Abstract
The many biological and biomedical effects of selenium are relatively unknown outside the selenium field. This fascinating element, initially described as a toxin, was subsequently shown to be essential for health and development. By the mid 1990s, selenium emerged as one of the most promising cancer chemopreventive agents, but subsequent human clinical trials yielded contradictory results. However, basic research on selenium continued to move at a rapid pace elucidating its many roles in health, development, and cancer prevention and promotion. Dietary selenium acts principally through selenoproteins, most of which are oxidoreductases involved in diverse cellular functions.
Keywords: Cancer, selenium, selenocysteine, selenoproteins
Selenium in animal nutrition and human health
Selenium has been linked to many health benefits in humans and other mammals such as decreasing the incidence of cancer, protecting against cardiovascular diseases, treating certain muscle disorders, and delaying the onset of AIDS in HIV-positive patients [1]. It also has roles in mammalian development and boosting immune function. Although small molecular weight selenocompounds have been implicated as beneficial agents in several cases, this is mostly due to high levels of selenium used in chemotherapy, and most attention in recent years has been given to selenoproteins being the primary responsible agents [1].
Selenium was initially considered a toxin, because it was responsible for a disorder in livestock that grazed on the plains of the Nebraska and Dakota territories. This disorder was described in 1856 by army surgeon T.C. Madison, stationed at Fort Randall in northern Nebraska [2]. The army horses that grazed freely around the fort suffered from a necrotic hoof malady and excessive losses of long hair in the tail and mane. Franke reported in the mid 1930s that the disease resulted from livestock eating seleniferous plants, which accumulated high levels of selenium from the soil [3].
Selenium continued to be regarded as a toxin, and even a carcinogen, until 1957 when Schwartz and Foltz found that it prevented liver necrosis in rats [4]. Thus, it became apparent that selenium was toxic at high levels but was an essential dietary micronutrient at low levels. The beneficial side of selenium was rapidly recognized in the livestock industry: the deficiency of this element was implicated in a number of disorders including white muscle disease, a myopathy affecting calves and lambs; pancreatic degeneration and exudative diathesis in birds and other livestock; hepatosis dietetica in swine; and ill thrift and reduced male fertility in sheep and cattle [5]. Supplementing the diets of livestock around the world with selenium was estimated to save this industry in the hundreds of millions of dollars. In the human population, low selenium status has been associated with Keshan disease, a cardiomyopathy found in rural areas of China, and Kashin-Beck disease, a chronic, endemic osteochondropathy found primarily in northeastern to southwestern China ([6] and references therein).
Of all the health benefits attributed to selenium, the one that has received the most attention is its role as a cancer preventive agent. In fact, hundreds of millions of dollars have been spent on human clinical trials examining the role of selenium in cancer prevention (e.g., see [7–12]). While at least one trial has shown a decreased incidence in prostate, colon and lung cancers [7], others found no positive health benefits (e.g., see [8,9]), and the largest cancer prevention trial ever undertaken was terminated early [8]. Questions arose whether these clinical trials were being carried out with insufficient understanding of how selenium functions at the molecular level [13,14]. Little consideration was also given to the selenium status of the participants or the potential negative consequences of administering selenium to such large numbers of individuals. Another concern was the recent evidence that certain selenoproteins manifest a dual personality in that they not only prevent, but can promote cancer (discussed later and reviewed in [15,16]).
Occurrence of selenocysteine (Sec) in protein and selenoprotein functions
Sec, the 21st amino acid in the genetic code
Sec is a selenium-containing amino acid that occurs in proteins in organisms representing the three domains of life (Eukarya, Archaea and Bacteria) as well as in viruses. Selenium was originally detected as a covalently-bound component in mammalian glutathione peroxidase [17] and the selenium-containing amino acid was subsequently identified as Sec in bacterial selenoprotein A [18]. Sequencing selenoprotein genes revealed that the UGA codon corresponded to the location of Sec in proteins [19,20], suggesting that Sec was the 21st proteinogenic amino acid.
Selenoproteins and selenoproteomes
To date, approximately 100 selenoprotein families have been discovered. There is a great diversity in the use of selenoproteins by organisms. The largest set of selenoproteins (selenoproteome) has been observed in a unicellular brown alga that has 59 selenoprotein genes [21]. With regard to common model organisms, zebrafish has 37, mouse has 24, Drosophila melanogaster and Escherichia coli have three, and Caenorhabditis elegans has one, whereas Saccharomyces cerevisiae and Arabidopsis do not encode selenoprotein genes.
Functions of mammalian selenoproteins
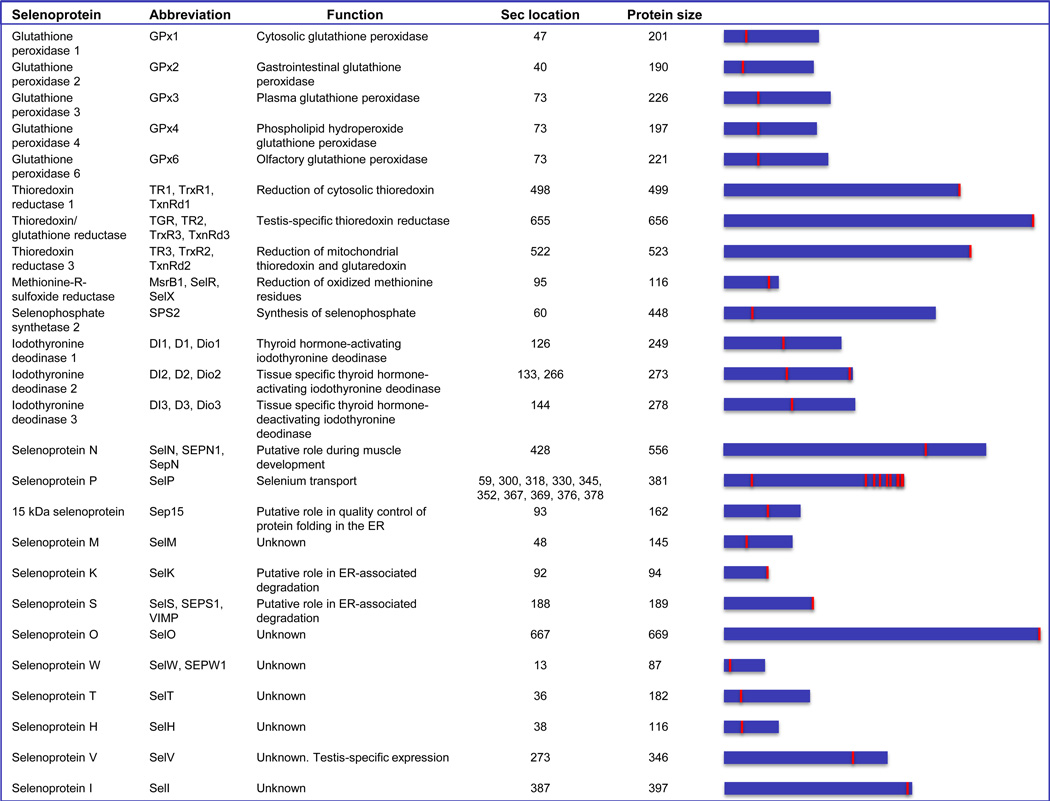
The human selenoproteome is encoded by 25 genes. Approximately half of these genes code for proteins with known functions [22] (Fig. 1). Humans have five selenoprotein glutathione peroxidases, which catalyze glutathione-dependent reduction of hydrogen peroxide or other peroxides; three thioredoxin reductases, which catalyze the reduction of thioredoxin or other proteins at the expense of NADPH; and three thyroid hormone deiodinases, which catalyze reductive deiodination of thyroid hormones, thereby activating or inactivating them [23]. There is also methionine-R-sulfoxide reductase 1 (MsrB1), which reduces oxidized methionine residues in proteins (repairing oxidatively damaged proteins and serving as a part of the redox regulation system involving reversible oxidation of particular methionine residues) [24]. In addition, human selenophosphate synthetase 2 is a selenoenzyme that catalyzes the ATP-dependent synthesis of selenophosphate, a selenium donor compound for Sec biosynthesis [25]. One human selenoprotein, Selenoprotein P, has 10 Sec residues. This plasma protein is synthesized primarily in the liver [26,27] and delivers selenium to other organs (see [28,29] and references therein).
Figure 1.
Human selenoproteome. The names, designations, protein size and location of Sec in human selenoproteins are shown. The length (blue panels) and location of Sec (red mark) in proteins are shown schematically on the right.
The specific functions of several other human selenoproteins are unknown, although many details of their biology have been established [30]. Among these, Selenoproteins S and K are ER membrane proteins involved in retrotranslocation of misfolded proteins from the ER to cytosol for subsequent degradation by the proteasome. The 15 kDa selenoprotein, Sep15, is an ER-resident protein implicated in quality control of protein folding. There is also a distant Sep15 homolog of unknown function, designated Selenoprotein M. Characteristic features of other selenoproteins include the nucleolar subcellular localization of Selenoprotein H and testis-specific expression of SelV. Selenoproteins T, V, H and W form a subfamily characterized by a thioredoxin-like fold with the Sec located in the N-terminal region of this domain, whereas Sec is located in the C-terminal region (typically the second or third amino acid from the end) of SelS, SelK, SelI and SelO. The latter protein is also the largest mammalian selenoprotein, and it is located in mitochondria. The physiological and biochemical functions of SelN have been linked to muscle disorders and control of ryanodine receptor, respectively. However, none of the specific functions of these selenoproteins are known.
At least three mammalian selenoproteins, thioredoxin reductase (TR) 1, TR3 and glutathione peroxidase (GPx) 4, are essential for development in mice, whereas knockout mice deficient in GPx1, GPx2, GPx3, Sep15, MsrB1, and SelM are viable and have only mild phenotypes in the absence of stress [31]. SelP knockout mice are associated with systemic selenium deficiency, and this phenotype can be rescued by supplementation with dietary selenium [32,33].
Selenoproteins are oxidoreductases
Essentially all functionally-characterized selenoproteins are oxidoreductases [30] in which Sec is the catalytic residue. Clearly, the unique catalytic properties of Sec are the reason selenium is used in these proteins. However, there is no consensus as to what these properties are. Suggestions include the nucleophilicity of Sec, its low pKa (e.g., compared to Cys), its ability to be a leaving group and its resistance to inactivation by overoxidation [34]. These properties can be partially compensated for by Cys (e.g., Cys mutants of most selenoproteins preserve ~1% activity, and in addition, Cys homologs are known for most selenoproteins), but not by any other residue.
Recent studies revealed an interesting regulatory role for a selenoprotein. MsrB1 and two Mical proteins, were found to regulate actin through reversible stereo specific methionine oxidation [35]. Actin polymers can be disassembled by Mical-catalyzed oxidation of Met41 and Met44 to methionine-R-sulfoxide residues, whereas reduction of these residues back to Met by MsrB1 promotes actin repolymerization [35]. This study also established a new type of regulatory posttranslational modification.
Occurrence of selenoproteins
Approximately half of eukaryotes have selenoproteins, whereas only about 25% of bacteria and 15% of archaea preserved these proteins during evolution [36]. Biosynthesis and insertion of Sec into proteins require several genes; therefore, once this trait is lost, it cannot be restored. The only known exception is the rare event of lateral transfer of an operon responsible for the Sec trait in prokaryotes [37].
Overall, there is currently a very good understanding of which organisms utilize selenoproteins, and which do not. Information is also available with regard to identification of selenoproteins and location of their Sec residues. The principal function of selenoproteins is their participation in redox homeostasis, although the specific functions are diverse. Functions of about half of human selenoproteins remain unknown.
Sec tRNA and biosynthesis of Sec
Being the 21st amino acid in the genetic code, Sec has its own tRNA, which is one of the major players in the Sec insertion machinery. It is also the only known tRNA that controls the expression of an entire class of proteins. Because this tRNA is initially aminoacylated with serine, Sec tRNA is often designated as tRNA[Ser]Sec. The tRNA[Ser]Sec population consists of two isoforms in mammals that differ from each other by a single 2’-O-methylribose at position 34 designated Um34 [38]. The synthesis of Um34 is dependent on selenium status and the resulting isoform, 5-methoxycarbonylmethyluracil-2’-O-methylribose (mcm5Um), is enriched under conditions of selenium adequacy and poorly expressed under conditions of selenium deficiency. The isoform lacking Um34, 5-methoxycarbonylmethyluracil (mcm5U), is less dependent on selenium status and is expressed under conditions of selenium deficiency [38].
These two Sec tRNA[Ser]Sec isoforms have different roles in mammalian selenoprotein synthesis [38], and are involved in the expression of different subclasses of selenoproteins. Mcm5U is involved in the synthesis of housekeeping selenoproteins and mcm5Um in the synthesis of stress-related selenoproteins (see section below “Roles of selenoproteins in health and development”).
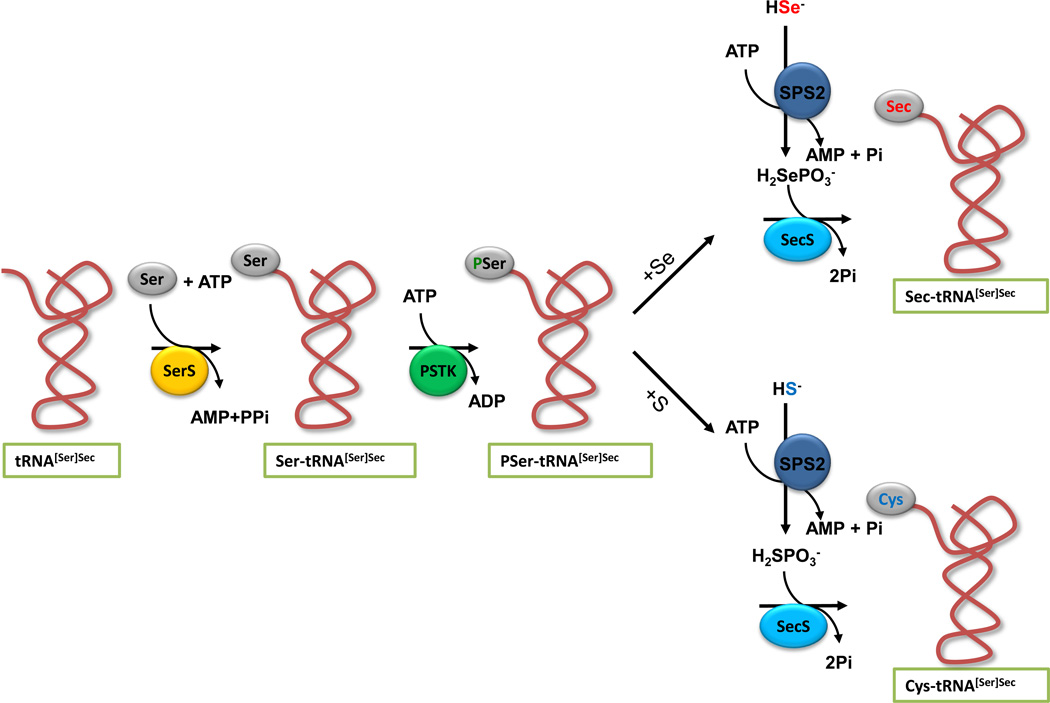
In all life forms that synthesize Sec on its tRNA, serine is initially attached to tRNA[Ser]Sec by seryl-tRNA synthetase (Fig. 2). In archaea and eukaryotes [39,40], seryl-tRNA[Ser]Sec is converted to phosphoseryl-tRNA[Ser]Sec by phosphoseryl-tRNA[Ser]Sec kinase (PSTK). This intermediate is then acted upon by selenocysteine synthase (SecS), wherein SecS converts the phosphoserine moiety to the acceptor molecule, likely aminoacrylyl-tRNA[Ser]Sec, for receiving the activated selenium donor, selenophosphate, resulting in the final product of the pathway, selenocysteyl-tRNA[Ser]Sec (Fig. 2). Selenophosphate is synthesized from selenide and ATP by selenophosphate synthetase 2 (SPS2). Since SPS2 is a selenoprotein, it may be involved in the auto regulation of selenoprotein synthesis [25]. In bacteria, SecS (designated SelA in bacteria) acts directly upon Ser-tRNA[Ser]Sec, converting the serine moiety to an intermediate, likely aminoacrylyl-tRNA[Ser]Sec as also occurs in eukaryotes, that in turn accepts selenophosphate, synthesized by selenophosphate synthetase SelD to yield Sec-tRNA[Ser]Sec [41].
Figure 2.
Biosynthesis of Sec and de novo biosynthesis of cysteine. The biosynthesis of Sec occurs on its tRNA and the pathway begins with the attachment of serine to Sec tRNA[Ser]Sec by seryl-tRNA synthetase (SerS) in the presence of ATP. Phosphoseryl-tRNA kinase (PSTK) phosphorylates the serine moiety to form an intermediate, phosphoseryl-tRNA[Ser]Sec, that in turn is acted upon by Sec synthase (SecS), converting the phosphoserine moiety to an intermediate, likely aminoacrylyl-tRNA[Ser]Sec. SecS accepts selenophosphate (H2SePO3−; upper pathway), converting the intermediate to Sec-tRNA[Ser]Sec. Selenophosphate synthetase 2 (SPS2) synthesizes selenophosphate from selenide in the presence of ATP. In the lower pathway, sulfide can replace selenide in the reaction with SPS2, generating thiophosphate (H2SPO3−) that in turn can interact with SecS and PSer-tRNA[Ser]Sec to yield Cys-tRNA[Ser]Sec.
Cys was also found to be incorporated into the selenoproteins TR1 and TR3, in place of Sec by a de novo pathway for Cys synthesis in mammalian cells and mouse liver [42]. The pathway involved replacing sulfide with selenide in the reaction with SPS2 to yield thiophosphate, which serves as a sulfur donor to yield Cys-tRNA[Ser]Sec that inserts Cys at UGA codons of selenoprotein mRNAs (Fig. 2).
Incorporation of Sec into protein
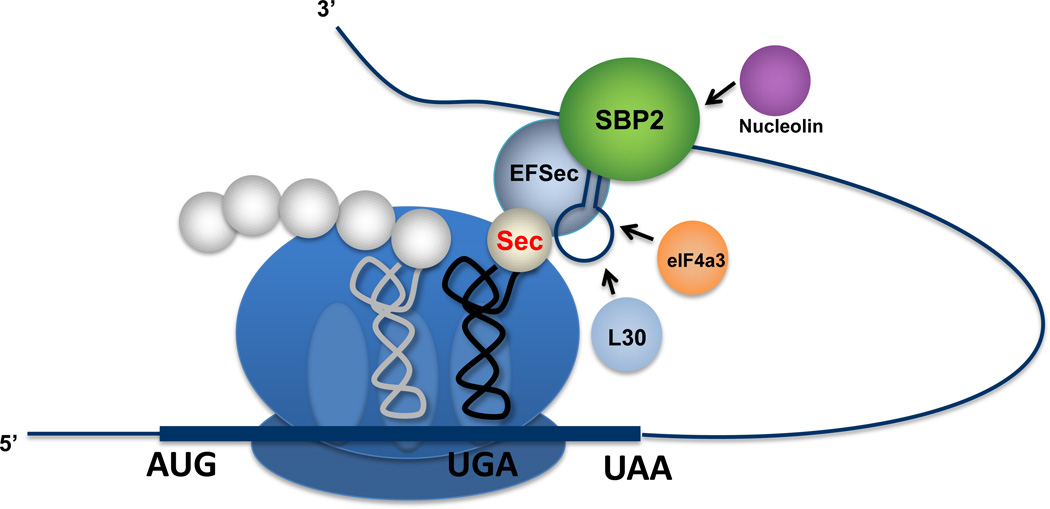
A specific cis-acting stem-loop structure, designated the Sec Insertion Sequence (SECIS) element, plays a major role in recoding UGA from stop to Sec [43]. The SECIS elements in archaea, bacteria and eukarya have completely different sequences, motifs and secondary structures [44]. Eukaryotic SECIS elements fall into two basic classes, designated Type I and II, and occur in the 3’-untranslated region of selenoprotein mRNAs. Archaeal SECIS elements have been found in both 3’-UTRs and 5’-UTRs of selenoprotein genes. In bacteria, SECIS elements are more simplified and occur within the coding regions immediately downstream of the UGA codon. They serve as the attachment site for the specific elongation factor, SelB, which is brought to the ribosome in a complex with Sec-tRNA[Ser]Sec [45]. The insertion of Sec in archaea and eukaryotes is more complex and involves additional factors. The specific elongation factor, EFsec, forms a complex with Sec-tRNA[Ser]Sec (EFsec-Sec-tRNA[Ser]Sec) that binds to the SECIS binding protein 2 (SBP2)-SECIS complex and the ribosome [45]. In addition, there are at least three other factors, ribosomal protein L30 [46], nucleolin [47] and eukaryotic initiation factor (eIF4a3) [48], that have roles in Sec insertion into protein. Although their precise roles in the incorporation process remain unclear, ribosomal protein L30 serves as part of the basic machinery responsible for Sec insertion, whereas nucleolin and eIF4a3 likely have regulatory roles tempering selenoprotein synthesis. A plausible mechanism of Sec incorporation into protein and how these factors are involved is shown in Figure 3.
Figure 3.
Incorporation of Sec into protein in mammals. The incorporation of Sec into protein involves a multifarious complex containing the Sec insertion sequence (SECIS) binding protein 2, SBP2, bound to the SECIS element that occurs in the selenoprotein mRNA 3’-untranslated region and the Sec elongation factor, EFsec, bound with Sect-RNA[Ser]Sec that is decoded at the ribosomal acceptor site. The other factors, L30, eIF4a3 and nucleolin, have regulatory roles in governing the insertion process. The decoded Sec-tRNA[Ser]Sec will be transferred to the peptidyl site, wherein the growing polypeptide will be covalently bound to Sec-tRNA[Ser]Sec.
Roles of selenoproteins in health and development
Mouse models involving Sec tRNA[Ser]Sec to elucidate the roles of selenoproteins in health and development
Various mouse models have been generated that use tRNA[Ser]Sec as a tool to providing a systems-level understanding of the roles of selenium and selenoproteins in health and development. These models encode: 1) transgenes carrying either wild type (designated Trspt) or mutant forms of tRNA[Ser]Sec; 2) conditional knockout of the tRNA[Ser]Sec gene (TrspΔ) targeting specific tissues and organs; and 3) TrspΔ complemented by Trspt or mutant tRNA[Ser]Sec transgenes, A37→G37 and T34→A34, designated TrsptG37 and TrsptA34, respectively [38]. TrsptG37 prevents N6-isopentyladenosine formation at position 37 that in turn prevents Um34 synthesis [38]. The Um34 isoform of tRNA[Ser]Sec, mcm5Um, is required to synthesize a subclass of stress-related selenoproteins (e.g., GPx1), whereas the non-Um34 isoform, mcm5U, synthesizes housekeeping selenoproteins (e.g., TR1) [49].
TrsptA34 has A in its wobble position that is converted to inosine and the resulting anticodon, ICA, decodes UGA and the Cys codons, UGU and UGC. Twenty and 40 copies of Trspt and TrsptG37, respectively, were used in transgenic mice [50]. The more copies of the mutant TrsptG37 transgene used, the less influence that the wild type Sec tRNA[Ser]Sec has on selenoprotein synthesis due to its dilution by the mutant transgene. However, in the case of TrsptA34, no more than 12 copies were tolerated by wild type mice, or two copies by liver-specific Trsp knockout, most likely, due to misreading Cys codons (inserting Sec in place of Cys) [49].
Mouse models involving TrsptG37
TrsptG37 mice had dramatic reductions in stress-related selenoproteins and were used to demonstrate roles of these proteins in muscle function [51] and protection from DNA damage [52]. Ribosome profiling showed that selenoprotein synthesis in the liver of TrsptG37 mice on selenium-adequate diets mimicked that of wild type mice maintained on selenium-deficient diets [53]. TrsptG37 mice carrying a cancer driver gene or exposed to an organ-specific carcinogen were used to elucidate the role of stress-related selenoproteins in various cancers. Results suggested that these selenoproteins reduce the incidence of colon [54] and prostate [55] cancers. Bi-transgenic mice carrying transforming growth factor α (TGFα) transgenes, which serves as a liver cancer driving gene when over-expressed, and TrsptG37 had a much higher incidence of liver tumors irrespective of whether they were maintained on a selenium-adequate or selenium-supplemented diet [56]. These mice, maintained on the selenium-deficient diet either with or without TGFα, developed a severe neurological disorder and widespread pyogranuloma. TrsptG37 mice were also targeted with a liver carcinogen when placed on selenium-deficient, -adequate and -supplemented diets [57].
Mouse models involving targeted removal of TrspΔ
TrspΔ directed to specific organs in mice revealed roles of selenoproteins in endothelial cell [58], cartilage and bone [59] and skin development [60]; heart disease [58]; breast [61] and prostate cancer prevention [62]; and immune [63,64], thyroid [65] and neuronal function [66]. The observed phenotypes differed from relatively mild effects to lethality, exposing diverse roles of selenoproteins in different cell types.
Mouse models involving TrspΔ/Trspt
Rescuing selenoprotein loss with TrsptG37 in conditional and standard TrspΔ provided alternative approaches in developing mouse models to elucidate selenoprotein roles in health and development. TrsptG37/TrspΔ mice expressed housekeeping selenoproteins and synthesized stress-related selenoproteins poorly, but exhibited an apparent normal phenotype [67]. These mice also expressed GPx4 poorly in testes which likely accounted for the reduced fertility found in males.
Roles of selenoproteins in cancer prevention and promotion
Although the above studies involving Sec tRNA[Ser]Sec led to many insights into the overall functions of selenoproteins in health and developmental issues, these approaches cannot assess the roles of individual selenoproteins in these processes. Focusing on specific selenoproteins has the advantage of addressing specific health and disease states. Interestingly, using both in vivo and in vitro approaches, three selenoproteins have been particularly instructive for understanding the role of selenoproteins in cancer. These proteins, TR1 [16], Sep15 [16] and GPx2 [15,68], were found to exhibit a split “Dr. Jekyll and Mr. Hyde” personality, both preventing and promoting cancer.
TR1, Sep15 and GPx2 are important cellular redox-regulators. Therefore, given that these oxidireductase functions would be needed by both normal and cancer cells, these very same processes most certainly result in anti- and pro-tumorigenic effects at a tissue-specific cellular level. For example, and as further discussed below, the high expression of Sep15 in colon cancer cell lines [69,70] may very well be a response of a tumor’s increased need for the associated redox function, without which proliferation and metastasis of tumor cells would be inhibited [69–71]. In contrast, since down-regulation of Sep15 mRNA expression has been found in 60% of malignant mesotheliomas [72], it remains to be elucidated whether these anti- and pro-tumorigenic effects are tumor stage or grade-dependent.
Role of TR1 in cancer prevention and promotion
TR1 is one of key redox regulators in mammalian cells. Its principal function is to control the redox state of thioredoxin, which in turn keeps surface-exposed Cys residues in cytosolic and nuclear proteins in the reduced state [73]. An analogous system occurs in mitochondria, consisting of thioredoxin 2 (Trx2) and TR3 (interestingly, cytosolic and mitochondrial TRs and Trxs are essential proteins). TR1 is also known to activate the tumor suppressor p53, and other cellular proteins, and it can be specifically targeted by carcinogenic electrophilic compounds [74]. Liver tumor incidence was dramatically enhanced by chemical carcinogenesis in mice lacking TR1 in hepatocytes compared to controls [75]. These and other properties of TR1 suggested that this selenoenzyme is an anticancer protein [76]. However, TR1 also has roles in cancer promotion. It is over-expressed in many cancers and cancer cell lines, and the cancer-related properties of these cells can be reversed (making them more like normal cells) by using specific inhibitors and anticancer drugs that target TR1 activity [77]. In addition, removal of TR1 in lung cancer cells changed morphology and anchorage-independent growth properties and led to a dramatic reduction in tumor progression and metastasis [78]. Thus, TR1 is also a pro-cancer protein and a prime candidate for cancer therapy.
Other studies have shown a role for TR1 in cancer that is independent of its major role of maintaining Trx in the reduced state [79]. For example, TR1-deficient cells were far more sensitive to selenium toxicity than Trx1-deficient cells [80]. Additionally, only TR1-deficient cells, and not Trx1-deficient cells, increased production and secretion of glutathione that was associated with enhanced selenite toxicity. All these studies involving TR1 elucidated the direct role of this selenoenzyme in governing malignancy and suggested alternative avenues for inhibiting the cancer processes.
Role of Sep15 in cancer prevention and promotion
Sep15 was first characterized in human T-cells [81], and like TR1, it was proposed to function as an oxidoreductase [82,83]. Sep15 is regulated by ER stress and forms a strong complex with UDP-glucose: glycoprotein glucosyltransferase (UGT), an enzyme that glucosylates misfolded proteins in the ER. Through this association, Sep15 is thought to contribute to quality control of protein folding and maturation, and this function may especially be important in the eye [84]. Reduced Sep15 expression was reported in lung cancer patients [85], in malignant lung, breast, prostate and liver tissues [86], as well as in cell lines derived from malignant mesothelioma cells [72]. Differences in polymorphic alleles of sep15 were found to be associated with various cancers in different ethnic groups [87,88]. Many of these observations suggested a role for Sep15 in tumor suppression. However, studies on colon cancer in vitro [69,70] and in vivo [71] have revealed a role for this protein in cancer progression, possibly through effects on cell cycle regulation [69,70] and/or interferon-γ-regulated inflammation [71]. Therefore, even though much of its biological function remains unclear, Sep15 appears to be an important contributor to human health and disease and a plausible target for cancer therapy.
Role of GPx2 in cancer prevention and promotion
Among the five selenocysteine-containing human glutathione peroxidases (GPx), GPx2, termed the intestinal GPx, is, like other selenoprotein peroxidases, considered an antioxidant protein [89]. GPx2 has been found to affect apoptosis and regulate self-renewal of the intestinal epithelium [15], making GPx2 an important contributor to healthy intestinal epithelia. Its protective function is further seen in a model of chemically-induced colon carcinogenesis, where GPx2 protected mice from developing colon pre-cancerous lesions [90] and tumors [91]. Furthermore, GPx2 contributes to the detoxification of carcinogens through its up-regulation by Nrf2/Keap1 [68], which also can be considered beneficial in terms of cancer prevention. However, this role of GPx2 may depend on the stage of cancer, as up-regulation of Nrf2-targets also has been described to provide hepatic cancer cells with an anti-oxidative advantage [92]. Additionally, since GPx2 is a target for the Wnt-pathway [93], which is associated with cell proliferation, and is up-regulated by β-catenin [94], it may also play a role in promoting tumor growth. Therefore, GPx2 appears to have roles in preventing and promoting cancer similar to TR1 and Sep15.
Concluding remarks
The biological functions of the micronutrient selenium are mediated in large part by selenoproteins: proteins containing Sec in the active site. Among the human selenoproteins encoded by 25 selenoprotein genes, about half are oxidoreductases, whereas the specific functions of the remaining selenoproteins are unknown. Human selenoproteins are involved in glutathione-dependent hydroperoxide removal, reduction of thioredoxins, selenophosphate synthesis, activation and inactivation of thyroid hormones, thioredoxin-dependent repair of oxidized methionine residues, and ERassociated protein degradation. These and other functions are responsible for the role of selenium in human health, including its pro- and anticancer activities, roles in the immune system, and other functions. There are many aspects of selenium and selenoprotein metabolism that remain to be explored. Although many of the factors involved in the insertion of Sec into protein have been defined, the overall mechanism is poorly understood. Furthermore, despite extensive efforts to evaluate the beneficial and detrimental effects of selenium in human clinical trials, critical barriers remain, including significant gaps in our knowledge of how selenium and selenoproteins act metabolically to prevent and, in some cases, promote cancer. A better understanding of these basic mechanisms will facilitate the design and interpretation of safe and effective human trials and lead to new strategies for therapeutic intervention. Several of the major unresolved questions in the selenium field are given in Box 1.
Box 1. Outstanding Questions.
Other than their presumed redox-regulating activity, what are the biological roles of the approximately one-half of the human selenoproteins whose functions are unknown?
What is the identity of the Um34 methylase responsible for converting mcm5U to mcm5Um?
What are the specific functions of the orphan selenium and selenoprotein machinery such as SPS1, SBP2L, ribosomal protein L30, nucleolin and eukaryotic initiation factor, eIF4a3?
How does dietary selenium function in terms of being protective against disease other than through selenoproteins?
What are the cellular uptake mechanisms for the various chemical forms of selenium?
What other selenoproteins manifest split personalities in preventing and promoting cancer?
How does selenoprotein expression affect disease states, e.g., diabetes, different cancers, cardiovascular disease, neurological disorders; and how do disease states affect selenoprotein expression?
What are the effects of selenoprotein SNPs in the human population?
What is the contribution of dietary selenium to healthy aging?
Highlights.
Selenocysteine is the 21st amino acid in the genetic code.
Selenoproteins are largely responsible for the many health benefits of selenium.
Some selenoproteins exhibit a split personality in preventing and promoting cancer.
Acknowledgements
The authors express their sincere appreciation to Dr. Vyacheslav Labunskyy for his help in designing Figure 3. This work was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research to D. L. H. and NIH grants to V. N. G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its Molecular Biology and Role in Human Health. Springer Science+Business Media, LLC; 2012. [Google Scholar]
- 2.Madison TC. Sanitary report-Fort Randall. In: Coolidge RH, editor. Statistical report on the sickness and mortality in the Army of the United States; 36th Congress Senate Exchange Document; 1860. pp. 37–41. [Google Scholar]
- 3.Franke KW. A new toxicant occurring naturally in certain samples of plant foodstuffs I. Results obtained in preliminary feeding trials. J. Nutr. 1934;8:597–608. [Google Scholar]
- 4.Schwarz K, Foltz CM. Factor 3 activity of selenium compounds. J. Biol. Chem. 1958;233:245–251. [PubMed] [Google Scholar]
- 5.Reilly C, editor. Selenium in food and health. Chapman and Hall; 1996. [Google Scholar]
- 6.Combs GF, Combs SB, editors. The role of selenium in nutrition. Academic Press; 1986. [Google Scholar]
- 7.Clark LC, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276:1957–1963. [PubMed] [Google Scholar]
- 8.Lippman SM, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NCI chemoprevention study of selenium for non-small cell lung cancer. http://www.cancer.gov/clinicaltrials/ft-ECOG-5597.
- 10. http://archive.tobacco.org/news/100214.html. [Google Scholar]
- 11.Dunn PK, Taylor PR. Prostate cancer prevention and the Selenium and Vitamin E Cancer Prevention Trial (SELECT): A selenium perspective. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its molecular biology and role in human health. 3rd edn. Springer Science+Business Media, LLC; 2012. pp. 297–312. [Google Scholar]
- 12.Sheehan HB, et al. High rates of serum selenium deficiency among HIV-and HCV-infected and uninfected drug users in Buenos Aires, Argentina. Public Health Nutr. 2012;15:538–545. doi: 10.1017/S1368980011001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield DL, Gladyshev VN. The outcome of Selenium and Vitamin E Cancer Prevention Trial (SELECT) reveals a need for better understanding of selenium biology and trial-subject targeting. Mol. Interventions. 2009;9:18–21. doi: 10.1124/mi.9.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayman MP, et al. Selenium and vitamin E supplementation for cancer prevention. JAMA. 2009;301:1876. doi: 10.1001/jama.2009.625. [DOI] [PubMed] [Google Scholar]
- 15.Banning A, et al. Glutathione peroxidase 2 and its role in cancer. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its molecular biology and role in human health. 3rd edn. Springer Science+Business Media, LLC; 2012. pp. 271–282. [Google Scholar]
- 16.Yoo MH, et al. Selenoproteins harboring a split personality in both preventing and promoting cancer. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its molecular biology and role in human health. 3rd edn. Springer Science+Business Media, LLC; 2012. pp. 325–334. [Google Scholar]
- 17.Flohe L, et al. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 18.Cone JE, et al. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. U S A. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers I, et al. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the 'termination' codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinoni F, et al. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl. Acad. Sci. U S A. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobler CJ, et al. The central role of selenium in the biochemistry and ecology of the harmful pelagophyte, Aureococcus anophagefferens. ISME J. 2013;7:1333–1343. doi: 10.1038/ismej.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryukov GV, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 23.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell. Mol. Life Sci. 2009;66:2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kryukov GV, et al. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. U S A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guimarães MJ, et al. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in selenocysteine metabolism? Proc. Natl. Acad. Sci. U S A. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson BA, et al. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J. Biol. Chem. 2004;279(9):8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 27.Schweizer U, et al. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renko K, et al. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Seppknockout mice. Biochem. J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 29.Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobanov AV, et al. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys. Acta. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conrad M, Schweizer U. Unveiling the molecular mechanisms behind selenium-related diseases through knockout mouse studies. Antioxid. Redox Signal. 2010;12:851–865. doi: 10.1089/ars.2009.2912. [DOI] [PubMed] [Google Scholar]
- 32.Schomburg L, et al. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill KE, et al. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 34.Hondal RJ, et al. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid. Redox Signal. 2013;18:1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee BC, et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereo selective methionine oxidation. Mol. Cell. 2013;51:397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Gladyshev VN. Comparative genomics of trace element dependence in biology. J. Biol. Chem. 2011;286:23623–23629. doi: 10.1074/jbc.R110.172833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero H, et al. Evolution of selenium utilization traits. Genome Biol. 2005;6:R66. doi: 10.1186/gb-2005-6-8-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatfield DL, et al. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J, et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. U S A. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu XM, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bock A, et al. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu XM, et al. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc. Natl. Acad. Sci. U S A. 2010;107:21430–21434. doi: 10.1073/pnas.1009947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berry MJ, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 44.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 45.Seeher S, et al. Post-transcriptional control of selenoprotein biosynthesis. Current Prot. Peptide Sc. 2012;13:337–346. doi: 10.2174/138920312801619448. [DOI] [PubMed] [Google Scholar]
- 46.Chavatte L, et al. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 47.Miniard AC, et al. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 2010;38:4807–4820. doi: 10.1093/nar/gkq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Budiman ME, et al. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell. 2009;35:479–489. doi: 10.1016/j.molcel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson BA, et al. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 50.Moustafa ME, et al. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol. Cell. Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornberger TA, et al. Selenoprotein-deficient transgenic mice exhibit enhanced exercise-induced muscle growth. J. Nutr. 2003;133:3091–3097. doi: 10.1093/jn/133.10.3091. [DOI] [PubMed] [Google Scholar]
- 52.Baliga MS, et al. Selenoprotein deficiency enhances radiation-induced micronuclei formation. Mol. Nutr. Food Res. 2008;52:1300–1304. doi: 10.1002/mnfr.200800020. [DOI] [PubMed] [Google Scholar]
- 53.Howard MT, et al. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013;288:19401–19413. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irons R, et al. Both selenoproteins and low molecular weight selenocompounds reduce colon cancer risk in mice with genetically impaired selenoprotein expression. J. Nutr. 2006;136:1311–1317. doi: 10.1093/jn/136.5.1311. [DOI] [PubMed] [Google Scholar]
- 55.Diwadkar-Navsariwala V, et al. Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model. Proc. Natl. Acad. Sci. U S A. 2006;103:8179–8184. doi: 10.1073/pnas.0508218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moustafa ME, et al. Selenium and selenoprotein deficiencies induce widespread pyogranuloma formation in mice, while high levels of dietary selenium decrease liver tumor size driven by TGFalpha. PLoS One. 2013;8:e57389. doi: 10.1371/journal.pone.0057389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasaikina MV, et al. Contrasting roles of dietary selenium and selenoproteins in chemically induced hepatocarcinogenesis. Carcinogenesis. 2013;34:1089–1095. doi: 10.1093/carcin/bgt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shrimali RK, et al. Selenoprotein expression is essential in endothelial cell development and cardiac muscle function. Neuromuscul. Disord. 2007;17:135–142. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Downey CM, et al. Osteo-chondroprogenitor-specific deletion of the selenocysteine tRNA gene, Trsp, leads to chondronecrosis and abnormal skeletal development: a putative model for Kashin-Beck disease. PLoS Genet. 2009;5:e1000616. doi: 10.1371/journal.pgen.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sengupta A, et al. Selenoproteins are essential for proper keratinocyte function and skin development. PLoS One. 2010;5:e12249. doi: 10.1371/journal.pone.0012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hudson TS, et al. Selenoproteins reduce susceptibility to DMBA-induced mammary carcinogenesis. Carcinogenesis. 2012;33:1225–1230. doi: 10.1093/carcin/bgs129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luchman HA, et al. Prostate epithelium-specific deletion of the selenocysteine tRNA gene, Trsp, leads to early-onset intraepithelial neoplasia. Am. J. Pathol. 2013 doi: 10.1016/j.ajpath.2013.11.025. DOI: In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shrimali RK, et al. Selenoproteins mediate T cell immunity through an antioxidant mechanism. J. Biol. Chem. 2008;283:20181–20185. doi: 10.1074/jbc.M802559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson BA, et al. Selenoproteins regulate macrophage invasiveness and extracellular matrix-related gene expression. BMC Immunol. 2009;10:57. doi: 10.1186/1471-2172-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiu-Ugalde J, et al. Thyroid function is maintained despite increased oxidative stress in mice lacking selenoprotein biosynthesis in thyroid epithelial cells. Antioxid. Redox Signal. 2012;17:902–913. doi: 10.1089/ars.2011.4055. [DOI] [PubMed] [Google Scholar]
- 66.Wirth EK, et al. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010;24:844–852. doi: 10.1096/fj.09-143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlson BA, et al. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J. Biol. Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 68.Brigelius-Flohe R, et al. The yin and yang of nrf2-regulated selenoproteins in carcinogenesis. Int. J. Cell Biol. 2012;2012:486147. doi: 10.1155/2012/486147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irons R, et al. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev. Res. (Phila) 2010;3:630–639. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuji PA, et al. Deficiency in the 15 kDa selenoprotein inhibits human colon cancer cell growth. Nutrients. 2011;3:805–817. doi: 10.3390/nu3090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsuji PA, et al. Knockout of the 15 kDa selenoprotein protects against chemically-induced aberrant crypt formation in mice. PLoS One. 2012;7:e50574. doi: 10.1371/journal.pone.0050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Apostolou S, et al. Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype. Oncogene. 2004;23:5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- 73.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2013 doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 74.Urig S, Becker K. On the potential of thioredoxin reductase inhibitors for cancer therapy. Semin. Cancer Biol. 2006;16:452–465. doi: 10.1016/j.semcancer.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 75.Carlson BA, et al. Thioredoxin reductase 1 protects against chemically induced hepatocarcinogenesis via control of cellular redox homeostasis. Carcinogenesis. 2012;33:1806–1813. doi: 10.1093/carcin/bgs230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem. Biophys. Res. Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, et al. Recent advances in the development of thioredoxin reductase inhibitors as anticancer agents. Curr. Drug Targets. 2012;13:1432–1444. doi: 10.2174/138945012803530224. [DOI] [PubMed] [Google Scholar]
- 78.Yoo MH, et al. Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J. Biol. Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 79.Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Tobe R, et al. Thioredoxin reductase 1 deficiency enhances selenite toxicity in cancer cells via a thioredoxin-independent mechanism. Biochem. J. 2012;445:423–430. doi: 10.1042/BJ20120618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gladyshev VN, et al. A new human selenium-containing protein. Purification, characterization, and cDNA sequence. J. Biol. Chem. 1998;273:8910–8915. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- 82.Kumaraswamy E, et al. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J. Biol. Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- 83.Labunskyy VM, et al. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 84.Kasaikina MV, et al. Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J. Biol. Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jablonska E, et al. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. Eur. J. Nutr. 2008;47:47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- 86.Wright ME, Diamond AM. Polymorphisms in selenoprotein genes and cancer. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its molecular biology and role in human health. 3rd edn. Springer Science+Business Media, LLC; 2012. pp. 345–354. [Google Scholar]
- 87.Nasr MA, et al. Allelic loss of the Sep15 locus in breast cancer. Cancer Ther. 2004;1:293–298. [Google Scholar]
- 88.Hu YJ, et al. Distribution and functional consequences of nucleotide polymorphisms in the 3'-untranslated region of the human Sep15 gene. Cancer Res. 2001;61:2307–2310. [PubMed] [Google Scholar]
- 89.Flohé L, Brigelius-Flohé R. Selenoproteins of the glutathione peroxidase family. In: Hatfield DL, Berry MJ, Gladyshev VN, editors. Selenium: Its molecular biology and role in human health. 3rd edn. Springer Science+Business Media, LLC; 2012. pp. 167–180. [Google Scholar]
- 90.Müller MF, et al. Deletion of glutathione peroxidase-2 inhibits azoxymethane-induced colon cancer development. PLoS One. 2013;8(8):e72055. doi: 10.1371/journal.pone.0072055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krehl S, et al. Glutathione peroxidase-2 and selenium decreased inflammation and tumors in a mouse model of inflammation-associated carcinogenesis whereas sulforaphane effects differed with selenium supply. Carcinogenesis. 2012;33:620–628. doi: 10.1093/carcin/bgr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dewa Y, et al. Molecular expression analysis of beta-naphthoflavone-induced hepatocellular tumors in rats. Toxicol. Pathol. 2009;37(4):446–455. doi: 10.1177/0192623309335062. [DOI] [PubMed] [Google Scholar]
- 93.Kipp AP, et al. The selenoproteins GPx2, TrxR2 and TrxR3 are regulated by Wnt signalling in the intestinal epithelium. Biochim. Biophys. Acta. 2012;1820:1588–1596. doi: 10.1016/j.bbagen.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 94.Kipp A, et al. Activation of the glutathione peroxidase 2 (GPx2) promoter by beta-catenin. Biol. Chem. 2007;388:1027–1033. doi: 10.1515/BC.2007.137. [DOI] [PubMed] [Google Scholar]