, we have cloned two SnRK1-encoding genes from the moss P. patens, where gene function can be studied directly by targeted knockouts in the haploid gametophyte. The two genes, PpSNF1a and PpSNF1b, encode highly similar proteins (Figure 1 and Supplementary Figure 2), and Southern blots indicate that there are no other closely related genes in the Physcomitrella genome (Supplementary Figure 1). PpSNF1a and PpSNF1b were knocked out by homologous recombination. Since neither single mutant exhibited any obvious phenotypes, we proceeded to make a double knockout mutant.
Significantly, we found that the double knockout mutant is completely devoid of Snf1-like protein kinase activity, as determined using the SAMS-peptide assay (Figure 4). The SAMS peptide is known to be efficiently phosphorylated by SnRK1 kinases from several plants (Halford et al, 2003), and our finding therefore suggests that PpSnf1a and PpSnf1b are the only two SnRK1 kinases that are expressed in Physcomitrella protonemata. This is the first time that a null mutant has been reported for Snf1-like kinase activity in any plant or animal. Characterization of the resulting phenotype may therefore shed some light on the role of the Snf1-like kinases not only in plants but also in other eukaryotes. We further note that the SAMS-peptide phosphorylating activity remained unaffected by any of the treatments that we tested, including different light conditions, external carbon sources and exposure to plant hormones (Supplementary Figure 4). This might indicate that the kinase activity is constitutive; however, it is also possible that its in vivo regulation is not retained in the extracts. In this context it should be noted that both the yeast Snf1 kinase (Wilson et al, 1996) and the mammalian AMPK (Davies et al, 1992) require rapid freezing after extraction to preserve the physiological activation state.
The snf1a snf1b double mutant exhibits pronounced developmental phenotypes that affect both protonemata and gametophores (Figure 5). In protonemata, there is a clear excess of caulonemal filaments, many of which grow up into the air. The few chloronemal filaments that are formed in the mutant are aberrant, with shorter cells and a high incidence of dead apical cells. Furthermore, there are fewer gametophores in the mutant, and those that are formed have small malformed leaves and an excess of rhizoids. Growth and development in Physcomitrella, as in other plants, is regulated by plant hormones. Thus, auxins promote caulonema and rhizoid formation but inhibit the formation of secondary chloronema and leafy shoots, while cytokinins induce bud formation (Ashton et al, 1979). Plant hormone signalling is also known to interact with sugar sensing and signalling, although the mechanisms involved are still poorly understood (Gibson, 2004). Significantly, we found that the snf1a snf1b double mutant exhibits altered sensitivities to the plant hormones auxin and cytokinin (Figure 6). We note that these results are consistent with the above-described developmental phenotypes of the double mutant. Thus, its increased sensitivity to auxin could explain the excess of caulonema and rhizoids, and the reduced sensitivity to cytokinin why less buds and leafy shoots are formed. We further note that the altered sensitivities of the double mutant, in particular its auxin hypersensitivity, could reflect a role of the SnRK1 kinase in coordinating growth and development with available energy supplies. According to this interpretation, the kinase would inhibit auxin-dependent signalling under low-energy conditions, thus preventing growth and morphogenesis that the plant cannot afford.
A further pronounced phenotype of the snf1a snf1b double mutant is premature senescence (Figure 5A). Snf1/AMPK has been implicated in ageing both in yeast (Ashrafi et al, 2000; Lin et al, 2003) and in human fibroblasts (Wang et al, 2003). However, in these cases, senescence was linked to increased kinase activity, which is the opposite of what we see. Interestingly, results from higher plants have also implicated hexokinase-dependent signalling in senescence (Yoshida, 2003). Thus, overexpression of hexokinase in tomato and Arabidopsis promotes senescence (Dai et al, 1999; Xiao et al, 2000), while an Arabidopsis hexokinase mutant instead shows delayed senescence (Moore et al, 2003). It should be noted that hexokinase functions upstream of Snf1 in yeast, where it generates a glucose-dependent signal that inhibits Snf1 activity (Rolland et al, 2002). Snf1 and hexokinase mutants therefore have opposite phenotypes in yeast. Notably, some of the other phenotypes that we see in the snf1a snf1b double mutant, such as increased auxin sensitivity and cytokinin resistance, are also opposite those of hexokinase mutants in Arabidopsis (Moore et al, 2003). Furthermore, a knockout of the major hexokinase gene in Physcomitrella, PpHXK1, produces a phenotype that is the opposite of the snf1a snf1b phenotype in the sense that less rather than more caulonemal filaments are formed (Olsson et al, 2003). Our finding that the snf1a snf1b double mutant shows premature senescence is therefore consistent with the notion that the SnRK1 kinases function downstream of hexokinase in plants. This does not necessarily mean that hexokinase functions as a sensor that regulates SnRK1 activity. It is also conceivable that the SnRK1 kinases are regulated by a metabolite downstream of hexose phosphorylation.
A striking phenotype of the snf1a snf1b double mutant is its inability to grow in conditions of reduced light (Figure 6C). Thus, it requires continuous high light, and is unable to grow either in continuous low light or in a normal day–night light cycle where 16 h of high light is followed by 8 h of darkness. Our finding that growth of the double mutant in both cases can be partially restored by the addition of glucose further suggests that it suffers from an inability to handle a reduced or variable energy supply rather than a light signalling defect. A possible interpretation is that the double mutant is locked in a high-energy growth mode, which is lethal when the energy supply is limited or variable. This would be in agreement with the proposed roles of the Snf1-related kinases as energy gauges (Hardie et al, 1998). It is also consistent with the excess of caulonemal filaments in the double mutant (Figure 5A), since caulonema formation in Physcomitrella requires a sufficient energy supply in the form of high light (Schumaker and Dietrich, 1997).
The inability of the snf1a snf1b double mutant to grow in a normal day–night light cycle is particularly intriguing, since an abundant energy supply in the form of high-intensity light is provided for 16 h each day. It is therefore unlikely that lack of energy alone can explain this phenotype. A more likely explanation is an inability to carry out metabolic changes that help the plant cope with the dark hours of the night. A key energy reserve in plants is starch, which accumulates in the chloroplasts during the day, and is mobilized to serve as an energy source during the night. Experiments in Arabidopsis have shown that this transient accumulation and breakdown of starch is important for growth in a normal day–night light cycle (Caspar et al, 1985, 1991; Lin et al, 1988), and several enzymes involved in starch mobilization are preferentially expressed during the night (Harmer et al, 2000). Furthermore, experiments in wheat have shown that the SnRK1 kinase is important for expression of α-amylase, a key enzyme in starch mobilization (Laurie et al, 2003). Interestingly, we found that the snf1a snf1b double mutant has only 41% of the starch content of the wild type after continuous growth for 9 days in high light (Figure 7). One possible reason for the reduced starch content is the fact that the double mutant has an excess of caulonemal filaments, since the latter contain fewer and less-developed chloroplasts. However, it is also possible that one or more enzymes involved in starch synthesis are regulated by PpSnf1a and PpSnf1b. In this context, we note that fructose-1,6-bisphosphatase, an enzyme needed for starch synthesis, is strictly dependent on Snf1 for its expression in yeast (Hardie et al, 1998). Further studies may help to clarify to what extent this or other enzymes involved in starch metabolism are regulated by the two SnRK1 kinases in Physcomitrella.
Materials and methods
Plant material and growth conditions
The standard growth conditions used were growth at 25°C in a Sanyo MLR-350 light chamber with constant irradiation (2000 lux) from the sides. Protonemal tissue was subcultured on cellophane (#325P, Cannings Packaging Ltd, UK)-covered BCD media (1 mM MgSO4, 1.85 mM KH2PO4, 10 mM KNO3, 45 μM FeSO4, 1 mM CaCl2, 1 × Hoagland's No. 2 solution and 0.8% agar) supplemented with 5 mM ammonium tartrate. For phenotypic analysis, protonemal tissue was precultured for 1 week on cellophane-covered ammonium-free BCD media. Small pieces of fresh protonemal tissue (approximately 2 mm in diameter) were then transferred onto ammonium-free BCD plates without cellophane. The colonies were cultivated for 3 or 4 weeks and then photographed.
Molecular cloning
Standard methods were used for all molecular cloning (Sambrook et al, 1989). The Codehop software (Rose et al, 1998) was used to design degenerate PCR primers based on SNF1-related plant sequences. Using P. patens genomic DNA as template, these primers amplified a 387 bp fragment. The sequence of this fragment was used to design primers for 5′- and 3′-RACE. The resulting RACE products were cloned and sequenced, providing a full-length 2488 bp sequence of the PpSNF1a cDNA. PpSNF1a-specific primers were then used to amplify a 5089 bp genomic fragment carrying the PpSNF1a gene, which was cloned into pCR®2.1 to produce pMT103 (Table I). In a low-stringency PCR, PpSNF1a-specific primers amplified a faint 648 bp fragment derived from the PpSNF1b gene. This fragment was cloned, and its sequence was used to design specific primers for PpSNF1b, which were used to obtain overlapping 5′- and 3′-RACE fragments. A unique Bpu1102I site within the overlap was used to splice the two fragments into a 2511 bp full-length PpSNF1b cDNA. Specific primers were then used to amplify a 5107 bp genomic fragment carrying the PpSNF1b gene, which was cloned into pCR®2.1 to produce pMT115. The PpSNF1a and PpSNF1b genomic and cDNA sequences have been deposited in GenBank with accession codes AY347743, AY347744, AY347745 and AY347746. The sequences of the oligonucleoties used will be provided on request.
Table 1.
Plasmids
| Plasmid | Insert |
|---|---|
| pJO177 | MET3 promoter, 2μ origin, URA3 |
| pUBW302 | Source of G418 resistance cassette |
| pMT102 | PpSNF1a ORF in pJO177 |
| pMT103 | PpSNF1a genomic clone |
| pMT104 | PpSNF1a targeting construct |
| pMT108 | ScSNF1 ORF in pJO177 |
| pMT115 | PpSNF1b genomic clone |
| pMT123 | Source of hygromycin resistance cassette |
| pMT126 | PpSNFb targeting construct |
| pMT139 | PpSNF1a RT–PCR internal standard template |
| pMT140 | PpSNF1b RT–PCR internal standard template |
| pMT141 | PpSNF1b ORF in pJO177 |
Southern blot analysis
P. patens genomic DNA prepared from young protonemal tissue was digested to completion with HindIII, BglII or EcoRI. Using standard blotting procedures (Sambrook et al, 1989), the DNA was transferred to nylon filters that were UV crosslinked. The filters were hybridized to 32P-labelled probes for 4 h at 63°C and were then washed in 0.5 × SSC/0.1% SDS at 63°C. For the PpSNF1a-specific probe, we used a 508 bp NsiI–BglII cDNA fragment, and for the PpSNF1b specific probe, a 526 bp XmnI–MscI cDNA fragment was used. Hybridizing bands were visualized in a phosphoimager.
Targeted gene disruptions
To disrupt the PpSNF1a gene, a 2446 bp internal BspEI–HpaI fragment of pMT103 encoding amino-acid residues 131–404 of PpSnf1a was replaced by a 1971 bp selection cassette containing the neomycin phosphotransferase (nptII) gene expressed from the 35S promoter followed by the OCS terminator. This selection cassette was excised with BamHI and HindIII from the plasmid pUBW302. Prior to transformation, the resulting targeting plasmid, pMT104, was linearized with SmaI and XbaI, thus producing a 4480 bp fragment having 1242 and 1269 bp of Physcomitrella DNA flanking the selection cassette on each side (Figure 4A). To disrupt the PpSNF1b gene, an internal 1140 bp AatII–BsgI fragment of pMT115 encoding amino-acid residues 200–323 of PpSnf1b was replaced by a 2578 bp selection cassette containing the hygromycin phosphotransferase (hpt) gene expressed from the NOS1 promoter followed by the OCS terminator. This selection cassette was excised with HindIII–XbaI from the plasmid pMT123. Prior to transformation, the resulting targeting plasmid pMT126 was linearized with KpnI and XmnI to produce a 3448 bp fragment with 264 and 606 bp of Physcomitrella DNA flanking the selection cassette on each side (Figure 4B). Moss protoplasts were transformed as described previously (Schaefer et al, 1991), and stable transformants were selected in the presence of 50 mg l−1 geneticin (Sigma G9516) or 30 mg l−1 hygromycin B (Sigma H3274).
Complementation in yeast
Yeast media and growth conditions were as described by Sherman et al (1986) but with twice as much leucine in the synthetic media. The vector pJO177, which was used for expression of Physcomitrella cDNAs in yeast, is a multicopy (2μ) shuttle vector carrying the auxotrophic URA3 marker and the MET3 promoter for expression of inserts. A 1764 bp cDNA fragment covering the PpSNF1a open reading frame was amplified by PCR and cloned into the SmaI site of pJO177 to produce the plasmid pMT102. A 1908 bp Esp3I–HindIII fragment of the PpSNF1b cDNA covering the open reading frame was subcloned into the SmaI site of pJO177 to produce plasmid pMT141. The S. cerevisiae SNF1 gene was PCR amplified from yeast genomic DNA and then cloned into the SmaI site of pJO177 to produce pMT108. The plasmids were transformed into yeast strains H316 (MATa snf1::LEU2) and H1307 (MATa snf4::LEU2). Both strains are congenic to W303-1A (Thomas and Rothstein, 1989) and therefore carry the ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 markers as well. Media lacking uracil was used to select transformants. The ability to complement snf1 knockout mutants was tested on media lacking methionine in order to derepress the MET3 promoter.
Protein extractions and kinase assay
Young protonemal tissue grown on cellophane-covered media was used as the starting material for protein extraction. The extraction procedure and subsequent ammonium sulphate fractionation were performed essentially as described by Man et al (1997). However, the Roche #1.697.498 proteinase inhibitor mix was used instead of benzamidine and PMSF, and PD10 columns (Amersham Biosciences) instead of G50 columns. Moreover, the extraction buffer was changed to fractionation buffer also in the crude extracts that were not subjected to ammonium sulphate precipitation. The SAMS peptide was purchased from Tocris (#1344) and the kinase assay was performed essentially as described (Man et al, 1997). We used a SAMS-peptide stock solution of 500 μM resulting in a final concentration of 100 μM in the assay, and a labelled ATP stock solution of 1 mM ATP with a specific activity of 1650 cpm pmol−1 resulting in a final concentration of 200 μM in the assay.
Quantitative RT–PCR
Young protonemal tissue grown on cellophane-covered BCD plates for 7 days under standard growth conditions were transferred by disc lifts to tester plates. Conditions tested included growth in the dark or under intense light (6000 lux), and growth in the presence of 0.2 M glucose or 0.2 M mannitol. After 24 h of treatment, the tissue was harvested and total RNA was prepared. Expression levels were measured as the ratio between the transcript-dependent RT–PCR product in 1 μg of RNA and an internal standard product. The internal standard was an in vitro-transcribed RNA, which was amplified with the same primer pair as the transcript, but producing a differently sized product (Siebert and Larrick, 1992). The primers used recognize stretches within the coding sequences with a limited sequence similarity, and were confirmed not to crossreact between the two transcripts (data not shown). Templates for the in vitro transcription of PpSNF1a and PpSNF1b internal standards were the plasmids pMT139 and pMT140, respectively. The pMT139 insert consists of a PpSNF1a genomic fragment deleted for an internal 376 bp HpaI/AflII fragment, while pMT140 has an insert consisting of a PpSNF1b genomic fragment deleted for an internal 908 bp BglII fragment.
Starch quantification
Protonemal tissue of the wild type and the snf1a snf1b double mutant was grown on BCD plates under standard conditions for 9 days. The plates were then transferred to darkness. Samples were harvested and frozen in liquid nitrogen after 0, 2, 4, 8, 16, 24 and 48 h in the dark. Extraction and quantification of starch were performed using a starch assay kit (Sigma, SA-20). Starch was extracted using the DMSO/HCl method described by the manufacturer but with 1/4 of the volumes in the protocol. The starch assay was performed according to the manufacturer's protocol but with sample volumes decreased to 1/5 of the original.
Supplementary Material
Supplementary Figure
Acknowledgments
We thank David Cove, Andrew Cuming and Celia Knight for advice and the Physcomitrella wild-type strain, and Lars Rask for helpful comments. This work was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), and the Swedish Research Council (VR).
References
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA 88: 8602–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Lin SS, Manchester JK, Gordon JI (2000) Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev 14: 1872–1885 [PMC free article] [PubMed] [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427–435 [DOI] [PubMed] [Google Scholar]
- Ball KL, Dale S, Weekes J, Hardie DG (1994) Biochemical characterization of two forms of 3-hydroxy-3-methylglutaryl-CoA reductase kinase from cauliflower (Brassica oleracia). Eur J Biochem 219: 743–750 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville CR (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1986) A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233: 1175–1180 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol 9: 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shabak Y, Giller Y, Ratner K, Levine A, Granot D (1999) Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11: 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Arro M, Becerra B, Morrice NG, Boronat A, Hardie DG, Ferrer A (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3- methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 233: 506–513 [DOI] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG (1989) Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studies using a specific and sensitive peptide assay. Eur J Biochem 186: 123–128 [DOI] [PubMed] [Google Scholar]
- Davies SP, Carling D, Munday MR, Hardie DG (1992) Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem 203: 615–623 [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M (1997) Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell 8: 1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI (2004) Sugar and phytohormone response pathways: navigating a signaling network. J Exp Bot 55: 253–264 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37: 735–748 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul M (2004) Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J Exp Bot 55: 35–42 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1997) The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M (1999) Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet 15: 29–33 [DOI] [PubMed] [Google Scholar]
- Laurie S, McKibbin RS, Halford NG (2003) Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an alpha-amylase (alpha-Amy2) gene promoter in cultured wheat embryos. J Exp Bot 54: 739–747 [DOI] [PubMed] [Google Scholar]
- Le Guen L, Thomas M, Bianchi M, Halford NG, Kreis M (1992) Structure and expression of a gene from Arabidopsis thaliana encoding a protein related to SNF1 protein kinase. Gene 120: 249–254 [DOI] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI (2003) Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J Biol Chem 278: 13390–13397 [DOI] [PubMed] [Google Scholar]
- Lin T-P, Caspar T, Somerville C, Preiss J (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphatase activity. Plant Physiol 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh RW, Davies SP, Clarke PR, Weekes J, Gillespie JG, Gibb BJ, Hardie DG (1992) Evidence for a protein kinase cascade in higher plants. 3-Hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 209: 923–931 [DOI] [PubMed] [Google Scholar]
- Man AL, Purcell PC, Hannappel U, Halford NG (1997) Potato SNF1-related protein kinase: molecular cloning, expression analysis and peptide kinase activity measurements. Plant Mol Biol 34: 31–43 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Nehlin JO, Ronne H (1990) Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J 9: 2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson T, Thelander M, Ronne H (2003) A novel type of chloroplast stromal hexokinase is the major glucose phosphorylating enzyme in the moss Physcomitrella patens. J Biol Chem 278: 44439–44447 [DOI] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG (1998) Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Reski R (1998) Development, genetics and molecular biology of mosses. Bot Acta 111: 1–15 [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H (1995) Glucose repression in fungi. Trends Genet 11: 12–17 [DOI] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26: 1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schaefer D (2002) A new moss genetics: targeted mutagenesis in Physcomitrella patens. Annu Rev Plant Biol 53: 477–501 [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP, Knight CD, Cove DJ (1991) Stable transformation of the moss Physcomitrella patens. Mol Gen Genet 226: 418–424 [DOI] [PubMed] [Google Scholar]
- Schumaker KS, Dietrich MA (1997) Programmed changes in form during moss development. Plant Cell 9: 1099–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB (1986) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Siebert PD, Larrick JW (1992) Competitive PCR. Nature 359: 557–558 [DOI] [PubMed] [Google Scholar]
- Smith FC, Davies SP, Wilson WA, Carling D, Hardie DG (1999) The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett 453: 219–223 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein RJ (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Lopez de Silanes I, Carling D, Gorospe M (2003) Increased AMP:ATP ratio and AMP-activated protein kinase activity during cellular senescence linked to reduced HuR function. J Biol Chem 278: 27016–27023 [DOI] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG (1996) Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28: 431–441 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure


