Abstract
The functional coupling of transcription and splicing has been reported both in vivo and in vitro, but the molecular mechanisms governing these interactions remain largely unknown. Here we show that p54nrb, a transcription/splicing factor, associates with the 5′ splice site (SS) within large complexes present in HeLa cell nuclear extracts, in which the hyperphosphorylated form of RNA polymerase II (RNAPIIO) is associated with U1 or U1 and U2 snRNPs. These RNAPIIO–snRNP complexes also contain other transcription/splicing factors, such as PSF and TLS, as well as transcription factors that interact with RNAPIIO during elongation, including P-TEFb, TAT-SF1 and TFIIF. The presence of these factors in functional elongation complexes, demonstrated using an immobilized DNA template assay, strongly suggests that the RNAPIIO–snRNP complexes reflect physiologically relevant interactions between the transcription and splicing machineries. Our finding that both p54nrb and PSF, which bind the C-terminal domain of the largest subunit of RNAPII, can interact directly with the 5′ SS indicates that these factors may mediate contacts between RNAPII and snRNPs during the coupled transcription/splicing process.
Keywords: p54nrb , PSF, RNA polymerase II, splicing complexes, transcription
Introduction
Splicing of mRNA precursors is an important step in eukaryotic gene expression, in which introns are removed from primary RNA polymerase II (RNAPII) transcripts. The majority of metazoan genes are interrupted by multiple introns, which comprise more than 95% of primary transcripts. Pre-mRNA splicing is executed by the spliceosome, whose assembly requires a complex series of binding and rearrangement events involving small nuclear ribonucleoprotein particles (snRNPs) (reviewed in Staley and Guthrie, 1998; Burge et al, 1999). In the first step of this assembly pathway, U1 snRNP binds the 5′ splice site (SS), in part due to the complementarity of the 5′ end of U1 snRNA to the 5′SS. Next, U2 snRNP binds to the branch site upstream of the 3′SS, generating pre-spliceosome complex A, which is subsequently converted into complex B upon joining of the U4/5/6 tri-snRNP. Additional rearrangements, involving significant changes in RNA:RNA interactions, yield the catalytically competent complex C, currently estimated to include five snRNAs and ∼150–300 proteins (Makarov et al, 2002; Rappsilber et al, 2002; Zhou et al, 2002). The early steps of spliceosome assembly can be bypassed in a simplified trans-splicing system, in which a DNA oligonucleotide complementary to the 5′ end of U1 snRNA (U1-blocking DNA) is used to block binding of a short 5′SS RNA substrate to U1 and promote its interaction with U4/5/6 snRNP and U2/4/5/6 splicing complexes (Konforti et al, 1993; Konforti and Konarska, 1994).
Early biochemical and cytological studies indicated that splicing can occur co-transcriptionally (Beyer and Osheim, 1988; Bauren and Wieslander, 1994) and that splicing factors are recruited to active transcription sites (Huang and Spector, 1991; Zhang et al, 1994). Subsequent functional studies showed that the identity of the RNAPII promoter affects splicing patterns of the nascent transcript (Cramer et al, 1997, 1999), further connecting transcription and splicing. Reciprocally, splicing appears to stimulate transcription, as both splicing factors (Fong and Zhou, 2001) and splicing signals (Furger et al, 2002) can enhance gene transcription. The C-terminal domain (CTD) of the largest subunit of RNAPII appears to provide a platform on which to link physically transcription to splicing and other gene expression steps, such as capping, polyadenylation and termination of transcription (reviewed in Hirose and Manley, 2000; Bentley, 2002; Proudfoot et al, 2002). The mammalian RNAPII CTD is composed of 52 tandem repeats of the YSPTSPS consensus peptide (Corden et al, 1985). Phosphorylation of Ser2 and/or Ser5 of this heptapeptide is a key event during the transcription cycle, with the hypophosphorylated form of RNAPII (RNAPIIA) being found in pre-initiation complexes and the hyperphosphorylated form (RNAPIIO) in elongation complexes (ECs). Several studies have reported the association of RNAPIIO with snRNP particles (Mortillaro et al, 1996; Vincent et al, 1996; Kim et al, 1997), as well as the direct interaction of the CTD with some splicing factors (Morris and Greenleaf, 2000; Emili et al, 2002). Yet, the functional specificity of these interactions has not been demonstrated. Moreover, both RNAPIIO (Hirose et al, 1999) or the CTD alone (Zeng and Berget, 2000) can enhance splicing, but the mechanisms underlying this stimulation are not known.
Polypyrimidine tract-binding protein (PTB)-associated splicing factor (PSF) and p54nrb are two closely related human nuclear proteins implicated in transcription and splicing, whose specific functions remain unclear. Several studies indicate that both factors are involved in transcriptional control (Basu et al, 1997; Urban et al, 2000; Mathur et al, 2001). However, while PSF has been shown to be essential for splicing in vitro (Patton et al, 1993), the direct involvement of p54nrb in pre-mRNA splicing has not been demonstrated. These multifunctional proteins have also been implicated in RNA editing and DNA recombination (reviewed in Shav-Tal and Zipori, 2002). Importantly, PSF and p54nrb were recently shown to interact with each other (Peng et al, 2002) and with the CTD (Emili et al, 2002).
Using protein–RNA crosslinking assays, we have identified a direct interaction between p54nrb and the 5′SS. The crosslink resides within large complexes, which contain RNAPII and snRNPs and are present in HeLa cell nuclear extracts (NE) independently of the ongoing transcription or splicing. One of these complexes contains RNAPIIO and U1 snRNP, while another additionally includes U2 snRNP. Besides p54nrb, both RNAPII–snRNP complexes contain other transcription/splicing factors (e.g. PSF and TLS), as well as those typically found in RNAPII ECs (e.g. P-TEFb and TAT-SF1). These factors are also present in functional ECs isolated using an immobilized DNA template assay, strongly suggesting that the analyzed complexes reflect physiologically relevant interactions between the transcription and splicing machineries. Furthermore, we confirmed the previously observed crosslinking of PSF to the 5′SS within the U2/4/5/6 spliceosome formed in the absence of ATP (Ismaili et al, 2001) and show that this complex also associates with RNAPIIO. The observed direct association of p54nrb and PSF with the 5′SS within snRNP complexes suggests their direct role in linking transcription to splicing.
Results
Crosslinking patterns of 5′SS RNA associated with U1 snRNP
To detect spliceosomal components interacting with the 5′SS, we previously analyzed crosslinking profiles of site-specifically modified, photoreactive 5′SS substrates (Ismaili et al, 2001). In the presence of U1-blocking DNA, the 5′SS RNA associates with U2, U4, U5 and U6 snRNPs (Konforti et al, 1993, and Figure 3A, lanes 1 and 2) and forms specific crosslinks with one of two splicing factors: PSF in the absence of ATP or hPrp28 in its presence (Ismaili et al, 2001, and Figure 1B, lanes 1 and 2).
Figure 3.
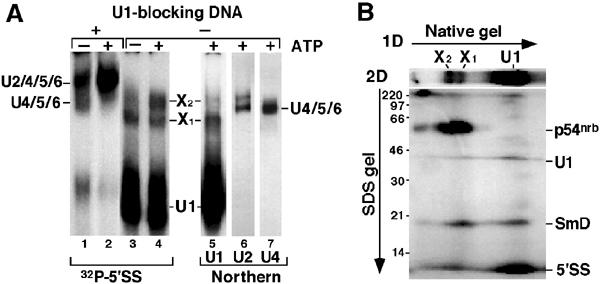
The 5′SS:p54nrb crosslink exists exclusively in large complexes containing U1 snRNP. (A) Standard binding reactions in the presence (lanes 1–4) or absence (lanes 5–7) of the 5′SS RNA, in the absence (lanes 1,3) or presence (lanes 2,4–7) of ATP and in the presence (lanes 1,2) or absence (lanes 3–7) of U1-blocking DNA were resolved in a native 4% gel. snRNP complexes were detected by autoradiography (lanes 1–4) or Northern hybridization with U1, U2 or U4 probes (lanes 5–7). Positions of complexes are indicated. (B) The 5′SS crosslinks were resolved in a native 4% gel in the first dimension and in an 11% SDS gel in the second. Positions of the free 5′SS and the crosslinks are indicated.
Figure 1.
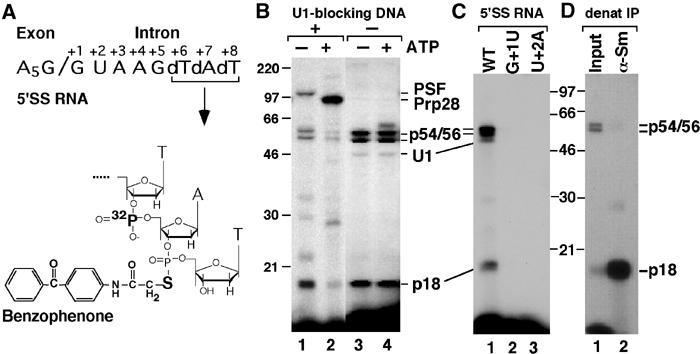
Crosslinking profile of the BP-derivatized 5′SS RNA. (A) Structure of the 32P-labeled, BP-derivatized 14-nt 5′SS RNA. (B) The 5′SS RNA was incubated with HeLa NE in the absence (lanes 1, 3) or presence (lanes 2, 4) of ATP and in the presence (lanes 1, 2) or absence (lanes 3, 4) of U1-blocking DNA. (C) Wild-type 5′SS RNA (WT) or 5′SS RNAs with single point mutations at intron pos. +1 or +2 was incubated with HeLa NE in the absence of ATP or U1-blocking DNA. (D) The 5′SS crosslinks were immunoprecipitated under denaturing conditions with α-Sm (Y12) antibodies. Crosslinked products were resolved in 11% SDS gels. Positions of the molecular weight markers and the 5′SS crosslinks are indicated.
Here, we have used an analogous approach to analyze 5′SS interactions in the U1 snRNP complex. A 14-nt synthetic 5′SS RNA containing a 32P group between intron pos. +6 and +7 and a photoreactive benzophenone (BP) group between intron pos. +7 and +8 (Figure 1A) was incubated with HeLa NE in the absence of U1-blocking DNA and irradiated with 302-nm UV light. Under these conditions, the BP-5′SS RNA stably binds U1 snRNP in an ATP-independent manner (confirmed by native gel analysis; see Figure 3A, lanes 3–5). This binding yields a 5′SS:U1 snRNA crosslink (Figure 1B, lanes 3 and 4) identified by its sensitivity to RNase H digestion in the presence of U1-specific DNA oligos (data not shown). In addition, two major protein crosslinks, p18 and a 54/56-kDa doublet, were detected in these reactions (the sizes of all crosslinked proteins were corrected for the contribution of the RNA component). Consistent with their association with U1 snRNP, these two crosslinks were precipitated by α-U1A antibodies under native conditions (see Figure 2B, lane 6). The observed interactions are specific to the 5′SS, as the crosslinks were not detected using 5′SS RNAs with single point mutations at either intron pos. +1 or +2 (Figure 1C).
Figure 2.
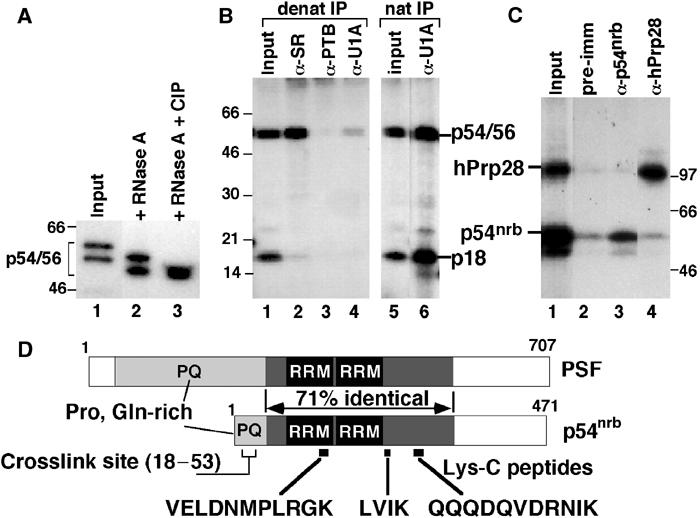
The 5′SS:p54/56 crosslink corresponds to p54nrb. (A) The 5′SS:p54/56 crosslinks were treated with RNase A (lane 2) or RNase A and phosphatase (CIP) (lane 3) and resolved in an 8% SDS gel. (B) The 5′SS crosslinks were immunoprecipitated under denaturing (lanes 1–4) or native (lanes 5 and 6) conditions using the indicated antibodies and resolved in an 11% SDS gel. (C) The 5′SS:p54nrb and 5′SS:hPrp28 crosslinks were purified from an SDS gel, mixed (lane 1), immunoprecipitated with pre-immune serum (lane 2) or the indicated antibodies (lanes 3 and 4) and resolved in an 8% SDS gel. (D) Diagram of the human PSF and p54nrb sequences. The two RRMs, the proline/glutamine-rich regions (PQ), the 293-aa internal region of high similarity between p54nrb and PSF, the sequences of three Lys-C peptides and the mapped region of the 5′SS crosslink within p54nrb (aa 18–53) are indicated.
Among the known U1 snRNP proteins, only U1C (17.5 kDa) and SmD (D1–13.5, D2–13.9 and D3–13.4 kDa) fall in the 18 kDa size range. p18 appears to represent one of the SmD proteins, as the 5′SS:p18 crosslink was efficiently recognized by the α-Sm antibody Y12 under denaturing conditions (Figure 1D). In yeast, the charged C-terminal tails of SmB, SmD1 and SmD3 directly contact the 5′SS bound to U1 snRNP, possibly stabilizing snRNP:intron interactions within the commitment complex (Zhang et al, 2001). This suggests that contacts between the 5′SS and Sm proteins within U1 snRNP are conserved between yeast and metazoans.
Thus, crosslinking analysis using a photoreactive 5′SS RNA bound to U1 snRNP revealed the formation of two products (5′SS:U1 snRNA and 5′SS:SmD) expected of U1 snRNP interactions and a 54/56 kDa doublet crosslink of unknown origin.
The p54/56 crosslinked to the 5′SS represents p54nrb, a transcription/splicing factor
There are no known U1 snRNP proteins in the 55 kDa size range, suggesting that the p54/56 crosslink represents a novel protein associated with this particle. The p54/56 doublet represents two differentially modified forms of a single protein, as digestion of the individually purified bands with several proteases yielded identical peptide patterns (data not shown). In addition, phosphatase treatment of the crosslink doublet generated a single product of ∼54 kDa (Figure 2A, lane 3), indicating that the two forms of the doublet differ in their phosphorylation state.
In an attempt to identify p54, we carried out immunoprecipitations under denaturing conditions using a battery of antibodies against selected splicing factors. To disrupt snRNP integrity, crosslinking reactions were first treated with RNase A, yielding faster migrating products due to the removal of eight 5′-terminal nucleotides of the 5′SS RNA in the crosslink (Figure 2A). Under these conditions, the α-SR antibody 16H3 immunoprecipitated the 5′SS:p54/56 doublet, but not the 5′SS:SmD crosslink (Figure 2B, lane 2). In contrast, antibodies against PTB (Figure 2B, lane 3), SF1, SRp54, U2AF65, hUAP56 or the 60K subunit of U2-SF3a (data not shown) did not recognize the 5′SS:p54/56 crosslink. Similarly, an α-U1A antibody, which efficiently precipitated both crosslinks under native conditions (Figure 2B, lane 6), failed to recognize them after the RNase A treatment (Figure 2B, lane 4), confirming that U1 snRNP was largely disrupted under these conditions.
To identify p54/56, complexes from a large-scale crosslinking reaction were RNase-digested and resolved by two-dimensional SDS gel electrophoresis. Positions of the labeled crosslinks were visualized by autoradiography and superimposed with signals of Western blot analysis using the α-SR antibody that immunoprecipitated the crosslink and the protein pattern detected by Coomassie staining (data not shown). Microsequencing analysis of the ∼54-kDa protein detected by these three methods yielded three peptide sequences (VELDNMPLRGK, LVIK and QQQDQVDRNIK) identified as fragments of p54nrb (Figure 2D), a protein originally identified due to its crossreactivity with an antibody raised against the yeast splicing factor Prp18 (Dong et al, 1993). p54nrb shows no significant homology to yPrp18, but is instead closely related to human splicing factor PSF, with which it shares a region of high similarity (Figure 2D). Analysis of the p54nrb primary sequence reveals an N-terminal P/Q-rich segment, followed by two RNA recognition motifs (RRMs) (Figure 2D). Although p54nrb does not contain RS repeats, it must contain SR-like epitopes, as both the 5′SS:p54nrb crosslink (Figure 2B, lane 2) and recombinant, bacterially expressed p54nrb (data not shown) were recognized by α-SR antibodies. Finally, crosslink identity was confirmed by specific precipitation of the 5′SS:p54nrb crosslink from a mixture of SDS gel-purified p54/56 and hPrp28 crosslinks using α-p54nrb antibody (Figure 2C). Preliminary characterization of the crosslink (data not shown) mapped its site within p54nrb to a 36-aa segment (aa 18–53) in the N-terminal region of the protein (Figure 2D).
We have therefore identified the p54/56 doublet as the transcription/splicing factor p54nrb, which crosslinks to the 5′SS via its N-terminal region. This protein shows significant homology with the splicing factor PSF, which also crosslinks to the 5′SS but within the U2/4/5/6 spliceosome complex formed in the absence of ATP.
The 5′SS:p54nrb crosslink resides in large complexes containing U1 and U2 snRNP particles
Crosslinking of p54nrb to the 5′SS RNA does not require ATP and is inhibited by blocking the 5′ end of U1 snRNA with a complementary DNA (Figure 1B) but not by equivalent amounts of other oligos (data not shown), suggesting an association dependent on binding of the 5′SS by U1 snRNP. To better characterize snRNP complexes associated with the 5′SS, crosslinking reactions were resolved in a native polyacrylamide gel (Figure 3A, lanes 1–4). As we have shown before (Konforti and Konarska, 1994), in the presence of U1-blocking DNA the 5′SS RNA associates with U2/4/5/6 and U4/5/6 snRNP complexes (Figure 3A, lanes 1 and 2), whereas in the absence of U1-blocking DNA it forms a characteristic, abundant complex with U1 snRNP (Konforti et al, 1993). In addition, two slowly migrating complexes, X1 and X2, were detected under these conditions (Figure 3A, lanes 3 and 4). The superimposable signals generated in reactions containing labeled 5′SS RNA and by Northern analysis of endogenous NE complexes using a U1 snRNA-specific probe (Figure 3A, lanes 3–5) suggested that both X1 and X2 contain U1 snRNP. This was confirmed by immunoprecipitation of glycerol gradient fractions (see below). While U4, U5 or U6 snRNA-specific signals were not detected in either X1 or X2 (Figure 3A, lanes 4 and 7), the most slowly migrating form of U2 snRNP and X2 exhibited similar mobility in the gel (Figure 3A, lanes 4 and 6, and data not shown). The formation of X2 requires incubation at 30°C, whereas X1 forms even on ice (data not shown). Moreover, X2 formation requires ATP, whereas the formation of X1 does not (Figure 3A, lanes 3 and 4). These features resemble requirements for the formation of splicing complexes involving U1 and U2 snRNPs.
The distribution of the 5′SS:p54nrb crosslink in these complexes was analyzed by resolving crosslinking reactions in a native gel in the first dimension and in an SDS gel in the second (Figure 3B). The 5′SS:p54nrb crosslink was detected exclusively in X1 and X2, but not in the most abundant 5′SS:U1 snRNP complex. In contrast, both the 5′SS:U1 snRNA and SmD crosslinks were present in all complexes, further confirming that they all contain U1 snRNP.
To better characterize these large complexes, X1 and X2 were separated by sedimentation through a 10–30% glycerol gradient (Figure 4A and B). Gradient fractions containing X1 (fraction 4) and X2 (fraction 7) were subjected to immunoprecipitation under native conditions using selected snRNP-specific antibodies (Figure 4C). As a control of their functionality, all antibodies were confirmed to precipitate the targeted protein, as shown by Western analysis of material immunoprecipitated from crosslinking reactions (data not shown). Anti-U1 70K antibody precipitated the 5′SS:p54nrb, SmD and U1 snRNA crosslinks from both fractions (Figure 4C, lanes 3 and 4), confirming that both X1 and X2 contain U1 snRNP. In contrast, the two U2 snRNP-specific proteins U2B″ and SAP155 (an SF3b component) were detected predominantly in fraction 7 (Figure 4C, lanes 5–8). We therefore conclude that X1 contains U1 snRNP, whereas X2 contains both U1 and U2 snRNPs. Consistent with this, hPrp5, FBP11 and FBP21 were found in both these complexes (Figure 4C, lanes 9–14). The higher level of these factors associated with X2 is consistent with their role in the U1–U2 snRNP interaction during pre-spliceosome assembly. X1 and X2 complexes do not appear to be associated with U4, U5 or U6 snRNPs, as α-U5-116K and α-hPrp4 antibodies (specific for components of U5 and U4/6 snRNPs, respectively) did not efficiently recognize any of the crosslinks (Figure 4C, lanes 15–18).
Figure 4.
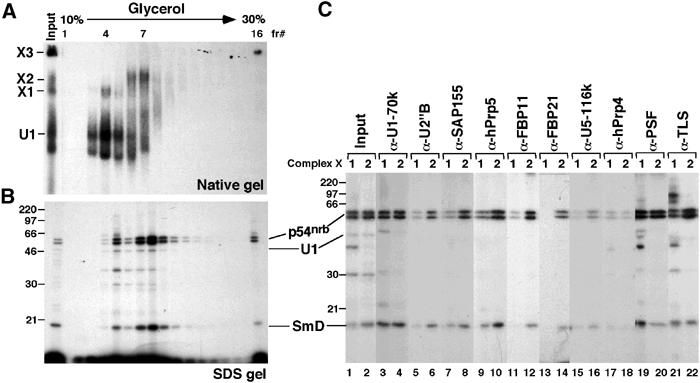
X1 and X2 complexes can be separated in a glycerol gradient. A 150-μl crosslinking reaction performed in the presence of ATP and absence of U1-blocking DNA was separated in a 10–30% glycerol gradient. In all, 10 μl of each fraction was resolved in either a 4% native gel (A) or an 11% SDS gel (B). (C) Fractions 4 and 7 of the glycerol gradient were immunoprecipitated under native conditions using the indicated antibodies, and the bound material was resolved in an 11% SDS gel. The identity of the ∼70 kDa signal generated with the α-TLS antibody (lane 21) has not been determined. Positions of the 5′SS crosslinks are indicated.
The splicing factor PSF was originally isolated in a stable interaction with PTB (Patton et al, 1993). Anti-PSF, but not α-PTB (data not shown), antibodies precipitated the U1 snRNA, p54nrb and SmD crosslinks from the gradient-separated X1 and X2 (Figure 4C, lanes 19 and 20), indicating that PSF is present in both complexes. X1 and X2 were also recognized by an antibody raised against TLS (Figure 4C, lanes 21 and 22), another transcription/splicing factor. As expected, α-SR antibodies precipitated the crosslinks from both fractions 4 and 7 (data not shown), possibly reflecting the reactivity of U1-70K, p54nrb or SR proteins associated with X1 and X2.
In summary, we have identified two large complexes, X1 and X2, within which the 5′SS crosslinks to p54nrb. Immunoprecipitation experiments using snRNP-specific antibodies show that both complexes contain U1 snRNP, whereas X2 also contains U2 snRNP. In addition to p54nrb, both X1 and X2 contain other transcription/splicing factors, such as PSF and TLS.
The 5′SS:p54nrb-containing snRNP complexes are associated with RNAPIIO and transcription factors
The electrophoretic and sedimentation properties of X1 and X2 indicated high molecular weight of these complexes (Figures 3A and 4A). Furthermore, the presence of PSF, p54nrb and TLS proteins, all of which can act as transcription factors and bind to both RNA and DNA, suggested a connection between X1, X2 and transcription. To test this possibility, immunoprecipitation of crosslinking reactions was carried out using several antibodies against RNAPII specific for the N-terminal segment (N-20) or the CTD (8WG16 recognizes the hypophosphorylated form, whereas H5 and H14 recognize the Ser2- and Ser5-phosphorylated forms, respectively). The N-20, H5 and H14 antibodies all precipitated the p54nrb, SmD and U1 snRNA crosslinks (Figure 5A, lanes 3, 5 and 6), indicating the presence of RNAPIIO. Similar experiments using gradient-separated X1 and X2 confirmed that both complexes are associated with RNAPIIO (data not shown). In contrast, the 8WG16 antibody did not efficiently recognize X1 or X2 complexes (data not shown and Figure 5A, lane 4), although it did precipitate large amounts of U1 snRNA (Figure 5C, lane 4). Therefore, hypophosphorylated RNAPIIA can bind both p54nrb (Emili et al, 2002) and U1 snRNPs, but this association does not produce the 5′SS:p54nrb crosslink, suggesting that rearrangement of RNAPII-bound factors takes place following CTD phosphorylation.
Figure 5.
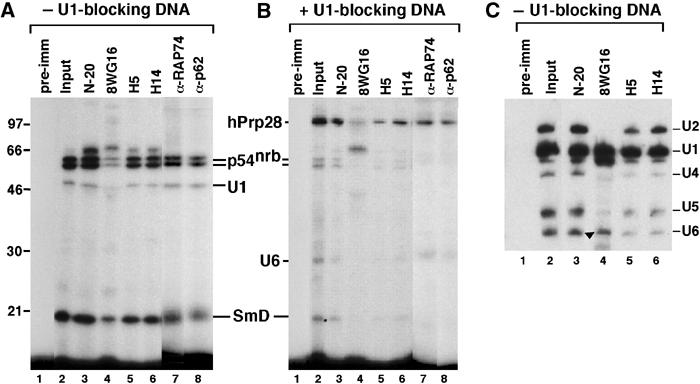
The p54nrb crosslink in X1 and X2 complexes is associated with RNAPIIO. A crosslinking reaction performed in the presence of ATP and in the absence (A) or presence (B) of U1-blocking DNA was immunoprecipitated under native conditions using the indicated α-RNAPII antibodies and the bound material resolved in an 11% SDS gel. The identity of the ∼65 kDa signal (e.g. lane 3 in panel A) has not been determined. (C) HeLa NE was incubated with ATP in the absence of the 5′SS RNA and immunoprecipitated as in (A). The bound RNA was analyzed by Northern hybridization with a mixture of U1, U2, U4, U5 and U6 snRNA probes. Positions of snRNAs are indicated. Although co-migrating with U6, the signal indicated by an arrow in lane 4 represents a degradation product of U1 snRNA.
The preferential association with RNAPIIO is characteristic not only of X1 and X2 but also of the spliceosome. The U2/4/5/6 complex, formed in the presence of U1-blocking DNA and ATP, binds RNAPIIO as evidenced by the precipitation of the 5′SS:hPrp28 crosslink with α-RNAPIIO antibodies (Figure 5B). Similarly, spliceosome complexes formed in the absence of ATP, in which the 5′SS RNA crosslinks to PSF (see Figure 1B, lane 1), are also associated with RNAPIIO (data not shown). These findings are consistent with earlier reports demonstrating a connection between spliceosome assembly and RNAPIIO (Hirose et al, 1999).
Additional immunoprecipitation experiments demonstrated the association of X1 and X2 with two basal transcription factors, TFIIF and TFIIH, as indicated by the presence of their RAP74 and p62 subunits, respectively (Figure 5A, lanes 7 and 8). In contrast, the initiation factor TFIIB was not detected in X1 or X2 (data not shown). Moreover, the transcription factor Sox6 and two elongation factors, P-TEFb (or its Cdk9 kinase subunit) and TAT-SF1, were detected in these complexes (Figure 6, lanes 7–12).
Figure 6.
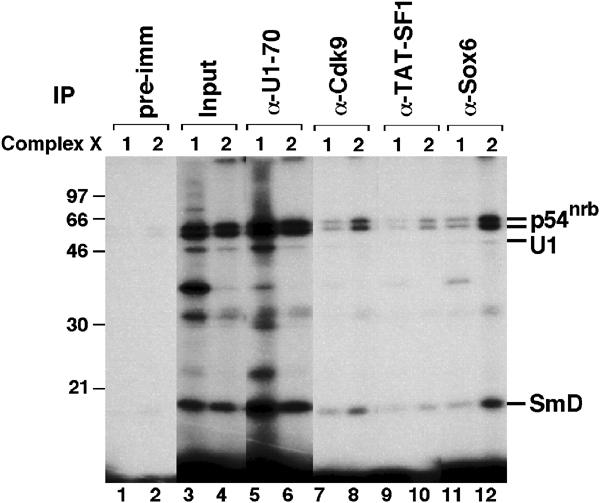
X1 and X2 complexes contain several transcription elongation factors. A crosslinking reaction performed in the presence of ATP was separated in a glycerol gradient, and fractions containing X1 (fraction 4) and X2 (fraction 7) complexes were immunoprecipitated under native conditions using the indicated antibodies and the bound material was resolved in an 11% SDS gel.
Both X1 and X2 are therefore associated with RNAPII preferentially in its hyperphosphorylated, elongating form. Consistently, X1 and X2 also contain transcription elongation factors, including P-TEFb or TAT-SF1, but lack factors typically involved only in the initiation phase of transcription, such as TFIIB.
The RNAPIIO–snRNP complexes contain a subset of functional EC factors
The association of X1 and X2 with RNAPIIO and multiple transcription/splicing and elongation factors suggests that these complexes reflect associations formed during transcriptional elongation. To compare X1 and X2 with functional complexes involved in active transcription, an immobilized DNA template assay (Ranish et al, 1999) was used to isolate ECs.
A biotinylated, 889-bp DNA transcription template containing the adenovirus major late promoter (AdMLP) was linked to magnetic streptavidin beads (Ad22, Figure 7A). Negative controls included a 490-bp DNA template identical to Ad22 but lacking the AdMLP (Ad20, Figure 7A) and reactions without any DNA bound to beads. After incubation of the immobilized DNA with NE to allow transcription, the templates were washed and cleaved at a unique PvuII site 32 bp downstream of the RNAPII start site (Figure 7A). This step liberates the formed ECs from the beads, while retaining initiation complexes assembled on the promoter-containing DNA fragments bound to the beads. The composition of the released ECs was then analyzed by SDS–PAGE and Western blotting. The Ad22 immobilized template yielded the expected 398-nt RNAPII transcript, whereas no transcript was detected with Ad20 (see Figure 7C), indicating that the complexes formed on the Ad22 template were active in transcription. As PvuII digestion released similar amounts of Ad20 and Ad22 DNA fragments (data not shown), the absence of signal from the Ad20 control reflects the lack of EC formation on this template.
Figure 7.
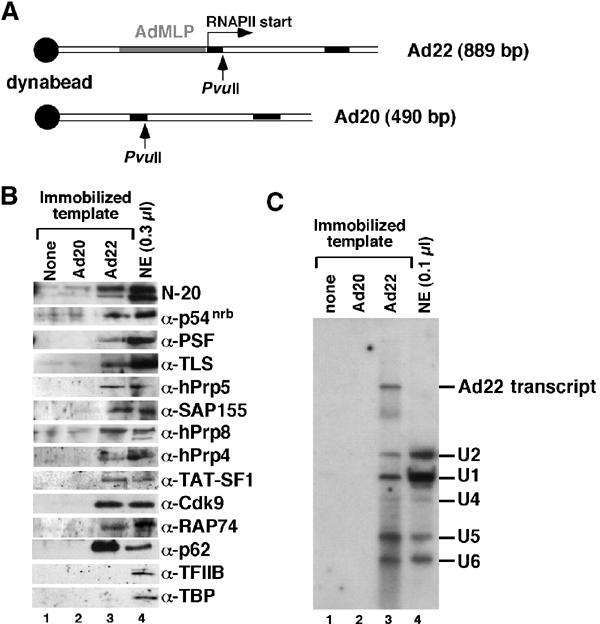
X1 and X2 complexes contain a subset of factors present in functional transcription ECs. (A) Schematic representation of the immobilized Ad20 and Ad22 DNA templates, showing the AdMLP, the two leading exons of the adenovirus major late transcription unit (black boxes), the start site for RNAPII transcription and the unique PvuII restriction sites. (B) Western blot analysis of protein factors bound to template fragments released from beads lacking DNA or linked to the Ad20 or Ad22 templates using the indicated antibodies. (C) Northern blot analysis of template fragments released from beads lacking DNA or linked to the Ad20 or Ad22 templates using probes for all the five U snRNAs and the Ad22 transcript.
Consistent with the formation of transcription ECs on the Ad22 template, the N-20 antibody detected RNAPII preferentially in the hyperphosphorylated form (Figure 7B). Moreover, the transcription/splicing factors p54nrb, PSF and TLS were detected at considerable levels. Other splicing factors, such as the U2 snRNP-associated hPrp5 or SAP155, were also associated with ECs formed on the Ad22, but not on Ad20 DNA or on beads without DNA. In addition, the U5 snRNP-associated hPrp8 and the U4/6-associated hPrp4 were detected, indicating the presence of U4, U5 and U6 snRNPs in functional ECs. This was confirmed by Northern blot analysis (Figure 7C), which detected all five snRNAs associated with the Ad22 template. Other EC-associated proteins (Figure 7B) included the transcription elongation factors TAT-SF1, P-TEFb (i.e. its Cdk9 subunit), TFIIF (or its RAP74 subunit) and the basal transcription factor TFIIH (detected by α-p62). In contrast, the initiation complex components TFIIB and TBP were not present at detectable levels on the Ad22 template, confirming the capture of ECs only.
These results demonstrate that ECs formed during active transcription contain numerous components of the splicing machinery and multiple proteins identified within X1 and X2 complexes. The fact that X1 and X2 include a large subset of functional EC components strongly suggests that these complexes reflect physiologically relevant interactions between the transcription and splicing machineries.
Discussion
The functional coupling of transcription and splicing, as well as the association of RNAPII with components of the splicing machinery, has been reported in recent years, but the detailed mechanisms and the functional specificity underlying these interactions remain mostly unknown. Here we report the direct association of the 5′SS with a transcription/splicing factor, p54nrb, in the context of large RNAPII–snRNP complexes. Besides the U2/4/5/6 snRNP spliceosome, at least two distinct, specific snRNP complexes associated with RNAPII can be identified in NE of HeLa cells. The smaller X1 complex consists of RNAPIIO associated with U1, while the larger X2 complex contains in addition U2 snRNP (Figure 8). X1 and X2 represent a direct RNAPIIO–snRNP interaction. Their components are not connected through DNA, as X1 and X2 formation is resistant to DNase digestion (data not shown). Moreover, the 5′ end of U1 snRNA in these complexes is available for binding the 5′SS RNA, arguing against a possible association via an RNA transcript.
Figure 8.
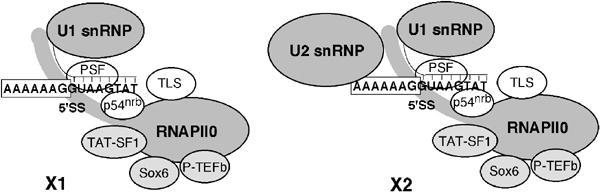
Schematic composition of X1 and X2 complexes. Relative positions of the indicated components are arbitrary.
The conditions required for X1 and X2 formation are strikingly similar to those characteristic of the early steps of spliceosome assembly in vitro. Similar to the binding of U1 snRNP to the 5′SS, the formation of X1 can take place on ice and in the absence of ATP. In contrast, the formation of X2 requires ATP and incubation at elevated temperatures, conditions that are also required for binding of U2 snRNP to the branch site in pre-mRNA. Accordingly, several factors linked to the early steps of spliceosome assembly are associated with X1 and X2, such as hPrp5, FBP11 and FBP21. Prp5 bridges between U1 and U2 snRNPs during pre-spliceosome assembly (Xu et al, 2004), while both FBP11 and FBP21 are known to interact in vitro with SF1, a protein binding the branch site in the commitment complex (Bedford et al, 1997, 1998). FBP11 is a putative mammalian ortholog of the yeast splicing factor Prp40, which interacts with the CTD (Morris and Greenleaf, 2000); FBP21 associates with U2 snRNP, splicing complex A and the U1C component of U1 snRNP (Bedford et al, 1998). While the detailed functions of these proteins are presently unknown, a possible network of interactions could involve the CTD, Prp40/FBP11, FBP21, U1C and SF1, linking RNAPII to the recognition of the 5′SS and branch site in its nascent transcript. Thus, X1 and X2 may resemble exon definition complexes associated with the elongating RNAPII.
The most abundant 5′SS crosslink detected in X1 and X2 complexes has been identified as p54nrb. The mouse homolog of p54nrb, NonO, activates transcription by binding an enhancer element in the intracisternal A particle (Basu et al, 1997) and induces the binding of other transcription factors to their response elements (Yang et al, 1997). In addition to p54nrb, the analyzed complexes contain the PSF and TLS proteins. PSF and p54nrb are highly related (71% identity) in the central region spanning the two RRMs and have been shown to interact with each other (Peng et al, 2002). These two proteins can bind both RNA and DNA (Patton et al, 1993; Yang et al, 1993; Zhang et al, 1993; Basu et al, 1997), act as transcriptional regulators (Basu et al, 1997; Urban et al, 2000; Mathur et al, 2001) and interact with the RNAPII CTD (Emili et al, 2002). Similarly, TLS binds DNA (Perrotti et al, 1998) and RNA (Lerga et al, 2001) and associates with RNAPII (Yang et al, 2000), snRNPs (Hackl et al, 1994) and spliceosomes (Calvio et al, 1995; Wu and Green, 1997). Interestingly, these three factors, p54nrb, PSF (this work and Ismaili et al, 2001) and TLS (Wu and Green, 1997), have been detected in direct proximity of either the 5′SS or the 3′SS. Thus, all the three proteins bind both DNA and RNA, have been implicated in splicing and transcriptional control, interact with RNAPII and can associate with snRNPs. Another transcription factor found to be associated with X1 and X2 is Sox6, also recently implicated in pre-mRNA splicing in vitro (Ohe et al, 2002).
Like X1 and X2, U2/4/5/6 complexes formed in HeLa NE in the presence of U1-blocking DNA are also associated with RNAPIIO and contain p54nrb and PSF. In fact, not only the spliceosomes formed in the presence of ATP (in which the 5′SS RNA crosslinks to hPrp28) but also the less stable U2/4/5/6 snRNP complexes formed in the absence of ATP (in which the 5′SS crosslinks to PSF) are preferentially associated with RNAPIIO. Previous studies have shown that RNAPIIO co-immunoprecipitates splicing factors and intermediates (Mortillaro et al, 1996; Kim et al, 1997), confirming that functional spliceosomes can interact with RNAPIIO even in the absence of ongoing transcription. Although U1 snRNP also binds RNAPIIA (as evidenced by efficient precipitation of U1 snRNA with 8WG16 antibody), X1 and X2 are preferentially associated with RNAPIIO. Similarly, while at least two transcription/splicing proteins, p54nrb and PSF, associate with both RNAPIIA and RNAPIIO, a significant rearrangement of these factors must occur upon CTD phosphorylation, allowing for crosslinking of the 5′SS to p54nrb exclusively in X1 and X2 complexes. The nature of this rearrangement is unclear and it is not known whether phosphorylation of proteins in addition to the CTD (e.g. p54nrb, PSF, U1–70K) may be required. It also remains to be established whether the described interactions of U1 snRNP, p54nrb and PSF with RNAPIIA reflect the events at the initiation and/or early elongation stages of transcription. Interestingly, PSF is also present in U4/5/6 tri-snRNP (Teigelkamp et al, 1997), suggesting that it may function as a recognition element for the joining of U4/5/6 to the pre-spliceosome.
In addition to RNAPIIO, X1 and X2 also contain several transcription factors typically found in ECs, such as P-TEFb, TAT-SF1 and TFIIF (Figure 8). Although X1 and X2 do not involve actively transcribing RNAPII, their composition suggests that they reflect natural interactions taking place during transcription/splicing. In fact, isolation and analysis of functional transcription ECs showed that numerous X1 and X2 components are associated with the transcribing RNAPII. In addition to RNAPII preferentially in the hyperphosphorylated form, functional ECs contain the transcription/splicing factors, p54nrb, PSF and TLS, as well as transcription elongation factors, such as P-TEFb and TAT-SF1. As expected, TFIIB (also absent from X1 and X2) and TBP, factors involved in the assembly of initiation complexes, were not found in ECs. Interestingly, the general transcription factor TFIIH was detected at high levels. It has recently been shown that TFIIH specifically associates with U1 snRNA, which in turn stimulates the rate of initiation of RNAPII (Kwek et al, 2002). This finding suggests a link between the initiation of transcription and mRNA splicing. In this context, it is interesting that we find U1 snRNP bound both to RNAPIIO in X1 and X2 complexes and to RNAPIIA. Also detectable in functional ECs were many snRNP-associated splicing factors, including those associated with U4/5/6 snRNPs, which are not X1 or X2 components. Accordingly, all the five snRNAs were also detected bound to the transcription template. Thus, the ECs isolated here include all the five snRNPs, pointing to the assembly of the entire spliceosome on the elongating RNAPIIO. Nevertheless, the fact that X1 and X2 contain a large subset of EC components strongly suggests that they include interactions present in functional transcription/splicing complexes.
Recently, p54nrb, PSF and matrin3 were found to bind inosine-containing RNAs specifically (Zhang and Carmichael, 2001). Like p54nrb and PSF, matrin 3 also contains two tandem RRMs and can regulate transcription (Hibino et al, 2000). Together, these observations suggest a model in which transcription/splicing factors like p54nrb, PSF, TLS, matrin 3, and, probably many others, participate in a complex structure, in the context of which the individual processes of gene expression—transcription, capping, editing, splicing, polyadenylation and possibly transport—are organized. Both PSF and p54nrb bind to the RNAPII CTD (Emili et al, 2002 and our data not shown). Our finding that these proteins can also directly interact with the 5′SS associated with RNAPII–snRNP complexes indicates that they may mediate contacts between the transcription and splicing machineries, perhaps forming a platform for post-transcriptional RNA processing.
Materials and methods
Antibodies
Several of the antibodies used were generously provided by M Bedford (α-FBP11, α-FBP21), R Burgess (8WG16), A Krainer (α-p54nrb used in Figure 2C), R Lührmann (α-U1-70k (H111), α-U5-116k, α-hPrp4), C Lutz (α-U1A), J Patton (α-PSF, α-PTB), C Query (α-hPrp5), R Reed (α-SAP155), R Roeder (α-TFIIB, α-p62), D Ron (α-TLS), M Roth (16H3) and W VanVenrooij (α-U2-B″).
The H5 and H14 antibodies were from Covance, α-TAT-SF1 from BD Biosciences, α-Sm (Y12) from Lab Vision and α-RNAPII (N-20), α-RAP74, α-Cdk9 and α-Sox6 from Santa Cruz Biotechnology. The α-p54nrb rabbit antibody used in Figure 7B was raised at Genemed Synthesis against the N-terminal (aa 1–15) peptide (QSNKTFNLEKQNHTP) of p54nrb. Rabbit α-hPrp28 antibody was generated at Covance against bacterially expressed hPrp28.
Binding reactions and UV crosslinking
A standard binding reaction (10 μl) contained 4 μl (∼10 μg protein) of a HeLa NE (Dignam et al, 1983), 2 mM Mg acetate, 20 mM KCl, 1 mM ATP and 5 mM creatine phosphate. To block the 5′ end of U1 snRNA, the U1-blocking DNA oligonucleotide (50 ng/μl, 5′-CAGGTAAGTAT-3′) was pre-incubated with NE (30°C for 20 min) prior to the addition of 5′SS RNA (5′-AAAAAGGUAAGdTdAdT-3′), prepared and derivatized with BP 4-iodoacetamide as previously described (Ismaili et al, 2001). After 15 min at 30°C, reactions were UV-irradiated (302 nm) for 3 min in a microtiter plate on ice. Typically, 1 × 104 and 1 × 106 cpm of the 32P-labeled 5′SS RNA were used for crosslinking and immunoprecipitation experiments, respectively.
Microsequencing of p54nrb
A large-scale binding reaction (4 × 500 μl) was centrifuged through a 200-μl cushion of buffer D (0.1 M KCl, 20 mM HEPES–KOH (pH 7.6), 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT) at 200 000 g for 30 min. The pellet was resuspended in 15 μl buffer D, treated with RNase A and T1 (100 μg/ml each) for 30 min at 30°C, solubilized in 80 μl of 9.5 M urea, 2% NP-40, 5% β-mercaptoethanol and 2% Bio-Lyte 3–10 (Bio-Rad) for 30 min at 25°C and resolved by isoelectrofocusing in the first dimension and by electrophoresis in a 9% SDS gel in the second. p54nrb was identified by superimposition of the Coomassie Brilliant Blue (G-250)-stained gel with the autoradiograph of the p54 crosslink and the α-SR antibody Western blot. Sequence analysis of the protein digested with Lys-C was performed at the Protein/DNA Technology Center of The Rockefeller University. A blast p search identified all the three obtained peptide sequences shown in Figure 2D within the human p54nrb.
Glycerol gradient fractionation
Crosslinking reactions (150 μl) containing ATP and 1 × 107 cpm of the 5′SS RNA were layered onto 10–30% glycerol gradients in 50 mM Tris-glycine and centrifuged for 3.5 h at 50 krpm at 4°C in an SW55 rotor. Gradients were divided into 16 300-μl fractions, of which 10 or 50 μl were used for native gel electrophoresis or immunoprecipitation, respectively.
Immunoprecipitation (IP)
For all IPs, beads were pre-blocked with 3% BSA for 30 min at RT and washed (3 × 500 μl) with IP-100 (100 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.05% NP-40, 2 mM MgCl2, 0.5 mM DTT). Subsequently, antibodies (5–20 μl) were bound to 15 μl of pre-blocked beads in 30 μl of IP-100. Protein A–trisacryl beads (Pierce) were used except for α-U2-B″ and Y12 antibodies, and H5 and H14, for which Protein G beads (Amersham) and Protein L beads (Santa Cruz Biotechnology), respectively, were used. The beads were then washed (3 × 500 μl) with IP-100.
For native IPs, the antibody-beads slurry was resuspended in 20 μl of buffer D containing protease inhibitors (1 mM PMSF, 3 μg/ml leupeptin and 5 μg/ml soybean trypsin inhibitor). The slurry was incubated for 1.5 h at 4°C with 10 μl of crosslinking reactions or 50 μl of gradient fractions and then washed gently (3 × 500 μl) with NETN50 (50 mM NaCl, 20 mM Tris–HCl (pH 7.5), 0.5% NP-40, 1 mM EDTA, 0.5 mM DTT) containing protease inhibitors at 4°C.
For the denaturing IP performed in Figure 2B, crosslinking reactions were pre-incubated with RNase A (0.1 μg/μl) and Empigen BB (1%) in 300 μl PBS containing 1 mM EDTA, 0.1 mM DTT and protease inhibitors at RT for 30 min and then sonicated for 3 × 5 s. The antibody-beads slurry was then added and the mix was incubated for 1.5 h at 4°C before washing (3 × 500 μl) with IP100.
For the denaturing IP performed in Figure 2C, gel-purified 5′SS:p54 and 5′SS:hPrp28 crosslinks were acetone-precipitated and dissolved in 1 ml of TBST (10 mM Tris–HCl (pH 8.0), 50 mM NaCl, 0.1% Tween 20). Anti-p54nrb or α-hPrp28 antibody beads were added and the slurry was incubated for 1.5 h at 4°C and washed (5 × 500 μl) with TBST at RT.
For all IPs, the input corresponded to 5% of the total reaction.
Immobilized template assay
For construction of the pBSAd20 plasmid, the first two leader exons of the adenovirus major late transcription unit, separated by a shortened version of the first intervening sequence, were cloned into the pBS- vector (Stratagene) under the T7 promoter. The pBSAd22 plasmid contains, in addition, a 399-nt sequence including the adenovirus major late promoter (AdMLP) immediately downstream of the T7 promoter.
Templates were synthesized by PCR from pBSAd20 or pBSAd22 using biotinylated, upstream primer Ade3 (5′-biotin-GTTGGGTAACGCCAGGG-3′) and downstream primer T3-L (5′-GCGCAATTAACCCTCACTAAA-3′). The resulting fragments were gel purified and bound to M-280 streptavidin Dynabeads (Dynal) according to the manufacturer's instructions. The immobilized templates were then blocked with 1% BSA in BC-100 buffer (20 mM HEPES–KOH (pH 7.6), 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 250 μM PMSF) for 20 min at RT. After washing with BC-100, 125 μg beads (∼800 ng immobilized DNA) were resuspended in 28 μl BC-100 containing 0.05% NP-40. Transcription reactions (105 μl total volume) were performed at 30°C for 30 min after the addition of 6.7 mM MgCl2, 400 μM NTPs and 35 μl (∼350 μg protein) HeLa NE (Dignam et al, 1983). The NE was pre-spun at 9 krpm for 2 min at 4°C and pre-incubated with 1 μg/100 μl sonicated Escherichia coli DNA for 10 min at RT. The bead-bound templates were washed gently (3 × 100 μl) with BC-100 containing 0.05% NP-40 and 0.05% BSA and then digested with PvuII (New England Biolabs) as previously described (Ranish et al, 1999). The supernatant was removed from the beads and subjected to 7 or 10% SDS–PAGE. Proteins were electroblotted onto nitrocellulose membranes and detected by ECL (Perkin-Elmer). As a negative control, beads lacking DNA were submitted to the same experimental procedure.
For RNA analysis, the material released from beads was phenol extracted, ethanol precipitated and resolved in a 10% polyacrylamide/8 M urea gel and probed by Northern hybridization.
Acknowledgments
We thank Drs M Bedford, R Burgess, A Krainer, R Lührmann, C Lutz, J Patton, C Query, R Reed, R Roeder, D Ron, M Roth and W Van Venrooij for several of the antibodies used in this work, J McCloskey for help at the initial stages of the project and Diana Colgan and Charles Query for many constructive discussions and helpful comments on the manuscript. This work was supported by NIH grant GM 49044 to MMK.
References
- Basu A, Dong B, Krainer AR, Howe CC (1997) The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol 17: 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L (1994) Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Chan DC, Leder P (1997) FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J 16: 2376–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Reed R, Leder P (1998) WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA 95: 10602–10607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol 14: 336–342 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN (1988) Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland RF, Cech TR, Atkins JF (eds), pp 525–560. New York: Cold Spring Harbor laboratory Press [Google Scholar]
- Calvio C, Neubauer G, Mann M, Lamond AI (1995) Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA 1: 724–733 [PMC free article] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM Jr, Dahmus ME (1985) A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA 82: 7934–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251–258 [DOI] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR (1997) Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA 94: 11456–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Horowitz DS, Kobayashi R, Krainer AR (1993) Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res 21: 4085–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ (2002) Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q (2001) Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933 [DOI] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl W, Fischer U, Luhrmann R (1994) A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol 124: 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino Y, Ohzeki H, Sugano N, Hiraga K (2000) Transcription modulation by a rat nuclear scaffold protein, P130, and a rat highly repetitive DNA component or various types of animal and plant matrix or scaffold attachment regions. Biochem Biophys Res Commun 279: 282–287 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL (1999) Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL (1991) Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev 5: 2288–2302 [DOI] [PubMed] [Google Scholar]
- Ismaili N, Sha M, Gustafson EH, Konarska MM (2001) The 100-kda U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA 7: 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL (1997) Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol 136: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti BB, Konarska MM (1994) U4/U5/U6 snRNP recognizes the 5′ splice site in the absence of U2 snRNP. Genes Dev 8: 1962–1973 [DOI] [PubMed] [Google Scholar]
- Konforti BB, Koziolkiewicz MJ, Konarska MM (1993) Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell 75: 863–873 [DOI] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A (2002) U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, Moreau-Gachelin F (2001) Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem 276: 6807–6816 [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH (2001) PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol 21: 2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL (2000) The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R (1996) A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA 93: 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohe K, Lalli E, Sassone-Corsi P (2002) A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc Natl Acad Sci USA 99: 1146–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B (1993) Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev 7: 393–406 [DOI] [PubMed] [Google Scholar]
- Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG (2002) PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Lozzo RV, Cooper DR, Calabretta B (1998) TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J 17: 4442–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S (1999) Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev 13: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D (2002) PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett 531: 109–114 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Teigelkamp S, Mundt C, Achsel T, Will CL, Luhrmann R (1997) The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3: 1313–1326 [PMC free article] [PubMed] [Google Scholar]
- Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S (2000) Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol 14: 774–782 [DOI] [PubMed] [Google Scholar]
- Vincent M, Lauriault P, Dubois MF, Lavoie S, Bensaude O, Chabot B (1996) The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res 24: 4649–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Green MR (1997) Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J 16: 4421–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC (2004) Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J 23: 376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Hickstein DD (2000) TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine–arginine proteins. Mol Cell Biol 20: 3345–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Hanke JH, Carayannopoulos L, Craft CM, Capra JD, Tucker PW (1993) NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol 13: 5593–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Yang MC, Tucker PW, Capra JD (1997) NonO enhances the association of many DNA-binding proteins to their targets. Nucleic Acids Res 25: 2284–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Berget SM (2000) Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol Cell Biol 20: 8290–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Abovich N, Rosbash M (2001) A biochemical function for the Sm complex. Mol Cell 7: 319–329 [DOI] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR (1994) Localization of pre-mRNA splicing in mammalian nuclei. Nature 372: 809–812 [DOI] [PubMed] [Google Scholar]
- Zhang WW, Zhang LX, Busch RK, Farres J, Busch H (1993) Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem J 290 (Part 1): 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG (2001) The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]


