Abstract
Iron is required for efficient oxygen transport, and hypoxia signaling links erythropoiesis with iron homeostasis. Hypoxia induces a highly conserved signaling pathway in cells under conditions of low O2. One component of this pathway, hypoxia-inducible factor (HIF), is a transcription factor that is highly active in hypoxic cells. The first HIF target gene characterized was EPO, which encodes erythropoietin—a glycoprotein hormone that controls erythropoiesis. The past decade has led to fundamental advances in our understanding of how hypoxia regulates iron levels to support erythropoiesis and maintain systemic iron homeostasis. We review the cell-type specific effects of hypoxia and HIFs in adaptive response to changes in oxygen and iron availability, as well as potential uses of HIF modulators for patients with iron-related disorders.
Keywords: Hypoxia, HIF, Iron, Erythropoiesis
Introduction
Iron, an essential nutrient, is required for oxygen delivery and is a cofactor in several enzymatic and redox reactions. In mammals, 70% of iron is found in red blood cells (RBCs), and 6% is a component of iron-containing proteins, which are required for respiration, energy metabolism, and endobiotic and xenobiotic metabolism. The remaining 24% is stored in ferritin. Iron levels are controlled by a multi-tissue homeostatic process in which dietary iron is absorbed through the proximal small intestine. Dietary iron (Fe+3) is reduced by an apical ferric reductase duodenal cytochrome b (DCYTB) to ferrous iron (Fe+2) and imported into the enterocyte via the apical iron transporter, divalent metal transporter-1 (DMT1, also known as NRAMP2 or DCT1 and encoded by SLC11A2) 1-3.
Tissues also have efficient mechanisms for use of heme iron—the major form of iron in meat. Although several transporters of heme have been identified, little is known about their in vivo functions 4-6. Once heme is transported in enterocytes, it is broken down by heme oxygenases to release ferrous iron 5. Iron can either be stored in ferritin or mobilized to circulation by the iron exporter ferroportin (encoded by SLC40A1) located on the basolateral surface of enterocytes 7-9. Once in circulation, iron is loaded onto transferrin 10, 11, where a large proportion of iron is used in RBC synthesis. Splenic macrophages recycle iron from senescent RBCs, limiting the amount of iron required from dietary sources to as little as 1–2 mg per day 12.
The liver is central to iron homeostasis because hepatocytes produce hepcidin, a circulating factor that regulates iron homeostasis. Furthermore, the liver can store and mobilize iron, depending on systemic demands13. We review how the hypoxia response affects systemic iron homeostasis and iron-related disorders.
Iron and Oxygen
Changes in atmospheric oxygen concentration have linked to key evolutionary events. A major hurdle in the evolution of multicellular organisms was the efficient delivery of O2. O2 has low solubility in water, resulting in selection for proteins that could efficiently transport oxygen. Hemoglobin, a highly conserved protein, can efficiently and reversibly bind oxygen and deliver it from the lungs to peripheral tissues. Hemoglobin is composed of 4 chains; each chain contains a heme group 14, 15 The charged iron ion in the heme group binds 1 molecule of O2, so a single hemoglobin protein has the capacity to bind 4 O2 molecules. The heme group provides a unique molecular characteristic—under a condition of high partial pressure of oxygen (pO2), such as in the lung alveoli, O2 binds with high affinity, whereas under a condition of low pO2, such as in peripheral tissues, O2 is released 16, 17. Most iron is used for RBC synthesis; iron deficiency reduces numbers of RBCs and oxygen transport, leading to tissue hypoxia.
Oxygen Sensing and Transcription
Tissues maintain specific levels of O2 for respiration and homeostasis. Cells respond to conditions of low O2 by altering gene expression patterns, which affects levels of several hundred proteins involved in cell survival. These changes are initiated by a heterodimeric nuclear transcription factor, hypoxia-inducible factor (HIF). HIF comprises an oxygen-dependent α-subunit (HIF1α 18, 19, HIF2α 20, or HIF3α 21) and a constitutively expressed β-subunit, aryl hydrocarbon nuclear translocator (ARNT) 22. In cells, adequate levels of oxygen cause rapid degradation of HIFα subunits by prolyl hydroxylase domain-containing enzymes (PHD). Three isoforms have been reported 23-25.
PHDs are 2-oxoglutarate-dependent dioxygenases; this activity requires iron binding at an active site and oxygen as a co-substrate, so they are an important link between levels of oxygen and iron 26. At normal levels of oxygen, PHDs hydroxylate HIF α subunits at conserved prolines. Oxygen-dependent prolyl-hydroxylation is required for HIF to bind von Hippel-Lindau tumor suppressor protein (VHL), and then to the E3 ubiquitin ligase complex, leading to ubiquitination and degradation of HIF α subunits 27-29. Inactivation of VHL in normoxic cells results in HIF activity demonstrating VHL is required for HIF degradation30.
A decrease in cellular oxygen or iron availability inhibits PHD-dependent proline hydroxylation of HIF α subunits. VHL can no longer bind to HIF α subunits, resulting in its stabilization. Reactive oxygen species (ROS) in the mitochondria are also required for HIF stabilization. Mitochondrial electron transport chain inhibitors or cells depleted of mitochondrial DNA do not stabilize HIF under hypoxic conditions 31-33. Following stabilization, HIF α subunits interact with ARNT and other transcription factors, such as p300/CBP, to activate transcription of genes that regulate the response to hypoxia 34 (Figure 1). HIF1α and HIF2α regulate distinct yet overlapping target genes.
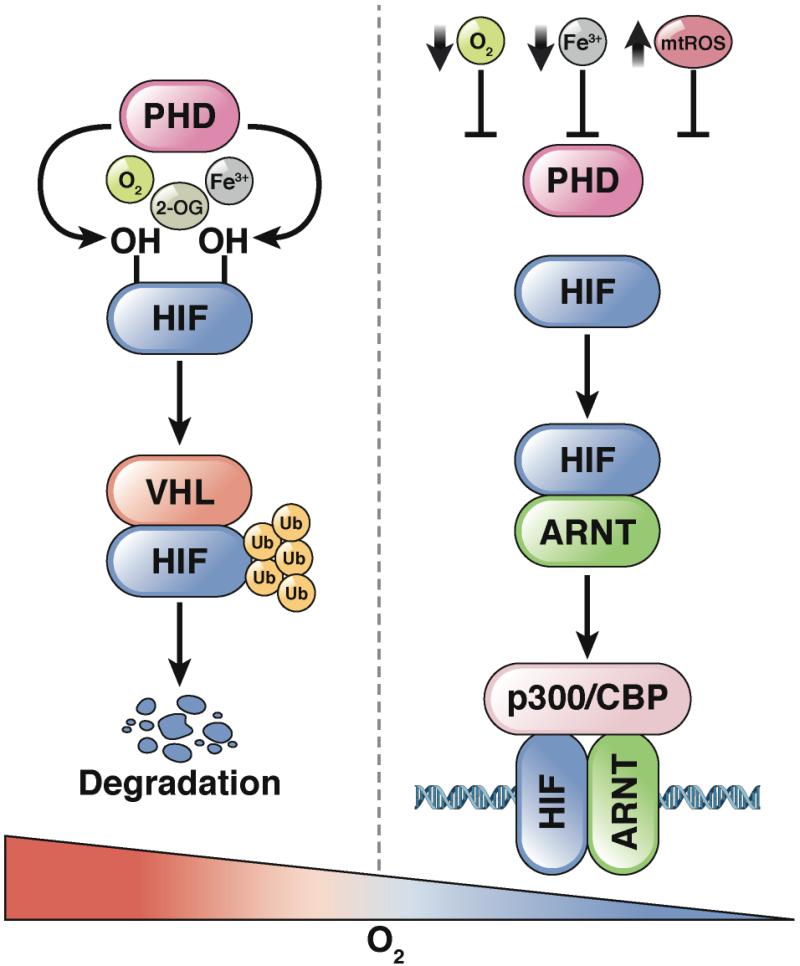
Figure 1. Iron- and Oxygen-dependent Regulation of HIF Protein Stability.
HIF1α and α2 subunits are hydroxylated on 2 proline residues by PHD enzymes, which require oxygen (O2), iron (Fe3+) and 2-oxogluatrate (2-OG). Hydroxylation leads to VHL binding and rapid ubiquitination and degradation. Under low iron concentrations, hypoxic conditions, or increase in mitochondrial ROS (mtROS), the HIFα subunit is no longer hydroxylated, and is therefore stabilized.
Specific stimuli can have different effects on HIF signaling. Low levels of iron, reductions in O2, or increases in mitochondrial ROS lead to activation of different subsets of HIF target genes. For example, low levels of iron in the intestine lead to activation of HIF2α and its target genes, including DMT1 and CYBRD1 (which encodes DCYTB), but do not alter expression of vascular endothelial growth factor (VEGF), which is frequently induced under conditions of hypoxia 35. The mechanisms of these differential gene expression patterns are not clear, might involve specific or relative inhibition of PHDs. In addition to regulating the transcriptional response to hypoxia, HIF2α binds to mRNAs and increases cap-mediated translation 36. Disruption of Hif1a, Hif2a, Arnt, or Vhl causes embryonic lethality in mice, demonstrating the importance of an appropriate hypoxic response in vivo 37-40.
Hypoxia and Regulation of Erythropoietin
Over a century ago, studies conducted during high-altitude expeditions associated levels of O2 with numbers of RBCs. Further studies into the mechanisms of erythropoiesis led to the discovery and isolation of erythropoietin, encoded by EPO 41,42. EPO expression is tightly regulated by developmental, physiological, and cellular mechanisms to maintain RBC homeostasis. The liver is the major source of erythropoietin during embryogenesis; after birth, production begins from the kidneys 43. Hypoxia and HIF signaling are the primary regulators of EPO in the kidney, but in response to anemia or hypoxia, EPO can be reactivated in hepatocytes 44, 45. In vitro promoter assays have shown that HIF1α and HIF2α bind and activate the EPO promoter 18 via canonical HIF response elements18, 46-48. However, subsequent studies in mice demonstrated that HIF2α specifically activates of EPO expression 49. In mice with disruption of Hif2a specifically in kidney produce lower levels of erythropoietin 44, 50. Moreover, in mice with hepatic disruption of Hif2a, extra-renal erythropoietin production is regulated by Hif2α (but not Hif1α)49. This demonstrated that HIF2α signaling is an important regulator of erythropoietin production.
Hepatocyte-specific disruption of Vhl in mice resulted in Hif1α and Hif2α activity and increased expression of Epo, leading to polycythemia 44, 49. Hif2α is therefore required and sufficient to activate expression of Epo. HIF2α is required not only for renal and hepatic expression of EPO, but also for its expression in neurons, astrocytes, and osteoblasts—these cellular sources regulate erythropoiesis independently of renal function. 51, 52
Hypoxia and Hematopoiesis
The discovery of EPO as a direct target of HIF18, 53, 54 provided the first evidence that HIF could regulate iron homeostasis by inducing erythropoietin and RBC production. Erythropoietin regulates RBC production by binding to the erythropoietin receptor on early and late erythroid progenitors, reducing apoptosis and increasing proliferation and differentiation, respectively 42. Disruption of Hif2a leads to severe anemia, pancytopenia, and hematopoietic defects 44, 50, 55. The alterations in the erythroid lineages result from decreased levels of erythropoietin. However, studies have demonstrated the role of hypoxia and HIF signaling in hematopoietic stem cell (HSC) maintenance. Quiescent HSCs are localized in hypoxic foci, and it has been proposed that O2 levels regulate their activity 56, 57. Deletion of ARNT, which prevents HIF1α and HIF2α function, reduces proliferation in hematopoietic progenitors 58. Exogenous VEGF rescues the hematopoietic proliferative defects in Arnt−/− mice 58. VEGF expression is also activated under conditions of hypoxia 59. The VEGF promoter contains canonical HIF response elements. Deletion of the VEGF receptor (such as in Flk1−/− mice) reduces numbers of hematopoietic progenitors, as observed in the Arnt−/− mice 60.
HIF2α was also shown to regulate hematopoiesis through a separate mechanism. In mice, disruption of Hif2a causes embryonic lethality. However crossing 129S6/SVEvTac and C57BL/6J mice results in survival of 20% of Hif2a−/− mice until 1 month after birth, and pancytopenia was observed in these mice 55. Interestingly, transplantation of bone marrow from Hif2a−/− mice into wild-type mice did not significantly alter differentiation or numbers of HSCs. A recent study of mice with hematopoietic cell-specific disruption of Hif2a demonstrated similar results 61. However, Hif2a−/− mice that received bone marrow transplants from wild-type mice had defects in hematopoiesis. HIF2α is therefore critical in maintaining a functional microenvironment during hematopoiesis, rather than a bone marrow cell factor, required for proliferation and differentiation of HSCs 55.
Hypoxia and Hepcidin
Hepcidin (encoded by HAMP) is a peptide hormone that is the master regulator of iron levels in humans and other mammals. It was initially discovered as an anti-microbial peptide that was mainly synthesized in the liver, could be detected in the urine, and was induced by inflammation 62-64. Hepcidin has antifungal activity against Candida albicans, Aspergillus fumigatus, and Aspergillus niger and antibacterial activity against Escherichia coli, Staphylococcus aureus, Staphylococcus epidermidis, and group B Streptococcus 62, 63. Shortly after its discovery, the link between hepcidin to iron homeostasis was uncovered when levels of hepcidin mRNA were found to change with levels of systemic iron. High systemic levels of iron, such as in patients with iron overload, increased hepcidin expression 64.
Usf2-knockout mice have multi-tissue iron overload (heart, pancreas, and liver)65. However, concentrations of iron in spleen were lower than in wild-type mice. Usf2 is a transcription factor, and analyses of gene expression patterns showed that hepcidin expression was virtually absent from livers of Usf2-knockout mice. However, Usf2 does not regulate hepcidin expression directly–the close proximity of Usf2 to Hamp resulted in both genes being disrupted in the Usf2-knockout mice. Mice with disruption of Hamp but intact Usf2 have demonstrated the role for hepcidin in progressive iron overload 66. Moreover, overexpression of Hamp leads to hyposidermia 67.
In subsequent years, the mechanisms by which hepcidin regulates iron homeostasis were uncovered. A landmark study demonstrated that hepcidin binds to the only known mammalian iron exporter, ferroportin 68, resulting in rapid internalization and degradation of hepcidin68, 69. Therefore, in the presence of hepcidin, small amounts of iron are mobilized from stores, whereas in the absence of hepcidin, iron is rapidly mobilized, leading to iron overload (such as in patients who produce low levels of hepcidin). The function of hepcidin correlates with its expression, which is regulated by systemic levels of iron. Low systemic levels of iron reduce hepcidin expression and increase iron mobilization. On the other hand, high systemic levels of iron lead to expression of hepcidin and reduce iron mobilization (Figure 2).
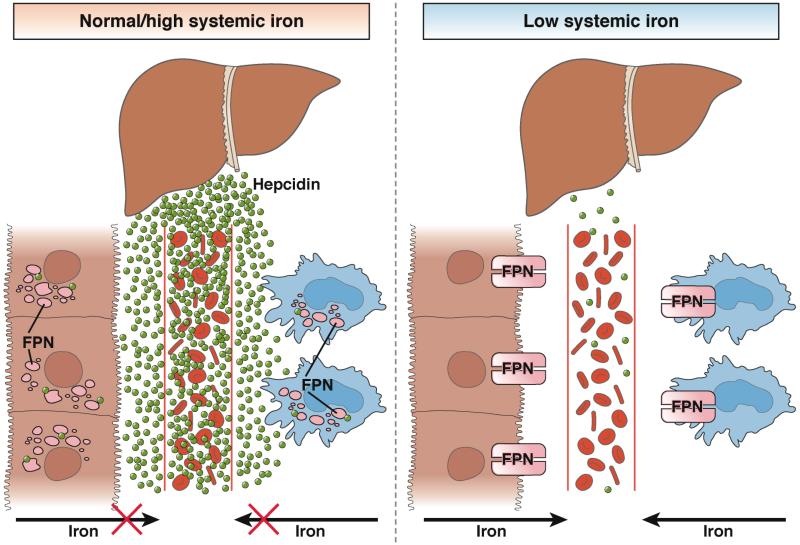
Figure 2. Hepcidin-mediated Mobilization of Iron.
During normal–high systemic levels of iron, hepcidin is expressed, leading to degradation of ferroportin and reduced mobilization of iron from the intestine and macrophage into circulation. Under conditions of low systemic iron, hepcidin expression is inhibited, stabilizing ferroportin and increasing mobilization of iron from the intestine and macrophage.
Hypoxic Repression of Hepcidin
Systemic hypoxia increases levels of erythropoietin and erythropoiesis. To maintain RBC synthesis, it is essential to have a reciprocal mechanism that increases systemic levels of iron. Over the last decade, we have greatly increased our understanding of hepcidin regulation. Several pathways have been shown to be involved in hepcidin regulation. There are reviews of these pathways in health and disease 70, 71. Here, we focus on hypoxic regulation of hepcidin.
Liver hypoxia is a strong repressor of hepcidin expression. Hypoxia can override the upregulation of hepcidin during liver inflammation and interleukin (IL)6 production 72. Shortly after hepcidin was discovered, it was shown in hepatoma-derived cell lines and mice that hypoxia could repress transcription of Hamp73. Moreover, prolonged hypoxia activates iron absorption, which coordinates with decreased Hamp expression in liver 74. In blood samples collected from healthy volunteers at sea level and then during acute and chronic exposure to high-altitude hypoxia, there was a rapid decrease in serum levels of hepcidin under the hypoxic conditions75.
In addition to hypoxia, inhibition of PHDs also represses hepcidin expression in hepatoma-derived cells, indicating that HIFs are involved in suppression 76. However, the precise mechanism by which hypoxia decreased hepcidin expression in the liver was not known. Activation of HIF in hepatocytes in mice decreased hepcidin expression and activated intestinal and macrophage expression of ferroportin 72. This demonstrated that HIF activation in the absence of hypoxia could repress hepcidin expression in vivo. In mouse hepatoma-derived cell lines, HIF1α bound and repressed the Hamp promoter 72.
However, this may not be true for the human HAMP promoter, and other mechanisms of gene regulation are likely involved 77-80.
Subsequent studies have identified erythropoietin-dependent and -independent pathways that affect HIF-induced repression of HAMP. Increases in liver or systemic expression of HIF2α led to robust activation of EPO and concomitant decrease in HAMP expression. Inhibition of erythropoietin with neutralizing antibodies, or disruption of Epo in mice, significantly reversed HIF2α-mediated repression of Hamp 78, 79. These findings are consistent with a recent study of indigenous populations who live at high altitudes 81. Although exposure of volunteers to high-altitude hypoxia significantly decreased levels of hepcidin and increased in erythropoietin and RBC production, 75 indigenous highlanders with stable erythropoietic iron requirements did not have reduced levels of hepcidin compared to their lowland counterparts81. These findings support the concept that erythropoietin and/or erythropoiesis are involved in hypoxia-mediated suppression of HAMP. Erythropoietin can directly inhibit hepcidin expression, by binding to erythropoietin receptors on hepatocytes 82. Moreover, several studies demonstrated that erythropoietin-induced erythropoiesis could inhibit hepcidin expression indirectly, through erythroid-derived factors (Figure 3) 83.
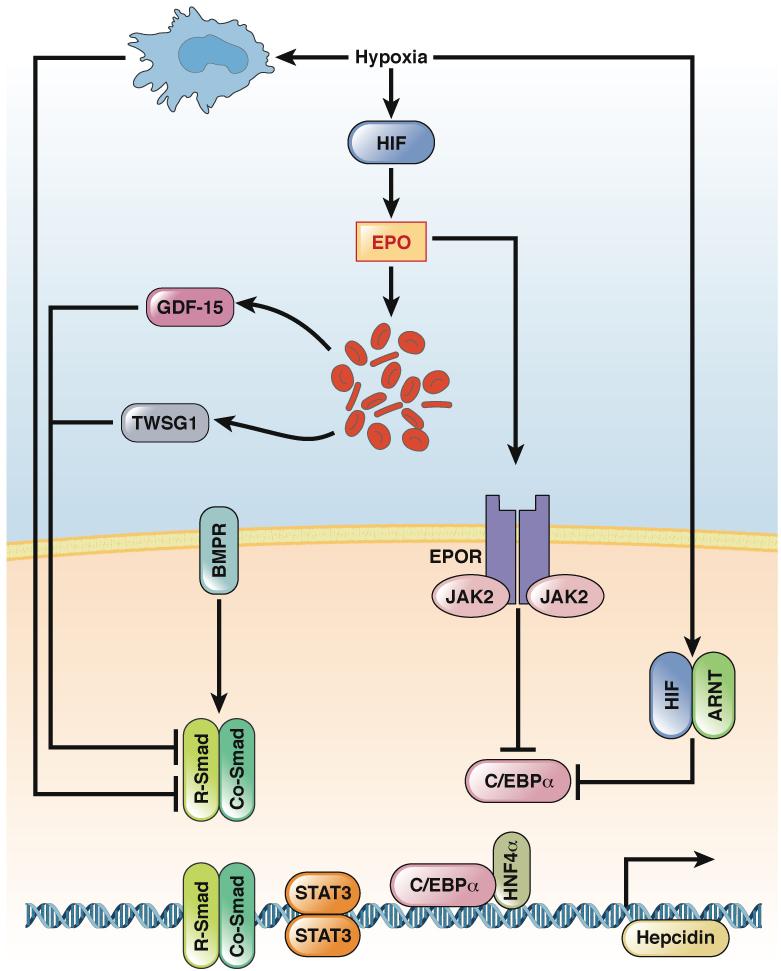
Figure 3. Transcriptional Regulation of HAMP by HIFs.
The gene encoding hepcidin (HAMP) is controlled by bone morphogenetic protein (BMP) signaling via its receptor (BMPR) to SMAD (R-SMAD) and CO-SMAD. The transcription factors STAT3, hepatic nuclear factor 4α (HNF4α), and C/EBPα are all required for basal expression of hepcidin. Hypoxia represses transcription of HAMP, via erythropoietin-dependent and erythropoietin-independent mechanisms. Systemic hypoxia leads to hepatic or renal activation of HIF, which increases expression of erythropoietin. Erythropoietin binding to its receptor, EPOR, reduces levels of C/EBPα mRNA and protein in hepatocytes. In addition, erythropoietin, by increasing erythropoiesis and erythroid-derived factors GDF15 and TWSG1, can prevent BMP signaling and activation of SMAD. Two erythropoietin-independent mechanisms have been described. Systemic hypoxia inhibits expression of C/EBPα and regulatory SMAD (R-SMAD) in hepatocytes through a macrophage-mediated mechanism; a local increase in HIF signaling in hepatocytes results C/EBPα degradation by proteasomes.
mRNA encoding growth differentiation factor 15 (GDF15) is highly induced during erythroblast differentiation 84. Clinical studies found that levels of GDF15 were increased in patients with disorders of erythropoiesis and hematopoiesis 83, 84. In cultured hepatocytes, high levels of GDF15 contribute to HAMP repression 84. However, when cells from serum samples of patients with erythropoiesis disorders were depleted of GDF15, HAMP was still repressed, indicating the involvement of other erythroid factors.
Similar to GDF15, twisted gastrulation (TWSG1) was identified as an erythroid-derived factor that can inhibit hepcidin expression 85. The mechanisms by which GDF15 and TWSG1 regulate hepcidin expression are not fully defined. However, TWSG1 can inhibit BMP-mediated activation of SMAD, an important transcriptional activator of HAMP 86.
Hypoxia can also inhibit hepcidin expression through an erythropoietin-independent pathway. Patients with Chuvash polycythemia have a specific mutation in Vhl that results in increased expression of HIF1α and HIF2α, increased numbers of RBCs (due to increased expression of erythropoietin) , and reduced expression of hepcidin 87. However, in linear regression analysis, after correcting for serum levels of ferritin, erythropoietin, and RBC count, the decrease in hepcidin levels persisted 87. This indicates that hypoxia-induced repression of hepcidin is independent of erythropoietin or RBC production. Consistent with these findings, mice that overexpress Hif1a only in hepatocytes do not have increased serum levels of erythropoietin or erythropoiesis. However, levels of hepcidin are reduced in these mice80. The erythropoietin-independent mechanism is initiated through a hypoxia-mediated degradation of C/EBPα, a critical transcription factor required for basal expression of hepcidin80. Hypoxic macrophages can also inhibit SMAD and C/EBPα in hepatocytes to reduce hepcidin expression 88 (Figure 3).
Because oxygen levels regulate iron levels and erythropoiesis, it is not surprising that hypoxia regulates hepcidin expression via several mechanisms. The goal is to understand under which physiologic and pathologic conditions these pathways are activated, and whether concomitant activation of these pathways has stronger effects on hepcidin expression.
Intestinal Oxygen Sensing and Iron Absorption
High systemic demand for iron leads to reductions in hepcidin and increases in apical and basolateral proteins involved in iron transport. In the intestine, hepcidin directly regulates degradation of ferroportin 68. However, 2 local pathways regulate ferroportin, DMT1, and DCYTB expression. The first pathway is mediated by the iron regulatory protein (IRP)1 and IRP2. The biochemical function and activity of IRPs, and their roles in vivo, were extensively reviewed 89.
IRP1 and IRP2 bind to specific elements of mRNAs called iron response elements (IRE). Binding of IRPs to IREs can block translation or increase mRNA stability, and affect levels of several proteins involved in iron homeostasis. Intracellular levels of iron regulate expression and activity of IRPs. Decreased cellular levels of iron activate IRPs, whereas high levels decrease IRP function (Figure 4). Mouse embryos that lack Irp1 and Irp2 die in utero, so these proteins are required for early development 90. Studies in which the genes encoding Irp1 or Irp2 were disrupted in mice demonstrated their redundancy, as well as their specific roles in cell physiology 91-94. Moreover, biochemical studies showed that the proteins have overlapping and specific mRNA targets 95.
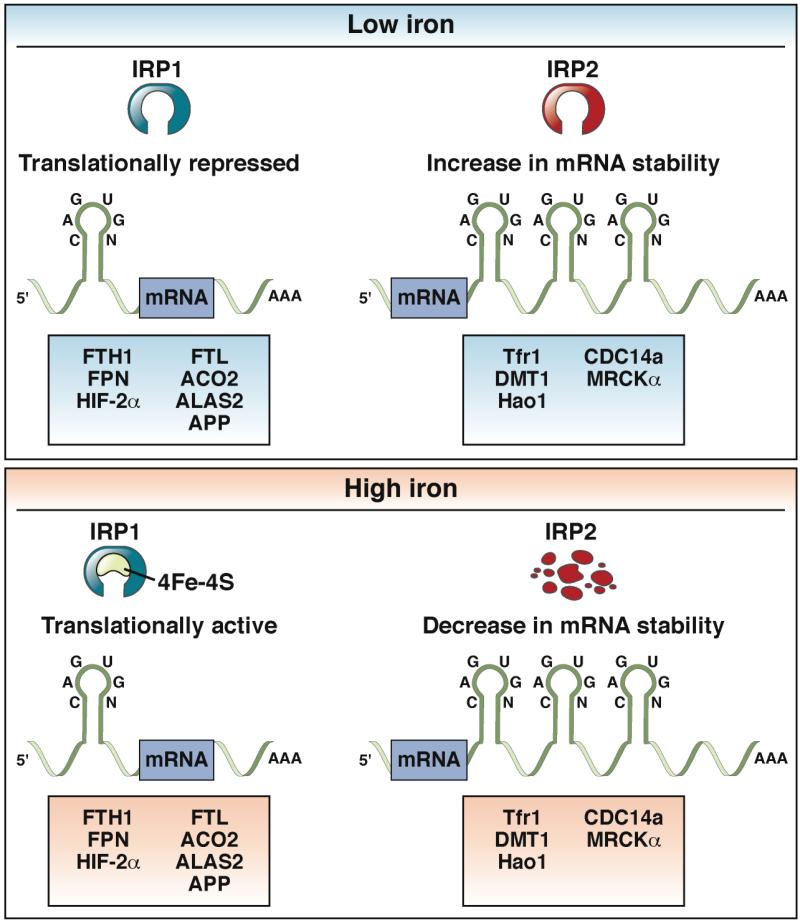
Figure 4. Regulation of IRE-containing Genes by IRP.
Low concentrations of iron increase binding of IRP1 and IRP2 to IRE-containing mRNAs. Transcripts containing an IRE in the 5’-UTR, such as FTH1(encodes H-ferritin), FTL (encodes L-ferritin), SLC40A1, ACO2 (encodes mitochondrial aconitase), ALAS2 (encodes aminolevulinate synthase 2), or APP (encodes amyloid precursor protein) bind IRPs to repress translation. Alternatively, transcripts with an IRE in the 3’-UTR, such as SLC11A2I (encodes DMT1), TFRC (encodes the transferrin receptor), HAO1 (encodes hydroxyacid oxidase 1), CDC14A (encodes cell division cycle 14A), or CDC42BPA (encodes myotonic dystrophy kinase-related Cdc42-binding kinase α) are stabilized by IRP binding. High cellular levels of iron lead to assembly of iron-sulfur clusters (4Fe-4S) in the IRE binding pocket of IRP1, which inhibit its interaction with IREs. IRP2 is degraded under high concentrations of cellular iron. Loss of IRP1 and IRP2 activity result in translation of mRNAs that contain IREs in the 5’-UTR and decreased stability of mRNAs with IREs in the 3’-UTR.
IRP1 and IRP2 target the mRNA that encodes ferroportin (SLC40A1 mRNA), binding to the IRE motif in its 5′-UTR to reduce its translation 8, 9. Intestinal disruption of genes encoding Irp1 and Irp2 in mice increased levels of ferroportin protein96. Activation of IRPs by iron deficiency would be expected to reduce translation of intestinal ferroportin. However, intestinal cells can express an SLC40A1 mRNA from an alternate promoter; this mRNA does not contain the IRE, allowing for its translation under conditions of iron deficiency 97.
Levels of DMT1 in the small intestine are also affected by intracellular iron levels. Four DMT1 isoforms have been identified, and are transcribed via alternate promoters98. These isoforms have similar transport activities, although they differ in the cells that express them, subcellular localization, and response to intracellular levels of iron 99. mRNAs encoding 2 isoforms that are regulated by iron level contain a functional IRE in the 3'-UTR and are stabilized by IRPs. Mice with a disruptions in genes encoding Irp1 or Irp2 have no changes in duodenal levels of Dmt1, likely due to their overlapping functions. However, intestine-specific deletion of Irp1 and Irp2 reduced levels of the mRNA encoding Dmt1 (Slc11a2 mRNA)96. The mechanism by which IRPs regulate levels of DMT1 is not clear, but is believed to involve a mechanism similar to IRP-mediated stabilization of mRNA encoding transferrin receptor-1 (TFRC) 100.
The ability of IRPs to stabilize the transcript and increase protein levels of DMT1 could be coordinated with hepcidin-mediated repression of SLC40A1. Under conditions of adequate systemic levels of iron, hepcidin degradation of ferroportin limits mobilization of dietary iron into the circulation. This could increase intracellular levels of iron in enterocytes, decreasing IRP function and reducing levels of DMT1. This model is consistent with what is observed during hepcidin deficiency. The increase in ferroportin leads to increased basolateral export of iron. This results in reduced iron within enterocytes, activation of intestinal IRPs, and increased levels of DMT1101.
HIF2α Regulation of Iron Transporters
Biochemical and knockout studies have characterized the roles of IRPs in local regulation of intestinal iron absorption. However, genes such as CYBRD1 (encodes DCYTB) have no IREs, yet levels of protein and mRNA greatly increase at low iron concentrations 3. In addition, transcription of SLC40A1 (encodes ferroportin) and SLC11A2 (encodes DMT1) are also induced following systemic increases in iron demand 102. These findings indicate that other mechanisms must be involved in regulating local iron absorption.
Studies have demonstrated the role of intestinal Hif2α in regulating iron absorption 35, 103, 104. Intestinal epithelial cells of mice that overexpress Hif2α have increased levels of ferroportin protein and mRNA 103. In mice with an intestine-specific disruption of Hif2a or littermate controls on normal or low-iron diets, Hif2α was required for increases in SLC40A1 mRNA in response to acute changes in iron levels. Surprisingly, low-iron induced expression of ferroportin was completely lost when Hif2a was disrupted intestines of mice103. Long-term deprivation of iron led to increases in Slc40a1 mRNA, in a Hif2α-dependent manner. However, mice with intestine-specific disruption of Hif2a and wild-type mice placed on continuous low-iron diets increased intestinal levels of ferroportin 103. This means that the acute, adaptive induction of ferroportin occurs via increased transcription of Slc40a1, and that long-term responses to systemic iron demand occur through a Hif2α-independent mechanism—most likely through a decrease in hepcidin-mediated degradation of ferroportin. This biphasic regulation provides an efficient system to maintain a rapid and sustained response to increase systemic iron requirements.
Overexpression of Hif2α greatly increases expression of Dmt1 and Dcytb. Upregulation of Dmt1 and Dcytb following acute or long-term iron deprivation is completely lost with disruption of intestinal Hif2α signaling 35, 104. Moreover, the adaptive increase in Dmt1 and Dcytb following increased erythropoiesis is also lost following disruption of Hif2α signaling in the intestine 105. Together, these findings demonstrate the role of HIF2α in the regulation of iron absorption following changes in systemic iron levels. Promoter regions of SLC40A1, SLC11A2, and CYBRD1 contain a canonical HIF response element, and chromatin immunoprecipitation assays have shown that HIF2α binds these promoters, to directly regulate these genes during increased demand for systemic iron 35, 103, 104.
Interactions Between IRP1 and HIF2α
The 5’-UTR of HIF2a mRNA contains a phylogenetically conserved IRE that binds IRP1 and IRP2 (although IRP1 appears to the major regulator of the HIF2a IRE ) 106, 107 to control iron-dependent regulation of the transcript. Translation of HIF2a mRNA was inhibited in iron-deficient cells, due to a high activity of the IRPs106, 107. Selective inhibitors of HIF2a translation have been developed, which promote binding of IRP1 binding to the 5’-UTR of the mRNA 107.
However, not all IRE-containing transcripts appear to be regulated by IRPs in vivo, so ancillary sequences could be involved. An in vivo study reported that iron deficiency in liver significantly increased activity of IRP1 and IRP2, and thereby decreased translation of HIF2a mRNA108. Two recent studies demonstrated the importance of IRPs in Hif2a regulation using knockout mice. Irp1- (but not Irp2-) knockout mice developed transient polycythemia, due to increased levels of Hif2a mRNA and renal Epo mRNA. In addition, Slc40a1, Dmt1, and Dcytb were induced in the small intestine of the Irp1-knockout mice 109, 110. These findings indicate that Irp1, through regulation of Hif2a mRNA, fine tunes the erythropoietic response to hypoxia.
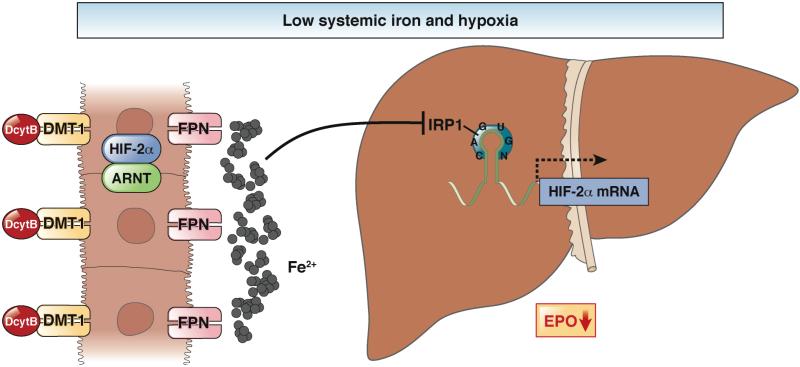
Hypoxia responsive erythropoietin-producing cells in the liver and kidney must be able to selectively decrease the level of HIF2α, depending on availability of oxygen and iron. In hepatic and renal cells that are hypoxic, activation of HIF2α leads increases production of erythropoietin. Under conditions of sufficient systemic iron, RBC production occurs. However, the increase in RBC production under conditions of hypoxia, during systemic iron deficiency, would result in hypochromic and microcytic RBCs. Therefore, the interaction between IRP1 and HIF2α could be an important mechanism that limits erythropoiesis under conditions of hypoxia and low systemic levels of iron. In the intestine, Hif2α translation is not regulated by Irps; levels of Hif2α increase on low-iron diets and is essential for the adaptive increase in ferroportin, Dmt1, and DcytB 35, 103, 104. This suggests an integrative mechanism where hypoxia and low level of iron might lead to a repression of HIF2α only in erythropoietin-producing cells. However, in the intestine, HIF2α increases iron absorption and systemic levels of iron. This results in release of the translational inhibition of hepatic and renal HIF2α, and an increase in erythropoietin and erythropoiesis (Figure 5). Studies are needed to determine how these pathways integrate into an optimal hypoxia and iron-regulated response.
Figure 5. Responses to Hypoxia and Low Systemic Levels of Iron in Liver and Intestine.
Hypoxia increases expression of erythropoietin and oxygen delivery. However, under conditions of low systemic iron, hypoxic activation of EPO would lead to microcytic RBCs. In the liver, low concentrations of iron reduce translation of HIF2α, to limit expression of EPO during times of hypoxia and low systemic iron. Coordinately, activation of intestinal HIF2α increases iron absorption. This increase in systemic level of iron relieves the translational block of HIF2α mRNA, increasing its protein levels in liver and/or kidney (not shown). This results in expression of EPO and erythropoiesis.
Hepcidin and Intestinal Hypoxia Signaling
Several in vivo studies have demonstrated that hepcidin deficiency (hepcidin-deficient mice) increases iron absorption, via increases in ferroportin, DMT1, and DCYTB proteins and mRNAs. Interestingly, these increases were lost when hepcidin-deficient mice were crossed to mice with intestinal disruption of Hif2a101. This was the first study to show that hepcidin and intestinal Hif2α signaling interact. However, it is unclear how hepcidin deficiency activates intestinal HIF2α signaling.
Researchers have identified 2 mechanisms important for activation of intestinal HIF2α following increased requirement for systemic iron 35, 103-105. During iron deficiency, it is thought that the low level of intestinal epithelial iron reduces the activity of PHDs, leading to increased expression of HIF2α 35, 103, 104. However, during increased erythropoiesis, intestinal levels of iron do not change, but intestinal levels of O2 decrease, resulting in activation of intestinal HIF2α 105. It is not clear if hepcidin affects either intestinal iron or O2 availability. These mechanisms require testing so we can better understand how HIF2α is activated during hepcidin deficiency.
HIFs and Iron-related Disorders
Heredity hemochromatosis is a genetic disorder that leads to iron overload. Mutations in genes such as HFE, TFR2, HFE2, SLC40A1, or HAMP reduce expression of hepcidin or, in the case of mutant forms of ferroportin, reduce hepcidin-mediated degradation of ferroportin 13. Although some patients have hereditary forms of hemochromatosis, others have secondary hemochromatosis, which mostly comprises acquired disorders in erythropoiesis 13. In these cases, iron overload can result from increased hemolysis, repeated blood transfusions, and hyper-absorption of iron. Similar to hereditary hemochromatosis, several types of secondary hemochromatosis are associated with reduced levels of hepcidin; correcting hepcidin levels reduces iron accumulation in tissues 111, 112. Moreover, secondary hemochromatosis disorders such as β-thalassemia are similar to hepcidin-deficient mice in that intestinal Hif2α signaling is activated 113. In patients with hereditary hemochromatosis or some forms of secondary hemochromatosis, iron overload is treated with several rounds of phlebotomy and/or administration with iron chelators 114, 115.
Intestinal HIF2α could be a therapeutic target for hereditary and secondary hemochromatosis; inhibiting its function could reduce iron absorption in the intestine. Since HIF2α regulates apical and basolateral iron transporters, blocking its action would shut down iron import and export, leading to a rapid response. Genetic deletion of Hif2a in the intestine improves iron overload in secondary hemochromatosis 113. To the best of our knowledge, no study has assessed the ability of chemical inhibitors of HIF2α to reduce iron overload, although several selective HIF2α inhibitors have been identified 107. In addition, HIF2α contains a pseudo ligand-binding pocket that can be used to allosterically inhibit HIF2α function 116. New specific inhibitors have been designed to block HIF2α function by targeting the pseudo ligand-binding pocket 117. Further studies are needed to determine the efficacy and potency of these compounds in vivo.
Anemia
Anemia, the most prevalent blood disorder worldwide, is defined as a decrease in RBCs and/or hemoglobin. Iron-deficient anemia is a common form of anemia caused by low dietary intake of iron, poor absorption, or loss of blood 118. Chronic anemia, also called anemia of inflammation, is a general anemic phenomenon observed in many patients with chronic diseases 119. The chronic inflammatory response can reduce serum levels of iron by increasing expression of hepcidin. This condition encompasses many distinct diseases, and mechanisms are specific for each disease. However, IL6 signaling via STAT3, which is induced by inflammation, increases hepcidin expression 120. This increase in hepcidin expression following acute inflammation protects tissues from pathogens, due low iron availability (pathogens require iron for proliferation). However, chronic inflammation thereby leads to anemia, and in certain cases can exacerbate progression of the patient’s primary disease.
In addition to an increase in hepcidin expression from chronic disorders, loss-of-function mutations in the TMPRSS6 gene increase expression of hepcidin, leading to iron-refractory iron-deficiency anemia (IRIDA) 121—a form of iron deficiency anemia that is unresponsive to oral iron therapy. Tmprss6-knockout mice and mice with a premature stop codon in Tmprss6 develop microcytic anemia, associated with high levels of hepcidin expression 122-124. Tmprss6 encodes a type II transmembrane serine protease that is highly expressed in liver and involved in membrane cleavage of hemojuvelin (HFE2)125, 126. Mutations in Tmprss6 reduce protease activity of the product and increase membrane hemojuvelin, leading to an increase in hepcidin expression 126. In most cases, anemia is efficiently treated with oral iron supplements. For cases such as chronic anemia of inflammation and IRIDA, more aggressive intravenous iron supplementation may be needed.
These disorders might be treated with reagents that activate HIF2α, which would decrease hepcidin in the liver and increase erythropoietin, leading to RBC production. Moreover, activation of intestinal HIF2α signaling could promote iron absorption. PHD inhibition activates HIF target genes; several PHD inhibitors exist and have been well characterized 127. These inhibitors are well tolerated by mice, and genetic disruption of PHDs increases HIF expression and reactivates hepatic expression of erythropoietin in mice 128. Several PHD inhibitors are being tested in clinical trials, and have shown some positive effects in patients with anemia 129. One major concern about PHD inhibitors is that constitutive activation of HIF can lead to inflammation and cancer 130, 131. Isoform-specific PHD inhibitors might be developed for selective modulation of HIF1α vs HIF2α activity to reduce these risks 132.
Polycythemia
Polycythemia is characterized by an increase in proportion of RBC volume, either from an increase in production of RBCs or a decrease of plasma volume 133. This affects iron homeostasis, because RBC production is tightly linked to systemic level of iron.
Polycythemia associated with high altitude is a physiologic response to hypoxia. However, several heart and lung diseases, which cause chronic systemic hypoxia, can lead to polycythemia 134, 135. Specific genetic alterations also activate HIF2α to cause polycythemia. Hemoglobin Chesapeake and Hemoglobin Kempsey are conditions in which a specific mutation in the α- or β-chain of hemoglobin greatly increases their affinity for oxygen, reducing oxygen delivery to the kidneys and causing polycythemia 136-138. Polycythemic diseases also arise through mutations in factors that regulate HIF activity. Chuvash polycythemia caused by a specific mutation in the VHL gene 139. In mouse models of this disorder, disruptions in Vhl increase Hif1α and Hif2α activity and production of erythropoietin. Mutations in PHD2 and gain-of-function mutation in HIF2α have also been found to cause polycythemia 140-142. Similar to HIF2α-based therapeutics for iron overload, inhibitors of HIF2α might be developed to treat polycythemia.
Future Directions
Hypoxia signaling is an important component of iron homeostasis and oxygen delivery. It regulates hepcidin, erythropoietin, and proteins that control intestinal iron absorption. However, many questions remain. Hypoxia, IRPs, and hepcidin mediate local and systemic iron responses. Disruption or overexpression of any these factors in cells or animals affects iron homeostasis. Studies are needed to determine how local and systemic hypoxic responses cooperate with hepcidin and IRP to control iron homeostasis.
Although HIF1α and HIF2α repress HAMP under hypoxic conditions, HIF2α specifically upregulates other genes that control iron absorption. However, we understand little about the precise mechanisms by which HIF1α and HIF2α control gene expression. HIF1α and HIF2α bind the same response element (a core sequence of A/G CGTG) but regulate different sets of genes. SLC40A1, SLC11A2, DCTYB, and EPO contain consensus HIF response elements in their proximal promoter regions that are specifically activated by HIF2α in vivo. It has been a challenge to identify the mechanisms that determine binding specificity. For example, in vitro reporter assays showed that the proximal promoters of SLC40A1, SLC11A2, and DCTYB specifically bind HIF2α, whereas the EPO proximal promoter is activated by HIF1α and HIF2α. However endogenous EPO expression is regulated by only HIF2α.
Increasing our understanding of the process of HIF gene regulation and its response to hypoxia could lead to development of reagents that control HIF activity with few side effects. It is also important to learn more about existing pharmacologic agents, as well as the new generation of HIF modulators, to determine if they might be effective therapeutics for iron-related disorders.
Funding
National Institutes of Health (grant CA148828 and DK095201); The University of Michigan Gastrointestinal Peptide Center; Jeffrey A. Colby Colon Cancer Research and the Tom Liu Memorial Funds of the University of Michigan Comprehensive Cancer Center to Y.M.S.
Abbreviations
- ARNT
aryl hydrocarbon nuclear translocator
- DMT1
divalent metal transporter-1
- DCYTB
duodenal cytochrome B
- GDF15
growth differentiation factor 15
- HIF
hypoxia inducible factor
- HIF1α
hypoxia inducible factor1-α
- HIF2α
hypoxia inducible factor-2α
- HJV
hemojuvelin
- HSC
hematopoietic stem cell
- IL6
interleukin-6
- IRE
iron response element
- IRIDA
iron-deficiency anemia
- IRP1/2
iron-response protein 1/2
- PHD
prolyl hydroxylase
- RBC
red blood cell
- ROS
reactive oxygen species
- TMPRSS6
Transmembrane protease, serine 6
- Tfr1
transferrin receptor-1
- TWSG1
twisted gastrulation 1
- USF2
upstream stimulating factor 2
- UTR
untranslated region
- VEGF
vascular endothelial growth factor
- VHL
von Hippel-Lindau
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conception and design: Y.M. Shah.
Development of methodology: N/A
Acquisition of data: N/A
Analysis and interpretation of data: Y.M. Shah and L. Xie
Writing, review, and/or revision of the manuscript: Y.M. Shah and L. Xie
Study supervision: Y.M. Shah
Conflicts of Interest: none declared.
References
- 1.Fleming MD, Trenor CC, 3rd, Su MA, et al. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–6. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 2.Gunshin H, Mackenzie B, Berger UV, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–8. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 3.McKie AT, Barrow D, Latunde-Dada GO, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–9. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 4.Qiu A, Jansen M, Sakaris A, et al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–28. doi: 10.1016/j.cell.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 5.Latunde-Dada GO, Simpson RJ, McKie AT. Recent advances in mammalian haem transport. Trends Biochem Sci. 2006;31:182–8. doi: 10.1016/j.tibs.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Latunde-Dada GO, Takeuchi K, Simpson RJ, et al. Haem carrier protein 1 (HCP1): Expression and functional studies in cultured cells. FEBS Lett. 2006;580:6865–70. doi: 10.1016/j.febslet.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 7.Donovan A, Lima CA, Pinkus JL, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 8.McKie AT, Marciani P, Rolfs A, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 9.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–12. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- 10.Aisen P, Brown EB. Structure and function of transferrin. Prog Hematol. 1975;9:25–56. [PubMed] [Google Scholar]
- 11.Aisen P, Leibman A, Zweier J. Stoichiometric and site characteristics of the binding of iron to human transferrin. J Biol Chem. 1978;253:1930–7. [PubMed] [Google Scholar]
- 12.Ganz T. Macrophages and systemic iron homeostasis. J Innate Immun. 2012;4:446–53. doi: 10.1159/000336423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson ER, Shah YM. Iron homeostasis in the liver. Compr Physiol. 2013;3:315–30. doi: 10.1002/cphy.c120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perutz MF. Structure of hemoglobin. Brookhaven Symp Biol. 1960;13:165–83. [PubMed] [Google Scholar]
- 15.Perutz MF, Rossmann MG, Cullis AF, et al. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185:416–22. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 16.Thom CS, Dickson CF, Gell DA, et al. Hemoglobin variants: biochemical properties and clinical correlates. Cold Spring Harb Perspect Med. 2013;3:a011858. doi: 10.1101/cshperspect.a011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perutz MF, Fermi G, Shih TB. Structure of deoxyhemoglobin Cowtown [His HC3(146) beta----Leu]: origin of the alkaline Bohr effect and electrostatic interactions in hemoglobin. Proc Natl Acad Sci U S A. 1984;81:4781–4. doi: 10.1073/pnas.81.15.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–8. [PubMed] [Google Scholar]
- 20.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 21.Makino Y, Cao R, Svensson K, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–4. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang BH, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 24.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 25.Schofield CJ, Zhang Z. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr Opin Struct Biol. 1999;9:722–31. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 26.Myllyharju J. Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 2013;208:148–65. doi: 10.1111/apha.12096. [DOI] [PubMed] [Google Scholar]
- 27.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 28.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 30.Kapitsinou PP, Haase VH. The VHL tumor suppressor and HIF: insights from genetic studies in mice. Cell Death Differ. 2008;15:650–9. doi: 10.1038/sj.cdd.4402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandel NS. Detection of oxygen-sensing properties of mitochondria. Methods Enzymol. 2002;352:31–40. doi: 10.1016/s0076-6879(02)52004-8. [DOI] [PubMed] [Google Scholar]
- 32.Chandel NS, Schumacker PT. Cellular oxygen sensing by mitochondria: old questions, new insight. J Appl Physiol (1985) 2000;88:1880–9. doi: 10.1152/jappl.2000.88.5.1880. [DOI] [PubMed] [Google Scholar]
- 33.Schroedl C, McClintock DS, Budinger GR, et al. Hypoxic but not anoxic stabilization of HIF-1alpha requires mitochondrial reactive oxygen species. Am J Physiol Lung Cell Mol Physiol. 2002;283:L922–31. doi: 10.1152/ajplung.00014.2002. [DOI] [PubMed] [Google Scholar]
- 34.Arany Z, Huang LE, Eckner R, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93:12969–73. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah YM, Matsubara T, Ito S, et al. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–64. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uniacke J, Holterman CE, Lachance G, et al. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–9. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–62. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian H, Hammer RE, Matsumoto AM, et al. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–4. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maltepe E, Schmidt JV, Baunoch D, et al. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–7. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 40.Gnarra JR, Ward JM, Porter FD, et al. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci U S A. 1997;94:9102–7. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyake T, Kung CK, Goldwasser E. Purification of human erythropoietin. J Biol Chem. 1977;252:5558–64. [PubMed] [Google Scholar]
- 42.Bunn HF. Erythropoietin. Cold Spring Harb Perspect Med. 2013;3:a011619. doi: 10.1101/cshperspect.a011619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson LO, Goldwasser E, Fried W, et al. Role of the kidney in erythropoiesis. Nature. 1957;179:633–4. doi: 10.1038/179633a0. [DOI] [PubMed] [Google Scholar]
- 44.Kapitsinou PP, Liu Q, Unger TL, et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood. 2010;116:3039–48. doi: 10.1182/blood-2010-02-270322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27:41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck I, Ramirez S, Weinmann R, et al. Enhancer element at the 3'-flanking region controls transcriptional response to hypoxia in the human erythropoietin gene. J Biol Chem. 1991;266:15563–6. [PubMed] [Google Scholar]
- 47.Semenza GL, Dureza RC, Traystman MD, et al. Human erythropoietin gene expression in transgenic mice: multiple transcription initiation sites and cis-acting regulatory elements. Mol Cell Biol. 1990;10:930–8. doi: 10.1128/mcb.10.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semenza GL, Koury ST, Nejfelt MK, et al. Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc Natl Acad Sci U S A. 1991;88:8725–9. doi: 10.1073/pnas.88.19.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rankin EB, Biju MP, Liu Q, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–77. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scortegagna M, Ding K, Zhang Q, et al. HIF-2alpha regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood. 2005;105:3133–40. doi: 10.1182/blood-2004-05-1695. [DOI] [PubMed] [Google Scholar]
- 51.Rankin EB, Wu C, Khatri R, et al. The HIF signaling pathway in osteoblasts directly modulates erythropoiesis through the production of EPO. Cell. 2012;149:63–74. doi: 10.1016/j.cell.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidemann A, Kerdiles YM, Knaup KX, et al. The glial cell response is an essential component of hypoxia-induced erythropoiesis in mice. J Clin Invest. 2009;119:3373–83. doi: 10.1172/JCI39378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang GL, Semenza GL. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood. 1993;82:3610–5. [PubMed] [Google Scholar]
- 54.Wang GL, Semenza GL. Molecular basis of hypoxia-induced erythropoietin expression. Curr Opin Hematol. 1996;3:156–62. doi: 10.1097/00062752-199603020-00009. [DOI] [PubMed] [Google Scholar]
- 55.Scortegagna M, Morris MA, Oktay Y, et al. The HIF family member EPAS1/HIF-2alpha is required for normal hematopoiesis in mice. Blood. 2003;102:1634–40. doi: 10.1182/blood-2003-02-0448. [DOI] [PubMed] [Google Scholar]
- 56.Parmar K, Mauch P, Vergilio JA, et al. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007;104:5431–6. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler IG, Barbier V, Wadley R, et al. Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: serially reconstituting hematopoietic stem cells reside in distinct nonperfused niches. Blood. 2010;116:375–85. doi: 10.1182/blood-2009-07-233437. [DOI] [PubMed] [Google Scholar]
- 58.Adelman DM, Maltepe E, Simon MC. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes Dev. 1999;13:2478–83. doi: 10.1101/gad.13.19.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forsythe JA, Jiang BH, Iyer NV, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–13. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuh AC, Faloon P, Hu QL, et al. In vitro hematopoietic and endothelial potential of flk-1(−/−) embryonic stem cells and embryos. Proc Natl Acad Sci U S A. 1999;96:2159–64. doi: 10.1073/pnas.96.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guitart AV, Subramani C, Armesilla-Diaz A, et al. Hif-2alpha is not essential for cell-autonomous hematopoietic stem cell maintenance. Blood. 2013;122:1741–5. doi: 10.1182/blood-2013-02-484923. [DOI] [PubMed] [Google Scholar]
- 62.Krause A, Neitz S, Magert HJ, et al. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480:147–50. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 63.Park CH, Valore EV, Waring AJ, et al. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–10. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 64.Pigeon C, Ilyin G, Courselaud B, et al. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–9. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 65.Nicolas G, Bennoun M, Devaux I, et al. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci U S A. 2001;98:8780–5. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lesbordes-Brion JC, Viatte L, Bennoun M, et al. Targeted disruption of the hepcidin 1 gene results in severe hemochromatosis. Blood. 2006;108:1402–5. doi: 10.1182/blood-2006-02-003376. [DOI] [PubMed] [Google Scholar]
- 67.Viatte L, Nicolas G, Lou DQ, et al. Chronic hepcidin induction causes hyposideremia and alters the pattern of cellular iron accumulation in hemochromatotic mice. Blood. 2006;107:2952–8. doi: 10.1182/blood-2005-10-4071. [DOI] [PubMed] [Google Scholar]
- 68.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 69.De Domenico I, Ward DM, Langelier C, et al. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18:2569–78. doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ganz T, Nemeth E. Hepcidin and iron homeostasis. Biochim Biophys Acta. 2012;1823:1434–43. doi: 10.1016/j.bbamcr.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finberg KE. Regulation of systemic iron homeostasis. Curr Opin Hematol. 2013;20:208–14. doi: 10.1097/MOH.0b013e32835f5a47. [DOI] [PubMed] [Google Scholar]
- 72.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–32. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicolas G, Chauvet C, Viatte L, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–44. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leung PS, Srai SK, Mascarenhas M, et al. Increased duodenal iron uptake and transfer in a rat model of chronic hypoxia is accompanied by reduced hepcidin expression. Gut. 2005;54:1391–5. doi: 10.1136/gut.2004.062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Piperno A, Galimberti S, Mariani R, et al. Modulation of hepcidin production during hypoxia-induced erythropoiesis in humans in vivo: data from the HIGHCARE project. Blood. 2011;117:2953–9. doi: 10.1182/blood-2010-08-299859. [DOI] [PubMed] [Google Scholar]
- 76.Braliou GG, Verga Falzacappa MV, Chachami G, et al. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J Hepatol. 2008;48:801–10. doi: 10.1016/j.jhep.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 77.Volke M, Gale DP, Maegdefrau U, et al. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS One. 2009;4:e7875. doi: 10.1371/journal.pone.0007875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Q, Davidoff O, Niss K, et al. Hypoxia-inducible factor regulates hepcidin via erythropoietin-induced erythropoiesis. J Clin Invest. 2012;122:4635–44. doi: 10.1172/JCI63924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mastrogiannaki M, Matak P, Mathieu JR, et al. Hepatic hypoxia-inducible factor-2 down-regulates hepcidin expression in mice through an erythropoietin-mediated increase in erythropoiesis. Haematologica. 2012;97:827–34. doi: 10.3324/haematol.2011.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson ER, Taylor M, Xue X, et al. The hypoxia-inducible factor-C/EBPalpha axis controls ethanol-mediated hepcidin repression. Mol Cell Biol. 2012;32:4068–77. doi: 10.1128/MCB.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lundgrin EL, Janocha AJ, Koch CD, et al. Plasma hepcidin of Ethiopian highlanders with steady-state hypoxia. Blood. 2013;122:1989–91. doi: 10.1182/blood-2013-03-491068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinto JP, Ribeiro S, Pontes H, et al. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111:5727–33. doi: 10.1182/blood-2007-08-106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanno T, Noel P, Miller JL. Growth differentiation factor 15 in erythroid health and disease. Curr Opin Hematol. 2010;17:184–90. doi: 10.1097/MOH.0b013e328337b52f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanno T, Bhanu NV, Oneal PA, et al. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096–101. doi: 10.1038/nm1629. [DOI] [PubMed] [Google Scholar]
- 85.Tanno T, Porayette P, Sripichai O, et al. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood. 2009;114:181–6. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang RH, Li C, Xu X, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 87.Gordeuk VR, Miasnikova GY, Sergueeva AI, et al. Chuvash polycythemia VHLR200W mutation is associated with down-regulation of hepcidin expression. Blood. 2011;118:5278–82. doi: 10.1182/blood-2011-03-345512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaston TB, Matak P, Pourvali K, et al. Hypoxia inhibits hepcidin expression in HuH7 hepatoma cells via decreased SMAD4 signaling. Am J Physiol Cell Physiol. 2011;300:C888–95. doi: 10.1152/ajpcell.00121.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 90.Smith SR, Ghosh MC, Ollivierre-Wilson H, et al. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–7. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 91.LaVaute T, Smith S, Cooperman S, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27:209–14. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 92.Meyron-Holtz EG, Ghosh MC, Iwai K, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–95. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, et al. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–91. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galy B, Ferring D, Hentze MW. Generation of conditional alleles of the murine Iron Regulatory Protein (IRP)-1 and -2 genes. Genesis. 2005;43:181–8. doi: 10.1002/gene.20169. [DOI] [PubMed] [Google Scholar]
- 95.Sanchez M, Galy B, Schwanhaeusser B, et al. Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood. 2011;118:e168–79. doi: 10.1182/blood-2011-04-343541. [DOI] [PubMed] [Google Scholar]
- 96.Galy B, Ferring-Appel D, Kaden S, et al. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 97.Zhang DL, Hughes RM, Ollivierre-Wilson H, et al. A ferroportin transcript that lacks an iron-responsive element enables duodenal and erythroid precursor cells to evade translational repression. Cell Metab. 2009;9:461–73. doi: 10.1016/j.cmet.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee PL, Gelbart T, West C, et al. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol Dis. 1998;24:199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 99.Shawki A, Knight PB, Maliken BD, et al. H(+)-coupled divalent metal-ion transporter-1: functional properties, physiological roles and therapeutics. Curr Top Membr. 2012;70:169–214. doi: 10.1016/B978-0-12-394316-3.00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–89. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mastrogiannaki M, Matak P, Delga S, et al. Deletion of HIF-2alpha in the enterocytes decreases the severity of tissue iron loading in hepcidin knockout mice. Blood. 2012;119:587–90. doi: 10.1182/blood-2011-09-380337. [DOI] [PubMed] [Google Scholar]
- 102.Zoller H, Theurl I, Koch R, et al. Mechanisms of iron mediated regulation of the duodenal iron transporters divalent metal transporter 1 and ferroportin 1. Blood Cells Mol Dis. 2002;29:488–97. doi: 10.1006/bcmd.2002.0587. [DOI] [PubMed] [Google Scholar]
- 103.Taylor M, Qu A, Anderson ER, et al. Hypoxia-inducible factor-2alpha mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology. 2011;140:2044–55. doi: 10.1053/j.gastro.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mastrogiannaki M, Matak P, Keith B, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest. 2009;119:1159–66. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anderson ER, Xue X, Shah YM. Intestinal hypoxia-inducible factor-2alpha (HIF-2alpha) is critical for efficient erythropoiesis. J Biol Chem. 2011;286:19533–40. doi: 10.1074/jbc.M111.238667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanchez M, Galy B, Muckenthaler MU, et al. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–6. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 107.Zimmer M, Ebert BL, Neil C, et al. Small-molecule inhibitors of HIF-2a translation link its 5'UTR iron-responsive element to oxygen sensing. Mol Cell. 2008;32:838–48. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davis MR, Shawron KM, Rendina E, et al. Hypoxia inducible factor-2 alpha is translationally repressed in response to dietary iron deficiency in Sprague-Dawley rats. J Nutr. 2011;141:1590–6. doi: 10.3945/jn.111.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson SA, Nizzi CP, Chang YI, et al. The IRP1-HIF-2alpha axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab. 2013;17:282–90. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wilkinson N, Pantopoulos K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2alpha mRNA translation. Blood. 2013;122:1658–68. doi: 10.1182/blood-2013-03-492454. [DOI] [PubMed] [Google Scholar]
- 111.Guo S, Casu C, Gardenghi S, et al. Reducing TMPRSS6 ameliorates hemochromatosis and beta-thalassemia in mice. J Clin Invest. 2013;123:1531–41. doi: 10.1172/JCI66969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gardenghi S, Ramos P, Marongiu MF, et al. Hepcidin as a therapeutic tool to limit iron overload and improve anemia in beta-thalassemic mice. J Clin Invest. 2010;120:4466–77. doi: 10.1172/JCI41717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Anderson ER, Taylor M, Xue X, et al. Intestinal HIF2alpha promotes tissue-iron accumulation in disorders of iron overload with anemia. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1314197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gattermann N. The treatment of secondary hemochromatosis. Dtsch Arztebl Int. 2009;106:499–504. doi: 10.3238/arztebl.2009.0499. I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanwar P, Kowdley KV. Diagnosis and treatment of hereditary hemochromatosis: an update. Expert Rev Gastroenterol Hepatol. 2013;7:517–30. doi: 10.1586/17474124.2013.816114. [DOI] [PubMed] [Google Scholar]
- 116.Scheuermann TH, Tomchick DR, Machius M, et al. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106:450–5. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scheuermann TH, Li Q, Ma HW, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9:271–6. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu K, Kaffes AJ. Iron deficiency anaemia: a review of diagnosis, investigation and management. Eur J Gastroenterol Hepatol. 2012;24:109–16. doi: 10.1097/MEG.0b013e32834f3140. [DOI] [PubMed] [Google Scholar]
- 119.Ganz T, Nemeth E. Iron sequestration and anemia of inflammation. Semin Hematol. 2009;46:387–93. doi: 10.1053/j.seminhematol.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nemeth E, Rivera S, Gabayan V, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–6. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Finberg KE, Heeney MM, Campagna DR, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat Genet. 2008;40:569–71. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Du X, She E, Gelbart T, et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088–92. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Finberg KE, Whittlesey RL, Fleming MD, et al. Down-regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood. 2010;115:3817–26. doi: 10.1182/blood-2009-05-224808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nai A, Pagani A, Silvestri L, et al. Increased susceptibility to iron deficiency of Tmprss6-haploinsufficient mice. Blood. 2010;116:851–2. doi: 10.1182/blood-2010-04-278655. [DOI] [PubMed] [Google Scholar]
- 125.Park TJ, Lee YJ, Kim HJ, et al. Cloning and characterization of TMPRSS6, a novel type 2 transmembrane serine protease. Mol Cells. 2005;19:223–7. [PubMed] [Google Scholar]
- 126.Silvestri L, Pagani A, Nai A, et al. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–11. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Selvaraju V, Parinandi NL, Adluri RS, et al. Molecular Mechanisms of Action and Therapeutic Uses of Pharmacological Inhibitors of HIF-Prolyl 4-Hydroxylases (PHDs) for Treatment of Ischemic Diseases. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Minamishima YA, Kaelin WG., Jr Reactivation of hepatic EPO synthesis in mice after PHD loss. Science. 2010;329:407. doi: 10.1126/science.1192811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan L, Colandrea VJ, Hale JJ. Prolyl hydroxylase domain-containing protein inhibitors as stabilizers of hypoxia-inducible factor: small molecule-based therapeutics for anemia. Expert Opin Ther Pat. 2010;20:1219–45. doi: 10.1517/13543776.2010.510836. [DOI] [PubMed] [Google Scholar]
- 130.Xue X, Taylor M, Anderson E, et al. Hypoxia-inducible factor-2alpha activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 2012;72:2285–93. doi: 10.1158/0008-5472.CAN-11-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Myllyharju J, Koivunen P. Hypoxia-inducible factor prolyl 4-hydroxylases: common and specific roles. Biol Chem. 2013;394:435–48. doi: 10.1515/hsz-2012-0328. [DOI] [PubMed] [Google Scholar]
- 133.McMullin MF. The classification and diagnosis of erythrocytosis. Int J Lab Hematol. 2008;30:447–59. doi: 10.1111/j.1751-553X.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 134.Rosove MH, Perloff JK, Hocking WG, et al. Chronic hypoxaemia and decompensated erythrocytosis in cyanotic congenital heart disease. Lancet. 1986;2:313–5. doi: 10.1016/s0140-6736(86)90005-x. [DOI] [PubMed] [Google Scholar]
- 135.Kent BD, Mitchell PD, McNicholas WT. Hypoxemia in patients with COPD: cause, effects, and disease progression. Int J Chron Obstruct Pulmon Dis. 2011;6:199–208. doi: 10.2147/COPD.S10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ogata RT, McConnell HM. Mechanism of cooperative oxygen binding to hemoglobin (spin-labeled triphosphate-concerted transition model-hemoglobin chesapeake) Proc Natl Acad Sci U S A. 1972;69:335–9. doi: 10.1073/pnas.69.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bunn HF. Subunit dissociation of certain abnormal human hemoglobins. J Clin Invest. 1969;48:126–38. doi: 10.1172/JCI105961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reed CS, Hampson R, Gordon S, et al. Erythrocytosis secondary to increased oxygen affinity of a mutant hemoglobin, hemoglobin Kempsey. Blood. 1968;31:623–32. [PubMed] [Google Scholar]
- 139.Ang SO, Chen H, Hirota K, et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat Genet. 2002;32:614–21. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 140.Percy MJ, Beer PA, Campbell G, et al. Novel exon 12 mutations in the HIF2A gene associated with erythrocytosis. Blood. 2008;111:5400–2. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Percy MJ, Furlow PW, Lucas GS, et al. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med. 2008;358:162–8. doi: 10.1056/NEJMoa073123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Percy MJ, Zhao Q, Flores A, et al. A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc Natl Acad Sci U S A. 2006;103:654–9. doi: 10.1073/pnas.0508423103. [DOI] [PMC free article] [PubMed] [Google Scholar]