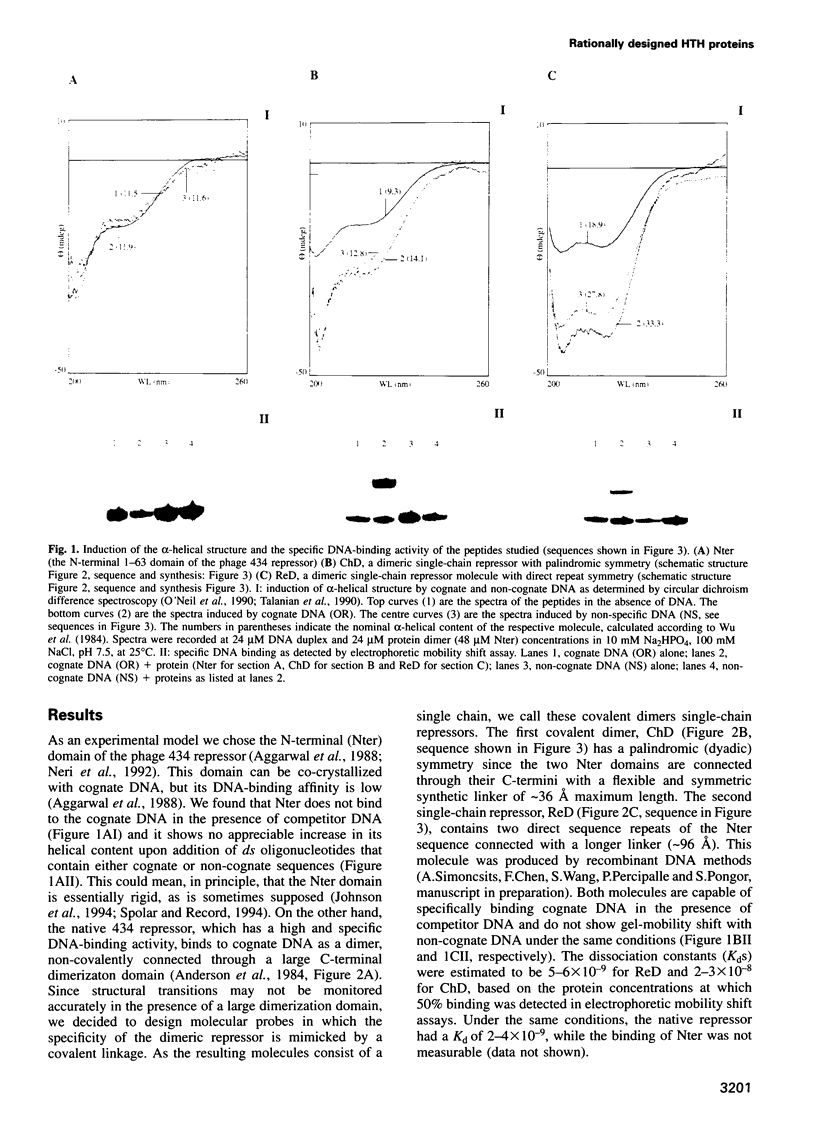
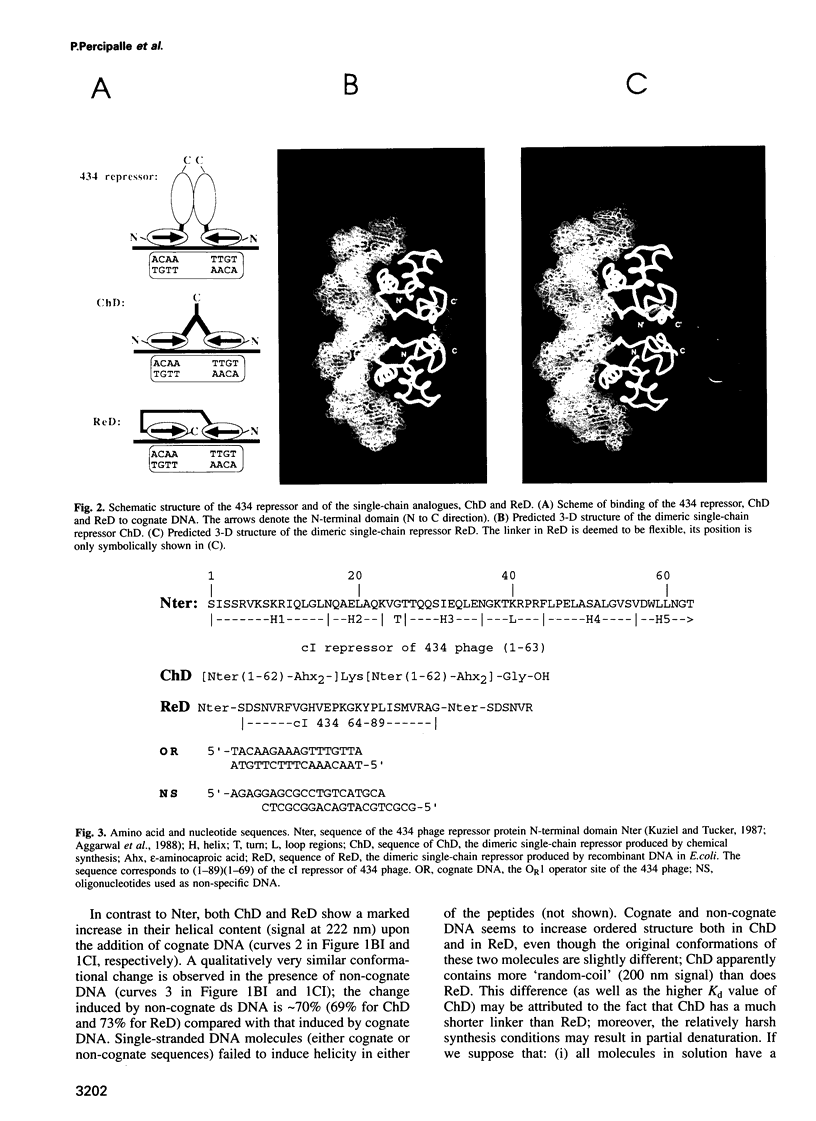
Abstract
Circular dichroism and electrophoretic mobility shift studies were performed to confirm that dimerized N-terminal domains of bacterial repressors containing helix-turn-helix motifs are capable of high-affinity and specific DNA recognition as opposed to the monomeric N-terminal domains. Specific, high-affinity DNA binding proteins were designed and produced in which two copies of the N-terminal 1-62 domain of the bacteriophage 434 repressor are connected either in a dyad-symmetric fashion, with a synthetic linker attached to the C-termini, or as direct sequence repeats. Both molecules bound to their presumptive cognate nearly as tightly as does the natural (full-length and non-covalently dimerized) 434 repressor, showing that covalent dimerization can be used to greatly enhance the binding activity of individual protein segments. Circular dichroism spectroscopy showed a pronounced increase in the alpha-helix content when these new proteins interacted with their cognate DNA and a similar, although 30% lower, increase was also seen upon their interaction with non-cognate DNA. These results imply that a gradual conformational change may occur when helix-turn-helix motifs bind to DNA, and that a scanning mechanism is just as plausible for this motif class as that which is proposed for the more flexible basic-leucine zipper and basic-helix-loop-helix motifs.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Anderson J., Ptashne M., Harrison S. C. Cocrystals of the DNA-binding domain of phage 434 repressor and a synthetic phage 434 operator. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1307–1311. doi: 10.1073/pnas.81.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony-Cahill S. J., Benfield P. A., Fairman R., Wasserman Z. R., Brenner S. L., Stafford W. F., 3rd, Altenbach C., Hubbell W. L., DeGrado W. F. Molecular characterization of helix-loop-helix peptides. Science. 1992 Feb 21;255(5047):979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Arrowsmith C. H., Pachter R., Altman R. B., Iyer S. B., Jardetzky O. Sequence-specific 1H NMR assignments and secondary structure in solution of Escherichia coli trp repressor. Biochemistry. 1990 Jul 10;29(27):6332–6341. doi: 10.1021/bi00479a002. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Castagnoli L., Vetriani C., Cesareni G. Linking an easily detectable phenotype to the folding of a common structural motif. Selection of rare turn mutations that prevent the folding of Rop. J Mol Biol. 1994 Apr 8;237(4):378–387. doi: 10.1006/jmbi.1994.1241. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hu J. C., Newell N. E., Tidor B., Sauer R. T. Probing the roles of residues at the e and g positions of the GCN4 leucine zipper by combinatorial mutagenesis. Protein Sci. 1993 Jul;2(7):1072–1084. doi: 10.1002/pro.5560020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. C., O'Shea E. K., Kim P. S., Sauer R. T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990 Dec 7;250(4986):1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Johnson N. P., Lindstrom J., Baase W. A., von Hippel P. H. Double-stranded DNA templates can induce alpha-helical conformation in peptides containing lysine and alanine: functional implications for leucine zipper and helix-loop-helix transcription factors. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4840–4844. doi: 10.1073/pnas.91.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziel W. A., Tucker P. W. Determination of vector: insert junctions in lambda gt10 cDNAs that do not recut with EcoRI. Nucleotide sequence of the lambda imm434 HindIII-EcoRI DNA fragment encoding part of the cI protein. Nucleic Acids Res. 1987 Apr 10;15(7):3181–3181. doi: 10.1093/nar/15.7.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P., Richmond T. J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wüthrich K. Determination of the nuclear magnetic resonance solution structure of the DNA-binding domain (residues 1 to 69) of the 434 repressor and comparison with the X-ray crystal structure. J Mol Biol. 1992 Feb 5;223(3):743–767. doi: 10.1016/0022-2836(92)90987-u. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Shuman J. D., Ampe C., DeGrado W. F. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991 Sep 17;30(37):9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- Percipalle P., Saletti R., Pongor S., Foti S., Tossi A., Fisichella S. Structural characterization of synthetic model peptides of the DNA-binding cI434 repressor by electrospray ionization and fast atom bombardment mass spectrometry. Biol Mass Spectrom. 1994 Dec;23(12):727–733. doi: 10.1002/bms.1200231203. [DOI] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. Dimerization of leucine zippers analyzed by random selection. Nucleic Acids Res. 1993 Sep 11;21(18):4348–4355. doi: 10.1093/nar/21.18.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudek V., Pasley H. S., Gibson T., Gausepohl H., Frank R., Pastore A. Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry. 1991 Feb 5;30(5):1310–1317. doi: 10.1021/bi00219a022. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dörr T., Oertel-Buchheit P., Pernelle C., Bracco L., Schnarr M., Granger-Schnarr M. Construction, purification, and characterization of a hybrid protein comprising the DNA binding domain of the LexA repressor and the Jun leucine zipper: a circular dichroism and mutagenesis study. Biochemistry. 1991 Oct 8;30(40):9657–9664. doi: 10.1021/bi00104a013. [DOI] [PubMed] [Google Scholar]
- Schoepfer R. The pRSET family of T7 promoter expression vectors for Escherichia coli. Gene. 1993 Feb 14;124(1):83–85. doi: 10.1016/0378-1119(93)90764-t. [DOI] [PubMed] [Google Scholar]
- Simoncsits A., Bristulf J., Tjörnhammar M. L., Cserzö M., Pongor S., Rybakina E., Gatti S., Bartfai T. Deletion mutants of human interleukin 1 beta with significantly reduced agonist properties: search for the agonist/antagonist switch in ligands to the interleukin 1 receptors. Cytokine. 1994 Mar;6(2):206–214. doi: 10.1016/1043-4666(94)90043-4. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994 Feb 11;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Talanian R. V., McKnight C. J., Kim P. S. Sequence-specific DNA binding by a short peptide dimer. Science. 1990 Aug 17;249(4970):769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- Tsao D. H., Gruschus J. M., Wang L. H., Nirenberg M., Ferretti J. A. Elongation of helix III of the NK-2 homeodomain upon binding to DNA: a secondary structure study by NMR. Biochemistry. 1994 Dec 20;33(50):15053–15060. doi: 10.1021/bi00254a014. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- Wu C. S., Ikeda K., Yang J. T. Ordered conformation of polypeptides and proteins in acidic dodecyl sulfate solution. Biochemistry. 1981 Feb 3;20(3):566–570. doi: 10.1021/bi00506a019. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao D., Revington M., Lee W., Jia X., Arrowsmith C., Jardetzky O. The solution structures of the trp repressor-operator DNA complex. J Mol Biol. 1994 May 13;238(4):592–614. doi: 10.1006/jmbi.1994.1317. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Berg O. G. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994 Feb 11;263(5148):769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]