Abstract
The periplasmic heme chaperone holoCcmE is essential for heme trafficking in the cytochrome c biosynthetic pathway in many bacteria, archaea, and plant mitochondria. This pathway, called system I, involves two steps: i) formation and release of holoCcmE (by the ABC-transporter complex CcmABCD), and ii) delivery of the heme in holoCcmE to the putative cytochrome c heme lyase complex, CcmFH. CcmFH is believed to facilitate the final covalent attachment of heme (from holoCcmE) to the apocytochrome c. Although most models for system I propose that holoCcmE delivers heme directly to CcmF, no interaction between holoCcmE and CcmF has been demonstrated. Here, a complex between holoCcmE and CcmF is “trapped”, purified, and characterized. HoloCcmE must be released from the ABC-transporter complex CcmABCD to interact with CcmF, and the holo-form of CcmE interacts with CcmF at levels at least 20-fold higher than apoCcmE. Two conserved histidines (here termed P-His1 and P-His2) in separate periplasmic loops in CcmF are required for interaction with holoCcmE, and evidence is presented that P-His1 and P-His2 function as heme-binding ligands. These results show that heme in holoCcmE is essential for complex formation with CcmF, and that the heme of holoCcmE is coordinated by P-His1 and P-His2 within the WWD domain of CcmF. These features are strikingly similar to formation of the CcmC:heme:CcmE ternary complex (Richard-Fogal and Kranz, JMB 2010), and suggest common mechanistic and structural aspects.
Keywords: Pathway, Biogenesis, Oxidation-Reduction, Heme trafficking, Cytochrome c Maturation
INTRODUCTION
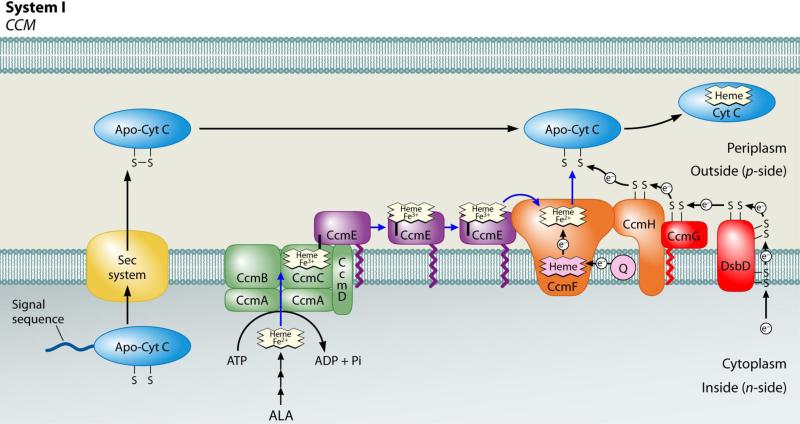
C-type cytochromes are heme proteins involved in vital electron transfer reactions in the cell. These cytochromes function outside of the cytoplasmic membrane in prokaryotes, in the lumen of chloroplasts, and in the intermembrane space of mitochondria. Cytochromes c are unique among heme proteins in that the heme is covalently attached to the protein (via thioether linkages between the 2- and 4-vinyls of heme and two thiols of a conserved Cys-Xxx-Xxx-Cys-His motif in the apoprotein). For covalent attachment to occur, both the apocytochrome thiols and the iron of heme must be reduced 1; 2. In many bacteria, plant and protozoal mitochondria, and archaea, holocytochrome c formation requires the cytochrome c maturation (ccm) pathway, called system I (see Fig 1), which comprises eight dedicated membrane proteins (in E. coli, CcmABCDEFGH) 3; 4; 5; 6; 7.
Fig 1.
Current working model of the system I cytochrome c biogenesis pathway. Model includes trafficking and oxidation states of heme as well as apocytochrome translocation and reduction. Adapted from (Kranz, Richard-Fogal et al. 2009).
System I can be conceptually described as two steps (see Fig 1): (i) formation and release of the heme chaperone protein, called holo (+ heme) CcmE, (from the ABC-transporter complex CcmABCD) and (ii) heme transfer from holoCcmE to the apocytochrome (putatively, via CcmFH) to yield a holocytochrome c. HoloCcmE binds heme through a unique covalent attachment between the β carbon of the heme 2-vinyl and a conserved histidine residue (His130, in E. coli CcmE) 8; 9; 10. Covalent attachment of heme to holoCcmE at His130 is mediated by the integral membrane proteins CcmC and CcmD, which form a stable complex with “trapped” holoCcmE (in the absence of CcmA and CcmB) 11; 12. HoloCcmE is released from its binding site in CcmCD upon ATP-hydrolysis by CcmA, which, together with CcmB, forms an ABC-transporter complex with CcmC and CcmD for release of covalent holoCcmE (see Fig 1) 13; 14; 15. CcmA, CcmB, and CcmC each have homology to individual subunits of components of ABC-transporter complexes, with the classic Walker domain found in CcmA 16. Released, oxidized (Fe3+) holoCcmE is proposed to chaperone its heme to the site of cytochrome c formation, the CcmFH complex (see Fig 1), but this has not been proven (see below) 12; 17; 18. CcmF, which forms an integral membrane complex with CcmH, is believed to be the site of thioether formation between the heme vinyls and the apocytochrome; thus, it has been termed the “cytochrome c heme lyase”. The CcmFH integral membrane complex has been purified 12 or co-immunoprecipitated 18; 19; 20. CcmF contains a separate and stable non-covalent heme b 12; 21, which we have hypothesized plays a role in reducing the incoming heme from holoCcmE 4; 21. Reduction of heme (to Fe2+) is a requirement for thioether formation 1; 2; 4, and would also favor the release of heme from CcmE His130 4; 11; 21. CcmG 22; 23; 24 and CcmH 25; 26 are thioredox-active proteins that maintain the apocytochrome thiols (in the Cys-Xxx-Xxx-Cys-His motif) in the reduced state 27; 28; 29. In some species, such as Rhodobacter capsulatus, ccmH is split into two open reading frames (called ccmH and ccmI). While there is significant evidence that the apocytochrome interacts with the CcmFH cytochrome c heme lyase complex (via CcmH) 25; 27; 30; 31, there has been no data demonstrating that holoCcmE interacts with CcmF (see Discussion for details). Nonetheless, heme delivery from holoCcmE to CcmF has been proposed in nearly every review on cytochrome c biogenesis during the last decade (e.g., 3; 4; 5; 6; 7). Thus, the proposal that holoCcmE trafficks heme directly to CcmF (see Fig 1) for holocytochrome c formation remains unproven.
Here, we describe purification and characterization of an intermediate complex between holoCcmE and CcmF, achieved by purifying CcmF from detergent-solubilized membranes lacking CcmG and CcmH. We show that holoCcmE must be released from CcmCD in order to interact with CcmF, and that holoCcmE forms a complex with CcmF at levels at least 20-fold higher than apoCcmE. We demonstrate that two periplasmic histidines (His173 and His303, here called P-His1 and P-His2, respectively) in separate periplasmic loops in CcmF are required for interaction with holoCcmE, with evidence that these residues function as heme-binding ligands. We propose that heme in holoCcmE is a critical component for interaction with CcmF and we discuss how these results mirror formation of the CcmC:heme:CcmE ternary complex 11 (see Fig 1).
RESULTS
The holoCcmE-CcmF complex
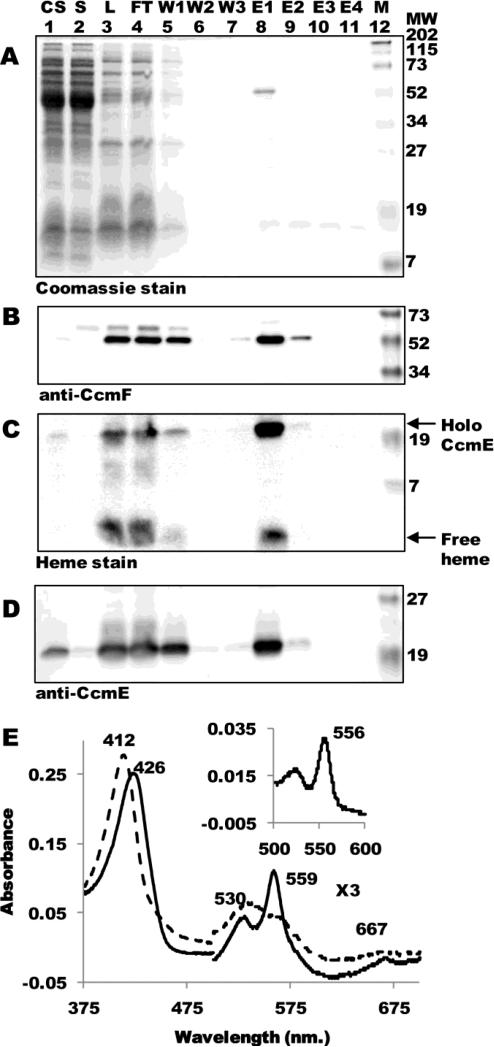
Although nearly all current in vivo models for the system I pathway presume a holoCcmE—CcmF intermediate during holocytochrome c biosynthesis, we have been unable to co-purify holoCcmE at detectable levels in our preparations of CcmFH 12. Our purifications of CcmF (and CcmFH complex) are typically from DDM-solubilized membranes that contain all Ccm proteins (CcmABCDEFGH). The inability to identify a complex between holoCcmE and CcmF could be due to a transient, short-lived interaction or current models for system I may be incorrect. In an attempt to detect an interaction between holoCcmE and CcmF, we expressed ccmF along with the minimal set of ccm components required for formation and release of holoCcmE from the CcmABCD complex (i.e., ccmABCDEF, or pGEX ΔGH; Fig 2 A-D). Note that the proteins CcmG and CcmH are absent in these cells. Full-length hexahistidine-tagged CcmF (54-kDa) was TALON-purified as a single polypeptide to greater than 90% purity (Fig 2A, lane 8) that reacted with CcmF antisera (Fig 2B, lane 8). Heme staining revealed that, in addition to the CcmF b-type heme (which dissociates from the protein and migrates as free heme during denaturing SDS-PAGE) preparations of CcmF from membranes lacking CcmG and CcmH contained 20-kDa holoCcmE (Fig 2C, lane 8). Immunodetection with CcmE antisera confirmed that the 20-kDa covalent heme species was holoCcmE (Fig 2D, lane 8).
Fig 2.
The CcmF-holoCcmE complex. (A) Coomassie blue staining of purified CcmF:His6 showing 54-kDa CcmF. (B) Anti-CcmF immunoblot of purified CcmF:His6 showing 54-kDa CcmF. (C) Heme staining of purified CcmF showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (D) Anti-CcmE immunoblot showing co-purified 20-kDa CcmE. For (A)-(D), abbreviations are CS, crude sonicate; S, soluble fraction; L, load (DDM-solubilized membranes); FT, flow through; W1, wash 1; W2, wash 2; W3, wash 3; E1, elution 1; E2, elution 2; E3, elution 3; E4, elution 4; M, molecular weight standards. (E) UV-Vis absorption spectra of CcmF-holoCcmE complex as purified (dotted line) or reduced with sodium dithionite (solid line). The region from 500-700 nm has been multiplied by a factor of 3. (Inset) Sodium dithionite-reduced pyridine hemochrome spectrum of purified CcmF-holoCcmE complex from 500-600 nm. Absorption maxima are indicated.
UV-Vis absorption spectra of the oxidized (as purified) CcmF-holoCcmE preparation showed a Soret maximum at 412 nm and broad α and β absorptions between 500 and 600 nm (Fig 2E). Chemical reduction with sodium dithionite yielded a Soret maximum at 426 nm and sharp α and β absorptions at 559 and 530, respectively. These spectral features, in addition to the 556 nm absorption in the reduced pyridine hemochrome (Fig 2E, inset) are characteristic of non-covalent, b-type hemes 32, and are indistinguishable from those of the CcmF b-heme 12; 21. Because the holoCcmE polypeptide is not readily detectable by Coomassie staining (Fig 2A, lane 8), it is likely that holoCcmE is less than stoichiometric with CcmF in the CcmF-holoCcmE complex. This result, together with only slight differences in the electronic absorptions of holoCcmE and CcmF, make it difficult to discern the spectral contributions of the heme from holoCcmE. We next wanted to analyze the role of other Ccm proteins as well as specific residues in CcmF in formation of the holoCcmE-CcmF complex.
HoloCcmE co-purifies with CcmF in the absence of CcmG and CcmH
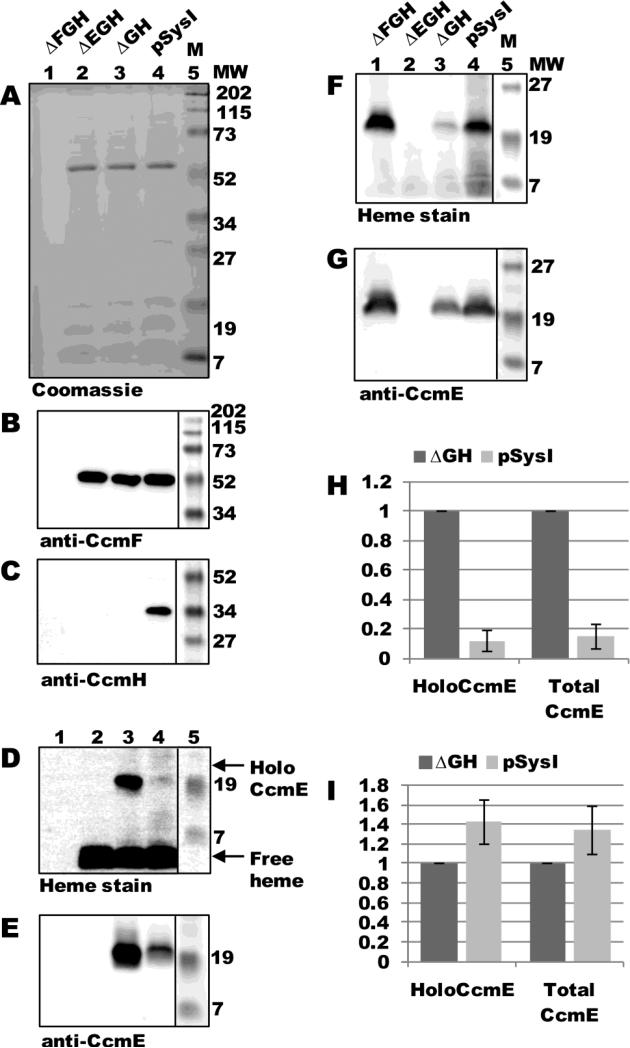
As mentioned above, purifications of CcmF from membranes containing all Ccm proteins (CcmABCDEFGH) typically do not yield detectable holoCcmE. Therefore, we directly compared levels of holoCcmE that co-purified with CcmF in the presence of all Ccm proteins (expressed from pSysI) to those that co-purified with CcmF in the absence of CcmG and CcmH (expressed from pSysI ΔGH). As controls, we included in this analysis constructs lacking ccmF (pSysI ΔFGH, or pGEX ccmABCDE) and ccmE (pSysI ΔEGH, or pGEX ccmABCDF:His6). With the exception of ΔFGH, CcmF was purified as a single full-length polypeptide (54-kDa; Fig 3A, lanes 2-4) that reacted with CcmF antisera (Fig 3B, lanes 2-4). Immunoblotting with CcmH antisera showed that co-purified 34-kDa CcmH was present only in CcmF purifications from the pSysI background, as expected (Fig 3C, lane 4). Heme staining and anti-CcmE immunoblotting of TALON-purified fractions revealed that, in the absence of CcmG and CcmH, CcmF co-purified with approximately 10-fold more holoCcmE than when CcmG and CcmH were present (Fig 3D and E, compare lanes 3 and 4; quantified in Fig 3H). In the absence of CcmF (pSysI ΔFGH), no holoCcmE was detected by heme stain or anti-CcmE immunoblot (Fig 3D and E, lane 1), showing that there was no detectable retention of holoCcmE on the TALON resin. Similarly, in the absence of CcmE (pSysI ΔEGH), no 20-kDa covalent heme species or reactivity with CcmE antisera were observed (Fig 3D and E, lane 2). Analysis of DDM-solubilized membrane fractions by heme staining and immunoblotting with CcmE antisera showed that all membranes (with the exception of those from pSysI ΔEGH) contained holoCcmE at levels at least as high as that of pSysI ΔGH (Fig 3F and G; quantified in Fig 3I). Therefore, the 10-fold higher levels of holoCcmE that co-purified with CcmF from pSysI ΔGH are not due to increased expression of holoCcmE from this construct. We suggest that CcmH prevents (controls) trapping of the holoCcmE/CcmF complex (see Discussion).
Fig 3.
HoloCcmE co-purifies with CcmF in the absence of CcmGH. (A) Coomassie blue staining of TALON-purified proteins from ΔFGH, ΔEGH, ΔGH, and pSysI backgrounds showing purified 54-kDa CcmF. (B) Anti-CcmF immunoblot of TALON-purified proteins showing 54-kDa CcmF. (C) Anti-CcmH immunoblot of TALON-purified proteins showing 34 kDa CcmH. (D) Heme staining of TALON-purified proteins showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (E) Anti-CcmE immunoblot of TALON-purified proteins showing co-purified 20-kDa CcmE. For (A)-(E), 6 ug purified protein was analyzed. (F) Heme staining of DDM-solubilized membrane fractions from ΔFGH, ΔEGH, ΔGH, and pSysI backgrounds showing 20 kDa holoCcmE. (G) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20 kDa CcmE. For (F) and (G), 70 ug total protein was analyzed. (H) Quantification of the results of heme staining (holoCcmE) and anti-CcmE immunoreactivity (total CcmE) from TALON-purified fractions from three independent experiments. (I) Quantification of the results of heme staining and anti-CcmE immunoreactivity from DDM-solubilized membrane fractions from three independent experiments. For (H) and (I), percent holoCcmE and total CcmE is relative to ΔGH, which has been set at 100%. Error bars denote SD.
HoloCcmE must be released (by the ABC-transporter complex CcmABCD) to interact with CcmF
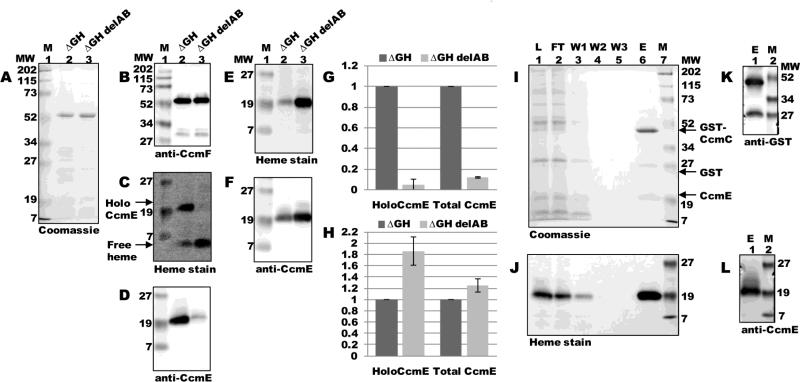
The first step in heme translocation in the system I (CCM) pathway involves formation of covalent (holo) CcmE, which occurs via complex formation with CcmC and CcmD 11; 12; 33. Subsequently, CcmA and CcmB form an ABC transporter “release complex” with CcmC and CcmD, which uses ATP hydrolysis to release holoCcmE for heme trafficking to (putatively) CcmFH 13; 14; 15. In the absence of CcmAB, holoCcmE becomes “trapped” with CcmCD in a very stable intermediate complex 11, and holocytochrome c formation is blocked at this step. We examined whether interaction between CcmF and holoCcmE is dependent on release of holoCcmE from CcmABCD. We engineered an in-frame deletion of ccmAB in pSysI ΔGH to yield pSysI ΔGH delAB (pGEX ccmCDEF:His6), with a GST translational fusion to CcmC rather than CcmA.
ccmF was expressed from pSysI ΔGH delAB and purified as a single full-length polypeptide (54-kDa; Fig 4A, lane 3) that reacted with CcmF antisera (Fig 4B, lane 3). Heme staining and anti-CcmE immunoblotting revealed that the absence of CcmAB resulted in at least a 10-fold decrease in the amount of holoCcmE that co-purified with CcmF (Fig 4C and D, compare lanes 2 and 3; quantified in Fig 4G). Analysis of DDM-solubilized membrane fractions by heme staining and immunoblotting with CcmE antisera showed that membranes from pSysI ΔGH delAB contained holoCcmE at levels at least as high as that of pSysI ΔGH (Fig 4E and F; quantified in Fig 4H). To confirm that release of holoCcmE from CcmCD was blocked by deletion of ccmAB, we purified GST:CcmC from the flow-through fraction of the TALON column (Fig 4 I-L). Purified full-length GST:CcmC (48-kDa) and free GST each reacted with GST antisera (Fig 4K, lane 1). Heme staining (Fig 4J, lane 6) and immunodetection with CcmE antisera (Fig 4L, lane 1) revealed that purified GST:CcmC contained high levels of trapped (unreleased) holoCcmE. Thus, in the absence of CcmAB, holoCcmE is trapped on CcmC, which effectively abolishes formation of the CcmF-holoCcmE complex. Only released holoCcmE interacts with CcmF, as previous models have hypothesized. This strongly suggests that the interaction we detect between holoCcmE and CcmF is a true intermediate during holocytochrome c formation.
Fig 4.
HoloCcmE must be released from CcmABCD to interact with CcmF. (A) Coomassie blue staining of TALON-purified proteins from ΔGH and ΔGH delAB backgrounds showing purified 54-kDa full-length CcmF. (B) Anti-CcmF immunoblot of purified CcmF proteins showing 54-kDa full-length CcmF. (C) Heme staining of purified CcmF proteins showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (D) Anti-CcmE immunoblot of purified CcmF proteins showing co-purified 20-kDa CcmE. For (A)-(D), 5 ug purified protein was analyzed. (E) Heme staining of DDM-solubilized membrane fractions from ΔGH and ΔGH delAB backgrounds showing 20 kDa holoCcmE. (F) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20 kDa CcmE. For (E) and (F), 70 ug total protein was analyzed. (G) Quantification of the results of heme staining (holoCcmE) and anti-CcmE immunoreactivity (total CcmE) from purified fractions from three independent experiments. (H) Quantification of the results of heme staining and anti-CcmE immunoreactivity from DDM-solubilized membrane fractions from three independent experiments. For (G) and (H), percent holoCcmE and total CcmE is relative to ΔGH, which has been set at 100%. Error bars denote SD. (I) Coomassie blue staining of purified GST-tagged CcmC from ΔGH delAB showing 48-kDa full-length GST-CcmC, 26-kDa GST, and 20 kDa CcmE. (J) Heme staining of purified GST-tagged CcmC showing co-purified 20-kDa holoCcmE. (K) Anti-GST immunoblot of purified GST-tagged CcmC showing 48-kDa full-length GST-CcmC and 26-kDa GST. (L) Anti-CcmE immunoblot of purified GST-tagged CcmC showing co-purified 20-kDa CcmE.
HoloCcmE, not apoCcmE, is essential for interaction with CcmF
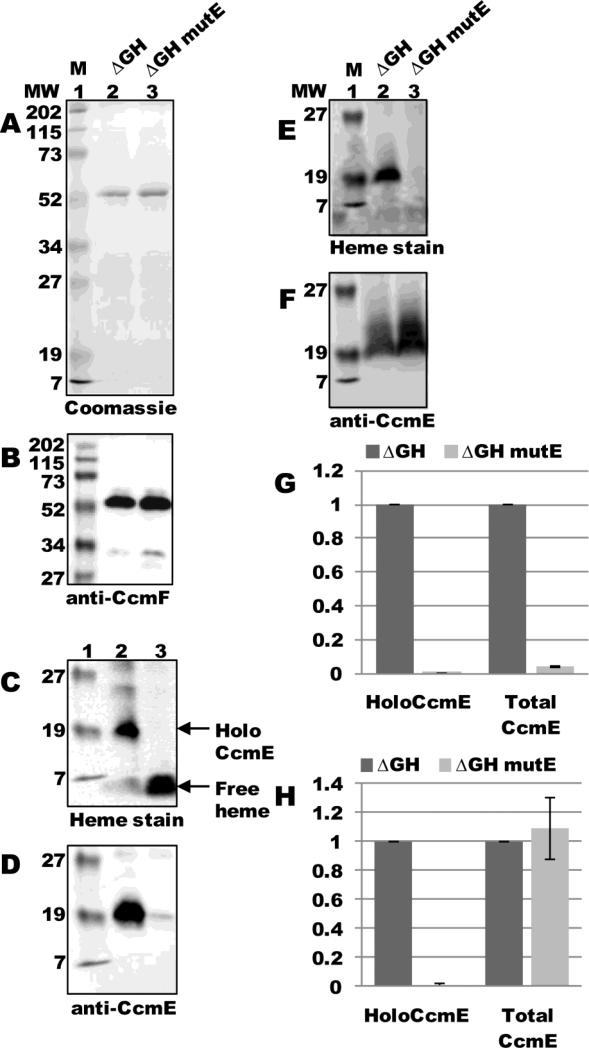
CcmE binds heme through a unique covalent bond between conserved His130 and the 2-vinyl of the heme 8; 9; 10. Previous work has shown that mutation of His130 to alanine abolishes covalent bond formation between heme and CcmE 34, although CcmE(His130Ala) still forms a complex with CcmCD and is likely released upon ATP hydrolysis by CcmAB 11. Since released CcmE(His130Ala) is completely apo- (lacking heme), holocytochrome c formation is blocked at this step (i.e., the covalent bond to heme in holoCcmE is necessary for CcmE to chaperone heme to, putatively, CcmFH). To test whether apo-CcmE could interact with CcmF, we engineered the His130Ala substitution in ccmE in pSysI ΔccmGH to yield pSysI ΔccmGH mutE (pGEX ccmABCDE(His130Ala)F:His6). ccmF was expressed from pSysI ΔGH mutE and purified as a single full-length polypeptide (54-kDa; Fig 5A, lane 3) that reacted with CcmF antisera (Fig 5B, lane 3). As expected, heme staining showed no evidence of holoCcmE in purified CcmF fractions (Fig 5C, lane 3; quantified in Fig 5G), since mutation of His130 results in only the apo-form of CcmE (Fig 5E, lane 3; quantified in Fig 5H). Immunoblotting with CcmE antisera revealed approximately a 20-fold decrease in the amount of apoCcmE that co-purified with CcmF (Fig 5D, compare lanes 2 and 3; quantified in Fig 5G), even though DDM-solubilized membranes from pSysI ΔGH mutE contained apoCcmE at levels at least as high as those of pSysI ΔGH (Fig 5F, compare lanes 2 and 3; quantified in Fig 5H). Thus, apoCcmE is not capable of interaction with CcmF, which suggests that holoCcmE—CcmF complex formation is heme-dependent (see Discussion).
Fig 5.
ApoCcmE does not co-purify with CcmF. (A) Coomassie blue staining of TALON-purified proteins from ΔGH and ΔGH mutE (CcmE His130Ala) backgrounds showing purified 54-kDa CcmF. (B) Anti-CcmF immunoblot of purified CcmF proteins showing 54-kDa CcmF. (C) Heme staining of purified CcmF proteins showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (D) Anti-CcmE immunoblot of purified CcmF proteins showing co-purified 20-kDa CcmE. For (A)-(D), 5 ug purified protein was analyzed. (E) Heme staining of DDM-solubilized membrane fractions from ΔGH and ΔGH mutE backgrounds showing 20 kDa holoCcmE. (F) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20 kDa CcmE. For (E) and (F), 70 ug total protein was analyzed. (G) Quantification of the results of heme staining (holoCcmE) and anti-CcmE immunoreactivity (total CcmE) from purified fractions from three independent experiments. (H) Quantification of the results of heme staining and anti-CcmE immunoreactivity from DDM-solubilized membrane fractions from three independent experiments. For (G) and (H), percent holoCcmE and total CcmE is relative to ΔGH, which has been set at 100%. Error bars denote SD.
CcmF P-His1 and P-His2 exhibit heme ligand activity
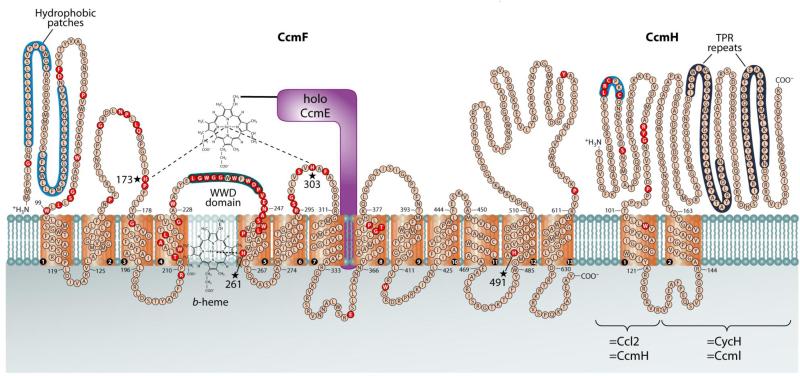
CcmF contains four conserved His residues (see Fig 6): His261 in TMD5 and His491 in TMD12 (here called TM-His1 and TM-His2, respectively); and His173 and His303 in periplasmic loops flanking the conserved WWD domain (here called P-His1 and P-His2, respectively). Alanine substitutions at any of the His residues in CcmF abolishes holocytochrome c formation 12; 18; 21. TM-His1 and TM-His2 are ligands to the b-heme in CcmF: mutation of either of these transmembrane His residues results in a loss of nearly all b-heme in the purified protein 12; 21. However, the roles of P-His1 and P-His2 are unknown. Based on their position flanking the WWD domain (which, in CcmC, has been shown to be the site of interaction with the holoCcmE heme 11), we hypothesize that P-His1 and P-His2 in CcmF may be ligands to the incoming heme from holoCcmE (see Fig 6 and Discussion).
Fig 6.
Topology of the CcmF and CcmH integral membrane proteins from E. coli. Possible histidine axial ligands to heme are starred (His173=P-His1; His303=P-His2; His261=TM-His1; His491=TM-His2). The highly conserved WWD domain is shaded as are the hydrophobic patches. Completely conserved amino acids (red) were identified by individual protein alignments using CcmF ORFs from selected organisms, as described in (Kranz, Richard-Fogal et al. 2009). Diagram is from (Kranz, Richard-Fogal et al. 2009).
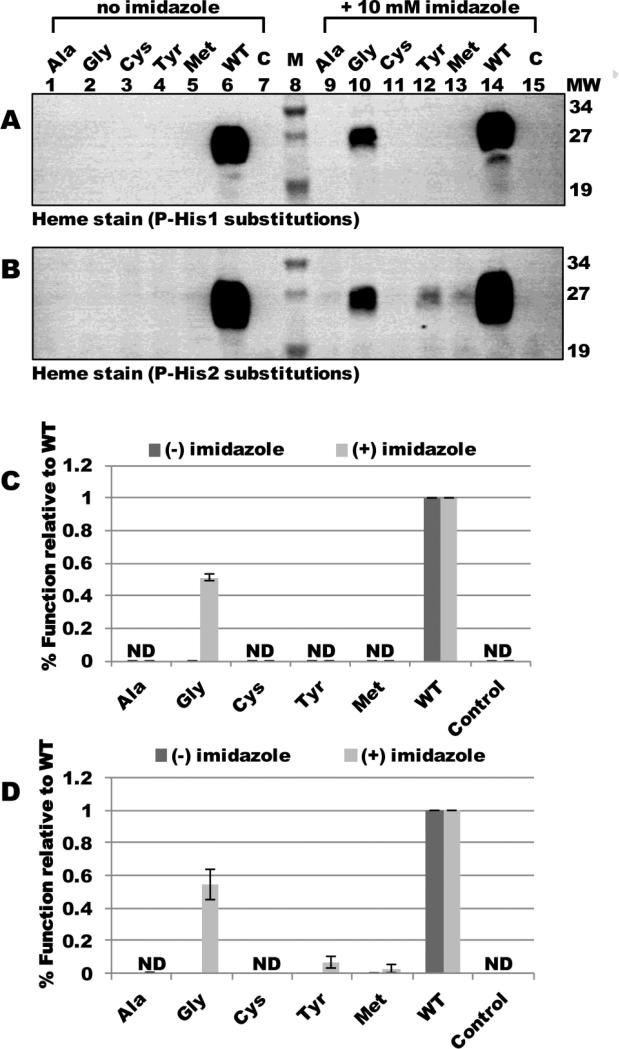
Previously, we showed that the cytochrome c assembly defects of glycine substitutions at TM-His1 and TM-His2 could be corrected in vivo by addition of 10 mM imidazole directly to culture 21. Conceptually, this is similar to the correction of heme binding in the myoglobin His93Gly “cavity” mutant by imidazole 35. Thus, functional correction of a histidine mutant by imidazole can be suggestive of a heme ligand function. To test whether P-His1 or P-His2 might exhibit a ligand function, we engineered alanine, glycine, cysteine, tyrosine, or methionine substitutions at each His residue and assayed for holocytochrome c4 formation in the absence and presence of imidazole. Heme staining of BPER fractions revealed that in the absence of imidazole, none of the engineered substitutions at P-His1 or P-His2 supported holocytochrome c formation (Fig 7A and B, lanes 1-7; quantified in Fig 7C and D). However, 10 mM imidazole corrected the holocytochrome c assembly defects of glycine substitutions at P-His1 and P-His2 to approximately 50% levels of WT (Fig 7A and B, lane 10; quantified in Fig 7C and D). Substitution of P-His1 with bulkier amino acids did not result in detectable holocytochrome c4 formation in the presence of imidazole (Fig 7A, lanes 9-15; quantified in Fig 7C). The tyrosine and methionine substitutions at P-His2 were corrected to approximately 10% and 5% levels of WT, respectively, suggesting that the P-His2 position may be more flexible than the P-His1 (i.e., for imidazole). We conclude that the glycine substitutions at P-His1 and P-His2 result in the formation of a cavity in which imidazole can bind and serve as a ligand to support holocytochrome c formation. However, bulkier amino acids (including perhaps the methyl side group of alanine) may present a steric hindrance to imidazole correction. Since we have previously shown that neither P-His1 nor P-His2 were ligands to the CcmF b-heme, we theorize that the apparent ligand function of these residues is related to the incoming heme from holoCcmE.
Fig 7.
In vivo heme attachment to cytochrome c4 in the presence or absence of imidazole. Heme staining of B-PER cell extracts showing 24-kDa holocytochrome c4 matured by full system I with the indicated substitutions at CcmF P-His1 (A) or P-His2 (B) in the presence or absence of 10 mM imidazole added to culture. P-His1 and P-His2 were each changed to the indicated residues. M, molecular weight standards; C, vector control. 100 ug total protein was analyzed. Quantification of chemiluminescent signal from heme staining of B-PER isolated proteins from three independent experiments for P-His1 substitutions (C) or P-His2 substitutions (D). Holocytochrome c4 signal is relative to WT, which has been set at 100%. Error bars denote SD. ND, no signal detected.
CcmF P-His1 and P-His2 are required for interaction with holoCcmE
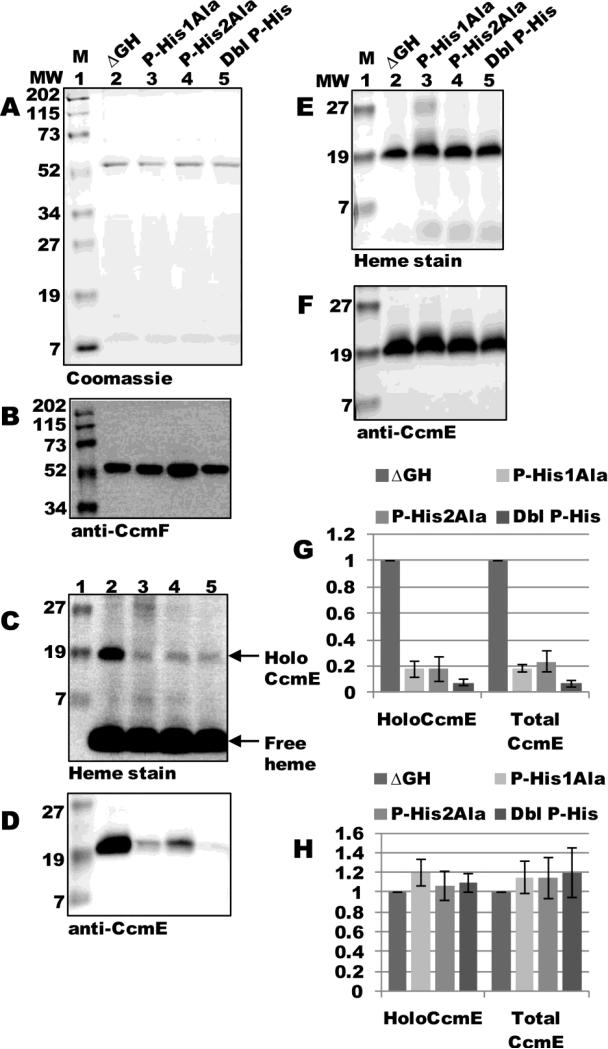
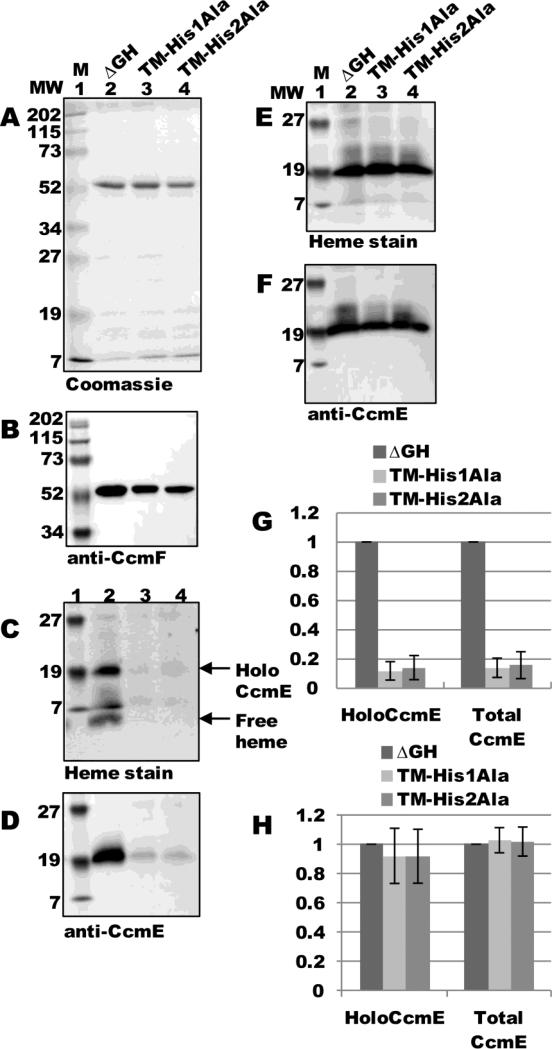
To directly test if P-His1 and P-His2 are required for interaction with holoCcmE, we engineered alanine substitutions at each residue, as well as a double alanine substitution, in the pSysI ΔGH background and assayed for holoCcmE in preparations of CcmF. ccmF was expressed from pSysI ΔGH(P-His1Ala), pSysI ΔGH(P-His2Ala), or pSysI ΔGH(P-His1Ala/P-His2Ala) and purified as a single full-length polypeptide (54-kDa; Fig 8A, lanes 3-5) that reacted with CcmF antisera (Fig 8B, lanes 3-5). Heme staining and immunoblotting with CcmE antisera showed approximately a 5-fold decrease in the amount of holoCcmE that co-purified with CcmF for each of the single substitutions (Fig 8C and D, compare lane 2 to 3 and 4; quantified in Fig 8G), and approximately a 10-fold decrease for the double mutant (Fig 8C and D, compare lanes 2 and 5; quantified in Fig 8G). DDM-solubilized membranes from all backgrounds contained similar levels of holoCcmE (Fig 8E and F; quantified in Fig 8H). Thus, P-His1 and P-His2 in CcmF are essential for interaction with holoCcmE, and are likely the ligands to the heme from holoCcmE.
Fig 8.
CcmF P-His1 and P-His2 are required for co-purification of holoCcmE. (A) Coomassie blue staining of TALON-purified proteins from ΔGH, ΔGH (P-His1Ala), ΔGH (P-His2Ala), and ΔGH (P-His1Ala/P-His2Ala) backgrounds showing 54-kDa CcmF. (B) Anti-CcmF immunoblot of purified CcmF proteins showing 54-kDa CcmF. (C) Heme staining of purified CcmF proteins showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (D) Anti-CcmE immunoblot of purified CcmF proteins showing co-purified 20-kDa CcmE. For (A)-(D), 6 ug purified protein was analyzed. (E) Heme staining of DDM-solubilized membrane fractions from ΔGH and each ΔGH mutant background showing 20 kDa holoCcmE. (F) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20 kDa CcmE. For (E) and (F), 70 ug total protein was analyzed. (G) Quantification of the results of heme staining (holoCcmE) and anti-CcmE immunoreactivity (total CcmE) from purified fractions from three independent experiments. (H) Quantification of the results of heme staining and anti-CcmE immunoreactivity from DDM-solubilized membrane fractions from three independent experiments. For (G) and (H), percent holoCcmE and total CcmE is relative to ΔGH, which has been set at 100%. Error bars denote SD.
The CcmF b-heme is required for interaction with holoCcmE
CcmF contains two conserved histidines in transmembrane domains (TM-His1 and TM-His2) that are ligands to the CcmF b-heme. Substitution of either histidine residue with alanine abolishes b-heme binding in CcmF to undetectable levels 21. To test if the CcmF b-heme is required for interaction with holoCcmE, we engineered alanine substitutions at TM-His1 and TM-His2 in the pSysI ΔGH background and assayed for holoCcmE in preparations of CcmF. ccmF was expressed from pSysI ΔGH(TM-His1Ala) or pSysI ΔGH(TM-His2Ala) and purified as a full-length polypeptide (54-kDa; Fig 9A, lanes 3-4) that reacted with CcmF antisera (Fig 9B, lanes 3-4). Heme staining and immunoblotting with CcmE antisera showed approximately an 8-fold decrease in the amount of holoCcmE that co-purified with CcmF for each substitution (Fig 9C and D, compare lane 2 to 3 and 4; quantified in Fig 9G). DDM-solubilized membranes from all backgrounds contained similar levels of holoCcmE (Fig 9E and F; quantified in Fig 9H). Note that purified CcmF (TM-His1Ala) and CcmF (TM-His2Ala) do not contain the b-heme (see “free heme” in Fig 9C, compare lane 2 to 3 and 4). This is in stark contrast to the P-His1 and P-His2 substitutions, which showed b-heme levels identical to WT CcmF (see “free heme” in Fig 8C). Thus, the b-heme in CcmF (with ligands from TM-His1 and TM-His2) is essential for interaction with holoCcmE.
Fig 9.
CcmF b-heme is required for co-purification of holoCcmE. (A) Coomassie blue staining of TALON-purified proteins from ΔGH, ΔGH (TM-His1Ala), and ΔGH (TM-His2Ala) backgrounds showing 54-kDa CcmF. (B) Anti-CcmF immunoblot of purified CcmF proteins showing 54-kDa CcmF. (C) Heme staining of purified CcmF proteins showing free heme (CcmF b-heme) and co-purified 20-kDa holoCcmE. (D) Anti-CcmE immunoblot of purified CcmF proteins showing co-purified 20-kDa CcmE. For (A)-(D), 5 ug purified protein was analyzed. (E) Heme staining of DDM-solubilized membrane fractions from ΔGH and each ΔGH mutant background showing 20 kDa holoCcmE. (F) Anti-CcmE immunoblot of DDM-solubilized membrane fractions showing 20 kDa CcmE. For (E) and (F), 70 ug total protein was analyzed. (G) Quantification of the results of heme staining (holoCcmE) and anti-CcmE immunoreactivity (total CcmE) from purified fractions from three independent experiments. (H) Quantification of the results of heme staining and anti-CcmE immunoreactivity from DDM-solubilized membrane fractions from three independent experiments. For (G) and (H), percent holoCcmE and total CcmE is relative to ΔGH, which has been set at 100%. Error bars denote SD.
DISCUSSION
Requirements for formation of the holoCcmE—CcmF complex
Here, we report isolation of an integral membrane protein complex between the system I heme chaperone, holoCcmE, and the putative cytochrome c synthetase, CcmF. Interaction between these two essential CCM proteins has long been suspected, but never shown directly. Ren and colleagues (2002) previously reported that CcmE could be immunoprecipitated from cell extracts using CcmF antisera (see Fig 4 of 18). However, since only the apo-form of CcmE was analyzed in that study (the strain utilized lacked the genes for ccmABCD altogether), the relevance of that finding to holocytochrome c formation is unclear. Despite the fact that only apoCcmE was analyzed, many CCM models have cited the findings of Ren and colleagues as evidence of interaction between holoCcmE and CcmF. In our study, using a strain expressing CcmABCDE and CcmF, we showed that covalent, released holoCcmE interacts with CcmF at levels at least 20-fold higher than the apo-form of CcmE (see Fig 5). Given that the predicted function of CcmF is to facilitate heme transfer from holoCcmE to the apocytochrome, it is not surprising that the holo-form of CcmE preferentially binds. Ren and colleagues also reported that point mutants in CcmF (including alanine substitutions at P-His1 and P-His2) showed unaltered interactions with (apo) CcmE, relative to WT CcmF. In contrast, we found that substitution of either P-His1 or P-His2 caused a 5-fold decrease in the levels of holoCcmE that co-purified with CcmF (relative to WT CcmF), and that the double mutant showed a 10-fold decrease (see Fig 8). The critical roles of P-His1 and P-His2 (which are in periplasmic loops adjacent to the CcmF WWD domain; see Fig 6) in binding holoCcmE had been proposed previously, and is further elaborated upon below. We suggest that the apoCcmE detected in preparations of CcmF represents a low background level (likely non-physiological), and that the holoCcmE “trapped” in complex with CcmF is the true physiological intermediate in system I.
Implications of P-His1 and P-His2 binding heme from holoCcmE
CcmF is a member of the heme-handling superfamily of proteins 36, which also includes CcmC and the system II cytochrome c synthetase, CcsBA. The hallmark of the heme-handling proteins is the “WWD domain” 16; 37; 38; 39, a conserved tryptophan-rich periplasmic loop that is flanked by two conserved histidine residues in adjacent periplasmic loops. CcmC, which is sometimes referred to as the holoCcmE synthase (due to its well-described role in formation of holoCcmE) 14; 33 forms a stable intermediate complex with holoCcmE (in the absence of CcmAB) 11. In the “trapped” holoCcmCDE complex, heme from holoCcmE is bound in the WWD domain of CcmC, and the two flanking histidines (CcmC His60 and His184) supply the 5th and 6th axial ligands to heme. Richard-Fogal and Kranz (2010) showed that substitution of either histidine, as well as certain tryptophans in the WWD domain of CcmC, caused perturbations in the absorption spectrum of the CcmCDE complex 11. In the absence of heme, there was no detectable interaction between (apo) CcmE and CcmC, indicating that the WWD domain of CcmC is a platform for interaction with heme rather than for the CcmE polypeptide. Thus, we proposed that heme, CcmC, and CcmE are each essential to form the stable “ternary” complex 11.
By analogy to the CcmCDE complex, we have hypothesized that the WWD domain of CcmF is the site of interaction for heme from holoCcmE, after it is released from CcmCD. Several of our findings here are in agreement with this hypothesis: i) CcmF P-His1 and P-His2 (which flank the WWD domain) exhibited ligand-type activity (Gly substitutions at these residues were functionally restored by adding exogeneous imidazole), ii) P-His1 and P-His2 were required for interaction with holoCcmE, iii) only the holo-form of CcmE (i.e., with heme), and not apoCcmE, interacted with CcmF, and iv) only holoCcmE released from CcmCD interacted with CcmF. The requirement for both heme (i.e., “holo” CcmE) and the WWD domain-flanking histidines for formation of the holoCcmE—CcmF complex is remarkably similar to formation of the holoCcmCDE complex.
Identifying the CcmF WWD domain as the binding site for holoCcmE also has implications for understanding the cytochrome c heme lyase function of CcmF. Experimentally established models for the membrane topology of CcmF 12 suggest that the b-heme (with ligands from TM-His1 and TM-His2) may be positioned spatially below the WWD domain (see Fig 6). Given this possible transmembrane “architecture,” a mechanism for reduction of the incoming heme from holoCcmE (in the WWD domain) by electron transfer directly from the b-heme can be readily envisioned. Further studies will be needed to test this. In particular, since the holoCcmE that co-purifies with CcmF is sub-stoichiometric, it will be necessary to further enrich for holoCcmE in the complex to begin these and other spectroscopic studies.
Absence of CcmGH is critical to formation of the holoCcmE—CcmF complex
Surprisingly, we found that levels of the “trapped” holoCcmE—CcmF complex increased 10-fold when CcmGH were absent (see Fig 3). In fact, purification of CcmF from this particular background (pSysI ΔGH) initially enabled us to detect the holoCcmE—CcmF complex. CcmG is a periplasmic thioredoxin that has been shown to reduce the cysteines of CcmH (in vitro) (e.g., 29), but it does not co-purify with the CcmFH complex 12; 20. By contrast, interaction between CcmF and CcmH is well established 12; 18; 19; 20. CcmH is a polytopic membrane protein with two transmembrane α helices and a large C-terminal periplasmic domain (see Fig 6). Apart from the two conserved cysteine residues in the N-terminal periplasmic domain, which have been shown to reduce the apocytochrome thiols (in the Cys-Xxx-Xxx-Cys-His motif) 25; 29; 40, discrete functional domains within CcmH have not been well defined. Recently, Verissimo and colleagues (2011) and Di Silvio and colleagues (2013) have shown that CcmI, which is analogous to the C-terminus of E. coli CcmH, interacts with the C-terminus of apocytochrome c (not at Cys-Xxx-Xxx-Cys-His), but not holocytochrome c 30; 31. Given our results here, it is likely that CcmH may also modulate the interaction between CcmF and holoCcmE. For example, in the absence of apocytochrome, CcmH may occlude the CcmF WWD domain from interaction with holoCcmE, thus only “permitting” holoCcmE binding and reduction (to Fe2+) when the apocytochrome c “acceptor” is present. Alternatively, but not mutually exclusively, CcmH may facilitate the rapid release of CcmE from the CcmF WWD domain. A recent study by Verissimo and colleagues (2013) used an in vitro approach to study interactions between apoCcmE, apocytochrome c2, and CcmI, and showed that apoCcmE interacts with CcmI (and CcmH) 41. They suggest a possible “supercomplex” involving all Ccm proteins, with CcmI at the center. Our approach differs in that we rely exclusively on interactions that form in vivo (in E. coli) in the appropriate cellular milieu. While our data is not consistent with formation of a supercomplex, as they have described, we cannot rule out that interactions between CcmI (CcmH) and apoCcmE may be physiologically important.
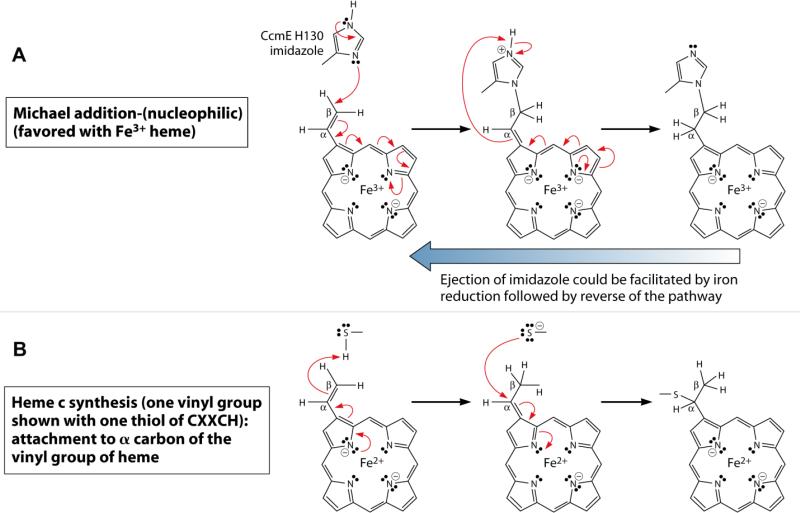
We have proposed that reduction of heme in holoCcmE accomplishes two purposes (see Fig 10). First, reduction of heme (to Fe2+) would favor ejection of the His130 imidazole from the β carbon of the heme 2-vinyl (see Fig 10A, reverse blue arrow). Second, reduced heme is required for the 2- and 4-vinyls of heme to form thioether linkages with the apocytochrome c thiols (at Cys-Xxx-Xxx-Cys-His; see Fig 10B). It makes ‘sense’ to only allow the first reaction to occur (ejection of CcmE His130) when the acceptor is properly positioned (by CcmH) for the second reaction. Thus, the synthetase reactions are elegantly orchestrated by the holoCcmE:CcmF:CcmH complex.
Fig 10.
Proposed reaction mechanisms for holoCcmE (His130) (A) and cytochrome c (B) linkage to heme vinyl groups. Noted are the oxidation states of iron (Fe3+ or Fe2+); red half arrows are one-electron transfers, and full red arrows are two-electron transfers. Transfer of the proton from the imidazolium to the alpha carbon in (A) is probably solvent- or protein-mediated (i.e., the proton may be abstracted at an early step, with a solvent- or protein-mediated protonation of the alpha carbon occurring at a later step). Reduction of heme (from Fe3+ to Fe2+) could favor ejection of the CcmE His130 imidazole adduct (reverse blue arrow) and is required for holocytochrome c formation. For simplicity, only a single vinyl of heme is shown. Adapted from (Kranz, Richard-Fogal et al. 2009).
MATERIALS AND METHODS
Bacterial Growth Conditions
Escherichia coli strains (Supplemental Table 1) were grown at 37°C by shaking at 230 rpm in Luria-Bertani broth (LB; Difco) supplemented with the appropriate antibiotics (Sigma-Aldrich) and other media additives at the following concentrations, unless otherwise noted: carbenicillin, 50 ug ml−1; chloramphenicol, 20 ug ml−1; gentamicin, 10 ug ml−1; IPTG (Gold Biotechnology), 1 mM; arabinose (Gold Biotechnology), 0.2 % (wt/vol).
Protein Expression and Purification
E. coli Δccm strain RK103 (Table S1) 17 was used for expression. Starter cultures were initiated from a single colony and grown overnight in 10 mL of LB with the appropriate antibiotics. 1 L of LB was inoculated with the 10 mL starter culture and grown to an OD600 of 1.8, then induced with 1 mM IPTG for 14-16 hr. Cells were harvested at 5,000 × g and frozen at −80°C. Cell pellets were thawed and resuspended in PBS (100 mM NaCl, 7.5 mM Na2HPO4, 2 mM NaH2PO4) and treated with 1 mM PMSF (Sigma-Aldrich) and 100 μg ml−1 egg white lysozyme (Sigma-Aldrich) for 30 min while shaking on ice. Cells were disrupted by repeated sonication for 30 sec bursts on a Branson 250 sonicator (50% duty, 8 output) until clearing of the suspension was observed. Crude sonicate was centrifuged at 24,000 × g for 20 min to clear cell debris, and membranes were isolated by centrifugation at 100,000 × g for 45 min. Membrane pellets were solubilized in a modified 1x TALON (Clontech) buffer (50 mM Tris-HCL, pH 7; 300 mM NaCl) with 1 % (wt/vol) dodecyl maltoside (DDM, Anatrace) on ice for 1 hr. DDM-solubilized membranes were centrifuged at 24,000 × g for 20 min to remove unsolubilized material. Solubilized membranes (L; load) were passed over TALON resin per the manufacturer's recommendations and washed in 1 × modified TALON buffer with 0.02 % DDM with increasing concentrations of imidazole (wash 1 (W1), 0 mM imidazole; wash 2 (W2), 2 mM imidazole; wash 3 (W3), 5 mM imidazole). Bound hexahistidine-tagged protein was eluted in 1 × modified TALON buffer containing 0.02 % dodecyl maltoside and 150 mM imidazole (E; elution). Total protein concentration was determined using the Nanodrop 1000 spectrophotometer (Thermo Scientific).
Cytochrome Reporter and Imidazole Complementation Assays
Cytochrome c4:His6 production was assayed in RK111 (Δccm carrying the arabinose-inducible chromosomal integrate of the cyt c4:His6 gene 21) harboring pRGK402 (pGEX ccmABCDE 21) and one of the following pBAD ccmF:His6GH plasmids (pRGK434, 435, 436, 437, 438, 439, 440, 441, 442, or 443; see Supplemental Table 1). Starter cultures were initiated from a single colony and grown overnight in LB with the appropriate antibiotics. 5 mL of LB was inoculated using 800 mL of starter culture and grown for 3 hr, then induced for 3 hr with 0.8 % (wt/vol) arabinose. Cells were harvested by centrifugation at 10,000 × g and the cell pellet was resuspended in 200 μL of BPER (Thermo Scientific) to lyse cells and extract protein. Total protein concentration was determined using the Nanodrop 1000 spectrophotometer (Thermo Scientific) and 100 μg was analyzed by SDS-PAGE followed by heme stain. Imidazole complementation assays were performed in the same way, with 10 mM imidazole (pH 7) added to the media prior to inoculation.
Production of antibodies to CcmF
E. coli CcmF was engineered with a hexahistidine tag as described in 12, and was expressed from pRGK386 in E. coli strain RK103 17. Cell growth, protein expression, and purification were carried out as described above. Eluted hexahistidine-tagged CcmF was analyzed by 12.5 % SDS-PAGE and electroeluted from gel fragments over 4-5 hr at 25 mV into 1x SDS-PAGE buffer (3.5 mM SDS, 50 mM Tris, 384 mM glycine). Purity of the preparations (assessed by Coomassie Blue staining) was greater than 95 %. Antiserum was generated in rabbits at a commercial facility (Cocalico Biologicals). Antibodies were purified from serum by ammonium sulfate precipitation and adsorbed against crude E. coli extract containing all other Ccm proteins.
Heme stains and other methods
Heme stains and immunoblots were performed as described previously 17; 42. Proteins were separated by 12.5 % SDS-PAGE and transferred to Hybond C nitrocellulose membranes (GE Healthcare). Anti-CcmF antibodies were used at a dilution of 1:10000, anti-CcmE antibodies at 1:10000, anti-GST antibodies at 1:10000, and anti-CcmH antibodies at 1:5000. Protein A peroxidase (Sigma-Aldrich) was used as the secondary label. The chemiluminescent signal for heme stains was developed using the SuperSignal Femto kit (Thermo Scientific) or, for immunoblots, the Immobilon Western kit (Millipore), and detected with an LAS-1000 Plus detection system (Fujifilm-GE Healthcare). The abundance of holoCcmE was determined by densitometry analysis of the chemiluminescent signal from heme stains and anti-CcmE immunoblots using the Science Lab 99-Image Gauge version 3.4 software (Fujifilm-GE Healthcare). Heme concentration in protein preparations was determined by pyridine extraction as described in 32 or heme staining as described in 43. Protein purity was assessed by Coomassie Blue staining of SDS-PAGE.
UV/Vis absorption spectroscopy
UV-visible absorption spectra were recorded with a Shimadzu UV-2101 PC UV-Vis scanning spectrophotometer at room temperature as described previously 44. All spectra were recorded in the same buffer (modified 1x TALON buffer) in which the proteins were purified. Chemically reduced spectra were generated by addition of sodium dithionite (sodium hydrosulfite).
Construction of plasmids
All oligonucleotide primer sequences, plasmids, and strains are given in Table S1. All oligonucleotides were synthesized by Sigma-Aldrich. To delete ccmAB from pRGK385 (pGEX ΔGH), primers “delABloop_BglII_Fwd” and “delABloop_BglII_Rev” were used to PCR amplify around pRGK385, excluding ccmAB. The resulting PCR product was gel purified, digested and re-circularized by ligation at the resulting BglII sites to yield pRGK427 (pGEX ΔGH delAB). All nucleotide substitutions were generated using the QuikChange I Site-Directed Mutagenesis Kit (Agilent Technologies) per the manufacturer's recommendations. Substitutions were engineered into pBAD-based pRGK388 for cytochrome reporter assays, or into pGEX-based pRGK385 (pGEX ΔGH) for protein expression and purification. To engineer the double P-His1Ala/P-His2Ala mutation, primers “ccmF_H303A_Fwd” and “ccmF_H303A_Rev” were used to introduce the P-His2Ala substitution into pRGK429 (pGEX ΔGH P-His1Ala). Each of the final constructs was sequenced to verify the mutation(s).
Supplementary Material
HIGHLIGHTS.
CcmF forms a complex with holoCcmE
HoloCcmE must be released from CcmABCD to interact with CcmF
Heme binding by holoCcmE is essential for complex formation with CcmF
CcmF P-His1 and P-His2 are required for binding holoCcmE
CcmH controls formation of the CcmF—holoCcmE complex
ACKNOWLEDGEMENTS
We thank Joseph P. Argus and Eric C. Bretsnyder for generation of antibodies to CcmF, and Joel Rankin for technical contributions. This work was supported by National Institutes of Health Grant R01 GM47909 to R.G.K.
ABBREVIATIONS
- TM
transmembrane
- P
p-side (of cytoplasmic membrane)
- ABC
ATP-binding cassette
- GST
glutathione S-transferase
- DDM
dodecyl maltoside
- CCM
cytochrome c maturation
- PMSF
phenylmethylsulfonylfluoride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Barker PD, Ferrer JC, Mylrajan M, Loehr TM, Feng R, Konishi Y, Funk WD, MacGillivray RT, Mauk AG. Transmutation of a heme protein. Proc Natl Acad Sci U S A. 1993;90:6542–6. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson DW, Neupert W. Import of cytochrome c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc Natl Acad Sci U S A. 1989;86:4340–4. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamel P, Corvest V, Giege P, Bonnard G. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim Biophys Acta. 2009;1793:125–38. doi: 10.1016/j.bbamcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73:510–28. doi: 10.1128/MMBR.00001-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mavridou DA, Ferguson SJ, Stevens JM. Cytochrome c assembly. IUBMB Life. 2013;65:209–16. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- 6.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18:266–74. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawyer EB, Barker PD. Continued surprises in the cytochrome c biogenesis story. Protein Cell. 2012;3:405–9. doi: 10.1007/s13238-012-2912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee D, Pervushin K, Bischof D, Braun M, Thony-Meyer L. Unusual heme-histidine bond in the active site of a chaperone. J Am Chem Soc. 2005;127:3716–7. doi: 10.1021/ja044658e. [DOI] [PubMed] [Google Scholar]
- 9.Stevens JM, Daltrop O, Higham CW, Ferguson SJ. Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J Biol Chem. 2003;278:20500–6. doi: 10.1074/jbc.M212925200. [DOI] [PubMed] [Google Scholar]
- 10.Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J Biol Chem. 2004;279:51981–8. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 11.Richard-Fogal C, Kranz RG. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol. 2010;401:350–62. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richard-Fogal CL, Frawley ER, Bonner ER, Zhu H, San Francisco B, Kranz RG. A conserved haem redox and trafficking pathway for cofactor attachment. Embo J. 2009;28:2349–59. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen O, Harvat EM, Thony-Meyer L, Ferguson SJ, Stevens JM. Loss of ATP hydrolysis activity by CcmAB results in loss of c-type cytochrome synthesis and incomplete processing of CcmE. Febs J. 2007;274:2322–32. doi: 10.1111/j.1742-4658.2007.05769.x. [DOI] [PubMed] [Google Scholar]
- 14.Feissner RE, Richard-Fogal CL, Frawley ER, Kranz RG. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol Microbiol. 2006;61:219–31. doi: 10.1111/j.1365-2958.2006.05221.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldman BS, Beckman DL, Bali A, Monika EM, Gabbert KK, Kranz RG. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J Mol Biol. 1997;268:724–38. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- 16.Goldman BS, Beck DL, Monika EM, Kranz RG. Transmembrane heme delivery systems. Proc Natl Acad Sci U S A. 1998;95:5003–8. doi: 10.1073/pnas.95.9.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feissner RE, Richard-Fogal CL, Frawley ER, Loughman JA, Earley KW, Kranz RG. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol Microbiol. 2006;60:563–77. doi: 10.1111/j.1365-2958.2006.05132.x. [DOI] [PubMed] [Google Scholar]
- 18.Ren Q, Ahuja U, Thony-Meyer L. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J Biol Chem. 2002;277:7657–63. doi: 10.1074/jbc.M110979200. [DOI] [PubMed] [Google Scholar]
- 19.Rayapuram N, Hagenmuller J, Grienenberger JM, Bonnard G, Giege P. The three mitochondrial encoded CcmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J Biol Chem. 2008;283:25200–8. doi: 10.1074/jbc.M802621200. [DOI] [PubMed] [Google Scholar]
- 20.Sanders C, Turkarslan S, Lee DW, Onder O, Kranz RG, Daldal F. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J Biol Chem. 2008;283:29715–22. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.San Francisco B, Bretsnyder EC, Rodgers KR, Kranz RG. Heme ligand identification and redox properties of the cytochrome c synthetase, CcmF. Biochemistry. 2011;50:10974–85. doi: 10.1021/bi201508t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Matteo A, Calosci N, Gianni S, Jemth P, Brunori M, Travaglini-Allocatelli C. Structural and functional characterization of CcmG from Pseudomonas aeruginosa, a key component of the bacterial cytochrome c maturation apparatus. Proteins. 2010;78:2213–21. doi: 10.1002/prot.22733. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang N, Gao YG, Hu HY, Xia ZX. Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region. Proteins. 2006;65:1021–31. doi: 10.1002/prot.21184. [DOI] [PubMed] [Google Scholar]
- 24.Beckman DL, Kranz RG. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993;90:2179–83. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Matteo A, Gianni S, Schinina ME, Giorgi A, Altieri F, Calosci N, Brunori M, Travaglini-Allocatelli C. A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c. J Biol Chem. 2007;282:27012–9. doi: 10.1074/jbc.M702702200. [DOI] [PubMed] [Google Scholar]
- 26.Zheng XM, Hong J, Li HY, Lin DH, Hu HY. Biochemical properties and catalytic domain structure of the CcmH protein from Escherichia coli. Biochim Biophys Acta. 2012;1824:1394–400. doi: 10.1016/j.bbapap.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Meyer EH, Giege P, Gelhaye E, Rayapuram N, Ahuja U, Thony-Meyer L, Grienenberger JM, Bonnard G. AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc Natl Acad Sci U S A. 2005;102:16113–8. doi: 10.1073/pnas.0503473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turkarslan S, Sanders C, Ekici S, Daldal F. Compensatory thioredox interactions between DsbA, CcdA and CcmG unveil the apocytochrome c holdase role of CcmG during cytochrome c maturation. Mol Microbiol. 2008;70:652–66. doi: 10.1111/j.1365-2958.2008.06441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monika EM, Goldman BS, Beckman DL, Kranz RG. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J Mol Biol. 1997;271:679–92. doi: 10.1006/jmbi.1997.1227. [DOI] [PubMed] [Google Scholar]
- 30.Verissimo AF, Yang H, Wu X, Sanders C, Daldal F. CcmI subunit of CcmFHI heme ligation complex functions as an apocytochrome c chaperone during c-type cytochrome maturation. J Biol Chem. 2011;286:40452–63. doi: 10.1074/jbc.M111.277764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Silvio E, Di Matteo A, Malatesta F, Travaglini-Allocatelli C. Recognition and binding of apocytochrome c to P. aeruginosa CcmI, a component of cytochrome c maturation machinery. Biochim Biophys Acta. 2013;1834:1554–61. doi: 10.1016/j.bbapap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 32.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 33.Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thony-Meyer L. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc Natl Acad Sci U S A. 1999;96:6462–7. doi: 10.1073/pnas.96.11.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvat EM, Redfield C, Stevens JM, Ferguson SJ. Probing the heme-binding site of the cytochrome c maturation protein CcmE. Biochemistry. 2009;48:1820–8. doi: 10.1021/bi801609a. [DOI] [PubMed] [Google Scholar]
- 35.Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93-->Gly. Biochemistry. 1994;33:6546–54. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH., Jr. Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Biochim Biophys Acta. 2007;1768:2164–81. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 37.Beckman DL, Trawick DR, Kranz RG. Bacterial cytochromes c biogenesis. Genes Dev. 1992;6:268–83. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- 38.Hamel PP, Dreyfuss BW, Xie Z, Gabilly ST, Merchant S. Essential histidine and tryptophan residues in CcsA, a system II polytopic cytochrome c biogenesis protein. J Biol Chem. 2003;278:2593–603. doi: 10.1074/jbc.M208651200. [DOI] [PubMed] [Google Scholar]
- 39.Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–96. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 40.Setterdahl AT, Goldman BS, Hirasawa M, Jacquot P, Smith AJ, Kranz RG, Knaff DB. Oxidation-reduction properties of disulfide-containing proteins of the Rhodobacter capsulatus cytochrome c biogenesis system. Biochemistry. 2000;39:10172–6. doi: 10.1021/bi000663t. [DOI] [PubMed] [Google Scholar]
- 41.Verissimo AF, Mohtar MA, Daldal F. The heme chaperone ApoCcmE forms a ternary complex with CcmI and apocytochrome c. J Biol Chem. 2013;288:6272–83. doi: 10.1074/jbc.M112.440024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315:90–4. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 43.Richard-Fogal CL, Frawley ER, Feissner RE, Kranz RG. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J Bacteriol. 2007;189:455–63. doi: 10.1128/JB.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–6. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.