Abstract
Background & Aims
The alimentary tract contains a diffuse endocrine system comprising enteroendocrine cells that secrete peptides or biogenic amines to regulate digestion, insulin secretion, food intake, and energy homeostasis. Lineage analysis in the stomach revealed that a significant fraction of endocrine cells in the gastric corpus did not arise from neurogenin3-expressing cells, unlike enteroendocrine cells elsewhere in the digestive tract. We aimed to isolate enriched serotonin-secreting and enterochromaffin-like (ECL) cells from the stomach and to clarify their cellular origin.
Methods
We used Neurod1 and Neurog3 lineage analysis, and examined differentiation of serotonin-producing and ECL cells in stomach tissues of Neurod1-cre;ROSAtdTom, Tph1-CFP, c-Kitwsh/wsh, and Neurog3Cre;ROSAtdTom mice, by immunohistochemistry. We used fluorescence-activated cell sorting to isolate each cell type for gene expression analysis. We performed RNA-seq analysis of ECL cells.
Results
Neither serotonin-secreting nor ECL cells of the corpus arose from cells expressing Neurod1. Serotonin-secreting cells expressed a number of mast cell genes, but not genes associated with endocrine differentiation; they did not develop in c-Kitwsh/wsh mice and were labeled with transplanted bone marrow cells. RNA-seq analysis of ECL cells revealed high expression levels of many genes common to endocrine cells including transcription factors, hormones, ion channels, and solute transporters but not markers of bone marrow cells.
Conclusions
Serotonin-expressing cells of the gastric corpus of mice appear to be bone marrow-derived mucosal mast cells. Gene expression analysis of ECL cells indicated that they are endocrine cells of epithelial origin that do not express the same transcription factors as their intestinal enteroendocrine cell counterparts.
Keywords: serotonin, Neurod1, enterochromaffin-like cell, histidine decarboxylase
INTRODUCTION
The alimentary tract is populated by a diffuse endocrine system comprised of enteroendocrine cells that secrete peptides or biogenic amines, accounting for 1–3% of the epithelial cells. Secreted products of enteroendocrine cells regulate digestive organ function, insulin secretion by pancreatic β cells, food intake, and energy homeostasis1, 2. Enteroendocrine cell subtypes are localized to distinct regions of the digestive tract. The histamine secreting ECL cell is the major enteroendocrine cell subtype in the acid-producing corpus along with ghrelin-, somatostatin-, and serotonin- secreting cells. As scattered as individual epithelial cells, enteroendocrine cells have been relatively inaccessible for biochemical, genetic, and functional studies that require enriched cells.
Transgenic mouse models have helped elucidate the function, origin, and differentiation of enteroendocrine cells, although gastric enteroendocrine cells, especially in the corpus have received relatively little attention3. Specification of gastric enteroendocrine cells shares some features with their intestinal counterparts. Expression of the bHLH transcription factor, Neurogenin3 (Neurog3) appears to be an early event in the specification of endocrine cells in the stomach, much like the intestine. Two studies examined gastric endocrine differentiation in Neurog3 null mice4, 5. Gastrin and somatostatin-expressing cells failed to develop in Neurog3−/− mice. However, unlike the complete absence of enteroendocrine cells in the intestine, small numbers of ghrelin, serotonin, and ECL cells were specified independently of Neurog34, 5 expression. The contribution of Neurog3 expressing cells to gastric enteroendocrine cell subtypes was difficult to determine since mice died by 2–3 days of age, when relatively small numbers of endocrine cells had differentiated.
We previously examined the cell fate of Neurog3+ cells in the digestive tract by recombination based cell lineage analysis3. Less than 50% of enteroendocrine cells in the corpus arose from Neurog3+ precursors in contrast to the pancreas, intestine, and antral stomach, where nearly all endocrine cells arose from Neurog3- expressing cells3. We found that 100% of serotonin cells in the corpus arose independently of Neurog3+ cells, as did a smaller fraction of ECL, ghrelin, and somatostatin cells. Another bHLH transcription factor, NeuroD is activated by Neurog3 and expressed in most intestinal EE cell precursors to further restrict them to the endocrine lineage6. The contribution of NeuroD+ cells in the stomach to EE cell subtypes has not been determined.
In the present work, we have further examined the origin of endocrine cells of the gastric corpus. Unexpectedly, the ECL lineage does not arise from NeuroD+ cells in the stomach unlike other digestive tract endocrine cells. We have harnessed the power of two relatively new approaches: isolation of labeled enteroendocrine cells by FACS combined with RNA sequencing. The results have provided an unprecedented in-depth view of the expressed transcriptome of histamine producing ECL cells. Isolation of enriched serotonin-expressing cells from the stomach corpus suggested that they arose from bone marrow cells in contrast to ECL cells that express many genes associated with endocrine differentiation.
MATERIALS AND METHODS
Additional Materials and Methods can be found in the Supplementary Material.
Transgenic mice
All studies using mice were approved by the IACUC at the University of Massachusetts Medical School. CFP coding sequences were inserted into BAC clones containing the genes for Tph1 (clone RP23-28A7) and Hdc (clone RP23-474H6) at the translational start site by homologous recombination in E. Coli7, 8 to generate Tph1-CFP and Hdc-CFP transgenes for pronuclear injection. The primers used for cloning are available upon request. Multiple pedigrees for each transgenic line were established.
RNA-seq sample preparation and Illumina Sequencing
For RNA sequencing, cDNA libraries were generated from RNA extracted from gastric HDC-CFP cells and HDC-CFP negative cell populations isolated by FACS. Quantity and integrity of obtained total RNAs were assessed on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). PolyA+ RNA was isolated from 100–400 ng of total RNA for RNA-seq library construction using the Illumina Truseq RNA sample preparation kit. Sequencing was performed on an Illumina HiSeq2000 instrument at the UMass deep Sequencing Core using multiplexed 50 bp single-end reads. Data output in fastq file format contained information on sequences and quality. The Sequence reads were analyzed with the FastQC program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) and all of the per-base quality scores were greater than 28. The RNA-seq reads were aligned to the Mus musculus Ensembl-NCBI37 reference genome with TopHat-v2.0.79. To quantify gene expression for the aligned reads, the HTSeq-count algorithm was applied to resulting sam files and the number of reads mapping to exons contained in the Ensembl-64 gtf were counted. Gene level count tables were processed by a custom R script loaded with the DESeq bioconductor package10. DESeq functions were used to normalize samples by their effective library size and to carry out differential expression analysis at the gene-level. For data visualization, the DESeq variance stabilization transformation was performed on read counts so that they were more amenable to plotting. Using the GOstatspackage to analyze Gene Ontology term associations, we analyzed the top 1200 up-regulated genes (using log2FC between base mean counts) to detect biological processes with over-represented genes11. To avoid inclusion of genes expressed at very low levels, we filtered the dataset to include genes with a base mean number of variance stabilized counts of greater than 100 between the two samples. Genes up regulated by >10-fold were included as differentially enriched.
Results
Serotonin and ECL cells in the corpus of the stomach do not arise from NeuroD expressing cells
In the intestine, Neurog3 expression initiates differentiation of early enteroendocrine precursors. Neurog3 induces NeuroD expression to restrict precursors to an endocrine cell fate as they mature6. To determine the contribution of NeuroD+ expressing cells to endocrine cells in the stomach, we crossed our NeuroD-Cre mice6 to the ROSA26RtdTom Cre reporter line to generate NeuroD-cre;ROSAtdTom mice.
We detected strong tdTom fluorescence at the base of the antral gastric glands exclusively in cells that expressed the endocrine marker chromograninA (ChgA), with virtually all ChgA staining cells expressing tdTom, much like the intestine6 (Fig. 1A). Similarly, all cells in the corpus that expressed NeuroD+ cells were endocrine-restricted. In contrast to the antral stomach, only 52% of the ChgA expressing endocrine cells of the corpus coexpressed tdTom fluorescence, indicating that 48% of ChgA cells did not arise from NeuroD expressing cells (Figure 1A, right).
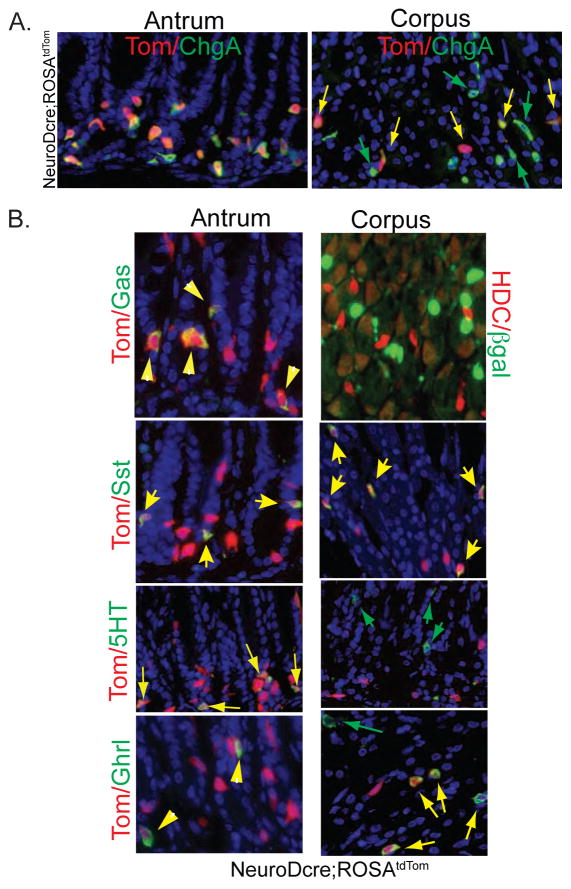
Figure 1. Contribution of NeuroD expressing cells to endocrine cells of the stomach.
(A) Expression of tdTomato in ChgA+ cells in antrum and corpus of NeuroD-cre;ROSAtdTomato mice. Absence of tdTom (green arrows) in many corpus ChgA+ cells. (B) Lineage tracing of NeuroD+ cells in gastric endocrine cell subtypes in NeuroDcre;ROSAtdTom or NeuroD-Cre:ROSA26R-LacZ mice. Yellow arrows denote endocrine cells coexpressing tdTom or LacZ; green arrows indicate endocrine cells that do not coexpress tdTom.
We further examined tdTom+ cells of NeuroD-Cre; ROSAtdTom mice to determine which endocrine subtypes arose from NeuroD expressing cells. In antral mucosa, nearly 100 % of each subtype, including gastrin-expressing G-cells, somatostatin-expressing D cells, serotonin-expressing EC cells and ghrelin-expressing cells expressed tdTom (Fig. 1B, left panels). In the corpus, all somatostatin cells and most ghrelin (65%)-cells expressed tdTom. However we were unable to identify tdTom expression in either HDC- or serotonin- expressing cells in the corpus (Figure 1B right panels), indicating that they did not arise from NeuroD expressing cells.
Serotonin cells in the corpus are related to bone marrow derived mast cells
Our prior studies examining the cell fate of Neurog 3 expressing cells in the corpus of Neurog3cre;ROSA26R-LacZ mice revealed that no cells expressing serotonin and approximately 40% of cells expressing ChgA, ghrelin, somatostatin, or HDC arose from Neurog3+ cells3. A recent study reported considerable differences in the recombination efficiency for different Cre reporter lines12. We re-examined the cell fate of Neurog3+ cells using ROSAtdTom reporter mice, considered to be the most sensitive indicator of Cre recombination. With the more efficient reporter we still failed to identify any serotonin cells in the corpus of Neurog3Cre;ROSAtdTom mice that expressed tdTom, confirming our earlier findings that they arose independently of Neurog3 (suppl. Fig. 1A). In the antrum, nearly all serotonin cells were labeled with tdTom versus 70% reported previously (Suppl. Fig. 1B). We also observed a much higher percentage of ChgA+ (67.4%) and ghrelin cells (70%), (Suppl. Fig. 1C, D) in the corpus arising from Neurog3+ cells, suggesting that earlier studies may have underestimated coexpression due to less efficient recombination.
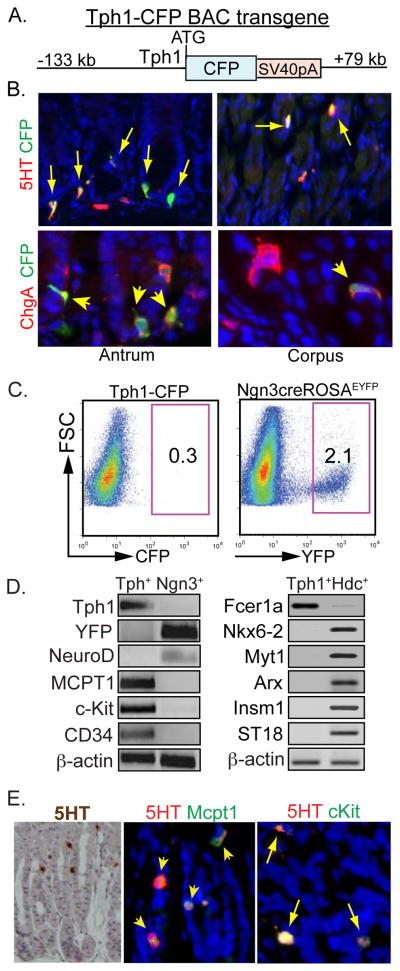
The identification of serotonin cells in the gastric corpus arising independently of Neurog3 expressing progenitor cells represents a major difference from all other enteroendocrine cells and implies they are specified by another pathway. In order to characterize this scattered cell population accounting for <0.3% of the corpus epithelium, we generated transgenic reporter mice expressing Cyan Fluorescent Protein (CFP) under the control of the gene encoding tryptophan hydroxylase 1 (Tph1), the rate-limiting enzyme in serotonin biosynthesis. The CFP coding sequences were inserted at the translational initiation site of the Tph1 gene in a BAC containing 133 kb of 5′ and 79 kb of 3′ flanking sequence (Fig. 2A). We identified CFP expression in all serotonin cells of the stomach, indicating that Tph1-CFP transgene driven expression was indistinguishable from the endogenous Tph1 gene (Fig. 2B). All CFP expressing cells expressed ChgA and as expected, ChgA+ cells that did not express CFP.
Figure 2. Expression of mast cell associated genes in serotonin cells of the corpus.
(A) Tph1-CFP BAC transgene. (B) Expression of CFP in stomach of Tph1-CFP mice stained with CFP and either 5HT or ChgA. Arrows denote double positive cells. (C) FACS enrichment of endocrine cells from the corpus of Tph1-CFP and Neurog3cre;ROSAEYFP mice (D) RT-PCR analysis of FACS-enriched cells from the corpus of Tph1-CFP compared to Neurog3Cre:ROSAEYFP or Hdc-CFP mice. (E) Coexpression of Mcpt1 and c-kit in mucosal serotonin cells.
To better characterize serotonin cells from the corpus, we prepared single cell suspensions from corpus mucosa13 of Tph1-CFP mice for subsequent enrichment by FACS. For comparison, we isolated enriched non serotonin-expressing endocrine cells from single cell suspensions from the corpus of Neurog3Cre; ROSAEYFP and from Hdc-CFP mice (described later). The endocrine cells derived from Neurog3 expressing progenitors in Neurog3Cre: ROSAEYFP mice express EYFP, accounting for approximately 2.1% of the total cell population whereas CFP+ EC cells represented only 0.3% of total (Fig. 2C).
Since we could only isolate ~3,000 live EC cells per mouse, our studies examined a limited number of transcripts by RT-PCR analysis, comparing their relative abundance in serotonin cells to other endocrine cells (Fig. 2D). The absence of Tph1 expression in the Neurog3Cre-ROSAEYFP+ cells confirmed our previous observation that serotonin cells did not arise from Neurog3 expressing cells. As expected, EYFP+ cells expressed NeuroD whereas NeuroD expression was negligible in CFP+ cells from Tph1-CFP mice. These observations indicate that corpus serotonin cells do not differentiate from cells that express either Neurog3 or NeuroD, a major difference from other enteroendocrine cells.
We next considered whether gastric corpus serotonin cells were mucosal mast cells, another cell type in the gastrointestinal tract known to produce serotonin and ChgA14–17. RNA isolated from FACS sorted serotonin cells was examined for expression of several markers of mast cell differentiation. Transcripts for Mcpt1, c-kit, Fcer1a, and CD34 were highly enriched in serotonin cells but not in the EYFP+ endocrine cells (Fig. 2D, left), or enriched ECL cells (Fig. 2D, Right), further suggesting that the serotonin-expressing cells were mucosal mast cells. To confirm that serotonin cells in the corpus were not endocrine cells, we examined enriched serotonin cells for expression of five additional transcription factors identified by RNAseq studies as highly expressed in ECL cells (described in a later section) and known to be associated with intestinal and pancreatic endocrine differentiation. Expression of these transcription factors (Nkx6-2, Myt1, Arx, Insm1, ST18) was negligible in Tph1-CFP+ cells in contrast to their expression in ECL cells (Fig 2 D) as will be described in the sections that follow.
To further characterize the gastric serotonin cells as mucosal mast cells, we examined serotonin cells in the corpus for coexpression of mast cell genes by immunohistochemistry (Fig. 2E). Most serotonin cells were embedded in the corpus epithelium (Fig. 2E, left). Nearly all serotonin cells coexpressed mast cell genes (Mcpt1, c-kit), suggesting that they were mucosal mast cells rather than connective tissue type mast cells that reside in the submucosa of the GI tract.
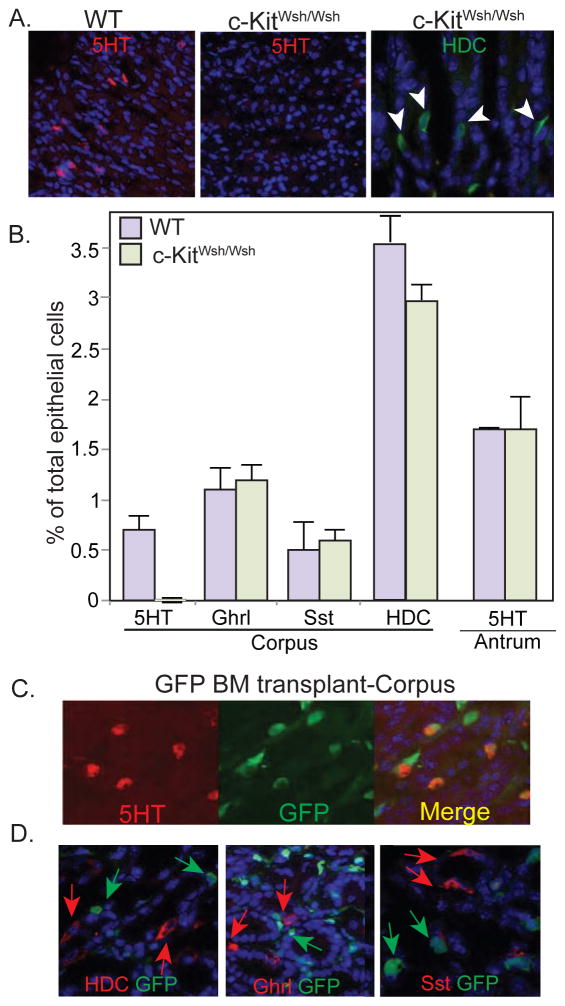
Expression of c-kit is a recognized feature of mature mast cells. A well characterized mutant c-kit allele; Wsh has a 2.8 MB genomic inversion upstream of the c-kit promoter. C-kitWsh/Wsh homozygotes show congenital absence of mast cells and melanocytes18. We examined the stomachs of c-KitWsh/Wsh mutant mice19, 20 for the presence of serotonin expressing cells in the corpus and were unable to identify serotonin cells (Fig. 3A, middle panel). Endocrine cells expressing ghrelin, somatostatin, and HDC developed normally in c-KitWsh/Wsh mice, suggesting that they were unrelated to the mast cell lineage (Fig. 3 A, B). Serotonin cells in the antral stomach were similarly unaffected in the c-KitWsh/Wsh mutants (Fig. 3B), confirming our suspicions that they were of different origin from those in the corpus. The absence of serotonin cells in the c-KitWsh/Wsh mutant mice strongly indicates that corpus serotonin cells are related to the mast cell lineage.
Figure 3. Gastric corpus serotonin cells are bone marrow derived mast cells.
(A) Corpus from WT and c-KitWsh/Wsh mutant mice immunostained for serotonin or HDC. (B) Presence of endocrine cell subtypes in c-KitWsh/Wsh mutant mice. Results shown as percentage of the total number of epithelial cells ± SD, for at least three mice. (C, D) Corpus from irradiated mice transplanted with EGFP donor bone marrow. (C) Separate and merged images showing coexpression of EGFP in serotonin cells. (D) Absence of EGFP expression in HDC, ghrelin, or somatostatin cells.
To further confirm that serotonin cells in the corpus originated from bone marrow, we transplanted lethally irradiated mice with EGFP labeled bone marrow donor cells. Twelve weeks later we examined the stomachs for the presence of EGFP labeled cells. The majority of EGFP+ cells were spindle shaped suggesting a mesenchymal origin. Approximately 78.3% of the serotonin cells in the corpus expressed EGFP suggesting that they arose from bone marrow derived donor cells (Fig. 3C). In contrast, endocrine cells arising from Neurog3+ cells including ECL cells, ghrelin cells and somatostatin cells never expressed EGFP indicating that the donor bone marrow labeling was specific for serotonin cells, confirming the c-kit mutant analysis (Fig. 3D).
Gastric ECL cells do not arise from bone marrow
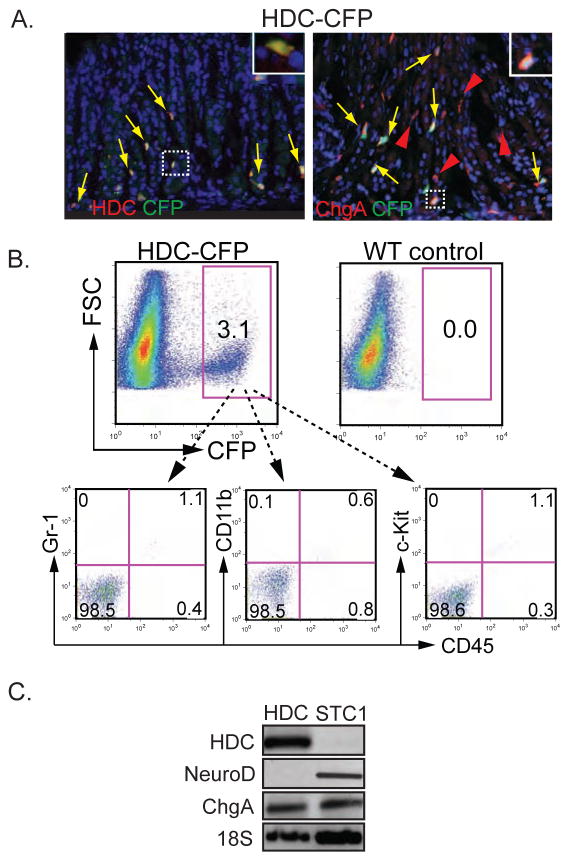
Histamine secreting ECL cells constitute a major enteroendocrine cell type in the corpus. In addition, some mast cells and immature myeloid cells express HDC to produce histamine8. We generated an Hdc transgenic reporter mouse that expressed CFP under the control of a previously described BAC spanning from −113 kb to +75 kb of the Hdc gene8 for isolating ECL cells by FACS for further analysis. CFP expressing cells in the corpus mucosa of Hdc-CFP mice coexpressed HDC protein and ChgA, indicating that transgene expression was directed to ECL cells and account for a fraction of ChgA+ cells as anticipated (Fig. 4A). FACS analysis of CFP+ ECL cells showed that ECL cells did not express c-kit. Likewise, most (>98%) of CFP+ cells in the stomach corpus did not express the bone marrow cell markers CD45, Gr-1 or CD11b (Figure 4B), unlike histamine producing CD11b+Ly6G+ immature bone marrow myeloid cells8. These observations suggest that unlike serotonin cells, ECL cells do not arise from bone marrow.
Figure 4. Enrichment and characterization of HDC+ ECL cells.
(A) Stomach of HDC-CFP mouse showing CFP expression in HDC+ and ChgA+ cells. (B) FACS analysis of CFP+ cells from the corpus of HDC-CFP mice with the indicated cell surface markers. (C) RT-PCR analysis of NeuroD gene expression in FACS-enriched HDC-CFP cells. NeuroD+ STC1 enteroendocrine cell line used as a positive control.
To confirm earlier lineage tracing studies that ECL cells did not express or arise from NeuroD+ cells, unlike most other enteroendocrine cells, we examined HDC-CFP+ cells isolated by FACS from gastric corpus mucosa for NeuroD expression by RT-PCR. In agreement with the lineage analysis, NeuroD transcripts were absent from enriched ECL cells but easily detected in the intestine endocrine STC1 cell line (Fig. 4C).
Sequencing the gastric ECL cell transcriptome
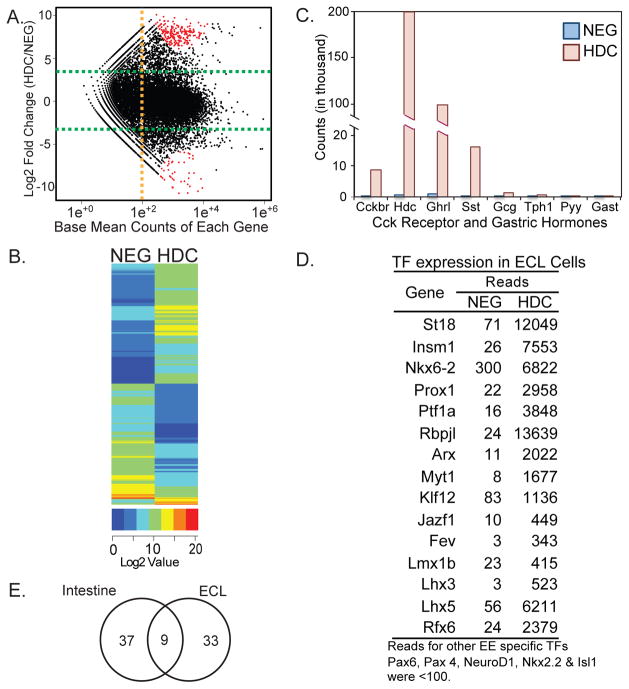
Overlapping expression of some “markers” in enteroendocrine cells, mucosal mast cells, and bone marrow derived cells complicates their use as markers for categorizing the origin of ECL and other endocrine cells of the stomach. To identify the characteristic features that define ECL cells as endocrine cells, we have sequenced the transcriptome of HDC expressing cells enriched from the corpus of HDC-CFP mice. Using Illumina RNA-seq technology we have compared the transcriptomes of HDC+ (CFP+) cells to HDC− (CFP−) control cells isolated by FACS. The abundance of HDC+ cells, which comprise up to 3.2% of sorted cells made it feasible to construct cDNA libraries from 150 ng (HDC-CFP+) and 400 ng (CFP negative) total RNAs pooled from 6 mice. Libraries were sequenced to a depth of up to 55 million variance normalized read counts mapped to 22,706 genes. Variance normalized sequence counts for several genes showed of good correlation with their expression measured by qPCR of RNA from HDC+ cells (Suppl. Fig. 2). To identify differentially expressed genes in enriched in HDC+ versus HDC− cells, we filtered out genes expressed at low levels. We identified 1,449 genes that were differentially expressed, upregulated by 10-fold to 830-fold in HDC+ versus HDC− cells (Fig. 5A). A map of the ~1,449 genes clustered by their expression values further illustrates the extensive differences in the gene expression profiles of HDC+ cells and their HDC− counterparts (Fig. 5B).
Figure 5. RNA-seq analysis of enriched ECL cells.
(A) Smear plot showing distribution of up- and down- regulated genes between Hdc-CFP+ and CFP− cells. (B) Heat map representation of genes differentially expressed between CFP+ and CFP− (NEG) cells. Scaled expression values of all 1,449 DEGs are shown for each group. (C) Variance normalized read counts for selected hormone and receptor genes in HDC+ and CFP− cells, (D) Transcription factors highly enriched in HDC+ cells. (E) Limited overlap of transcription factor expression in intestinal endocrine cells and HDC+ cells in the stomach. Only 9 ECL enriched TF genes were upregulated in intestinal enteroendocrine cells.
We also analyzed RNA-seq data for expression of other gastrointestinal hormones in HDC+ cells (Fig. 5C). Ghrelin (Ghrl) and somatostatin (Sst) were expressed at relatively high levels in HDC+ cells with 8–10 fold greater expression compared to HDC− cells. We confirmed expression of ghrelin and somatostatin in HDC+ cells by immunostaining and identified a subset of HDC+ cells that coexpressed ghrelin or somatostatin (Suppl. Fig. 3), indicating a potential relationship between these three enteroendocrine cell subtypes in the corpus. Other endocrine marker genes were expressed at low levels (Tph1, glucagon) or negligible levels (Pyy, substance P, secretin, CCK, and GIP), indicating relatively very limited overlap of hormone expression with enteroendocrine cells of the intestine (Fig. 5C). In addition, the absence of gastrin expression suggests minimal contamination with antral tissue.
Expression profile of HDC+ cells
Since HDC− cells are a heterogeneous population, comprised of parietal, chief, mucous and non-HDC endocrine cell types, we focused on the transcripts that showed the greatest absolute fold change in expression in HDC+ cells. Gene Ontology (GO) enrichment analysis showed significant overrepresentation of multiple GO terms in the HDC+ cells for multiple biological processes related to hormone synthesis and secretion, regulated secretion, neurotransmitter secretion, and neuronal differentiation and function (Suppl. Table 1, Suppl. Table 2). The enriched GO categories included upregulated expression of genes involved in endocrine function, including hormones and proteins associated with their secretion, members of the SNARE complex, ion channels and transporters, solute transporters, and transcription factors.
From the top 150 upregulated transcripts in HDC+ cells, we have listed 25 in Table 1 to illustrate the diversity of differentially expressed genes. The examples belong to categories of commonly expressed genes in cells with regulated secretory function like enteroendocrine cells. Three members of the granin family of regulated secretory proteins (chromogranin A, chromogranin B, and secretogranin 5) were expressed at very high and enriched levels in HDC+ cells.
Table 1.
Selected 25 differentially expressed genes from top 150 upregulated genes in HDC cells
| Gene | Description | Counts |
Fold Change | ||
|---|---|---|---|---|---|
| NEG | HDC | ||||
| 1 | Kcnmb2 | potassium large conductance calcium-activated channel, subfamily M, beta member 2 | 14 | 7425 | 532 |
| 2 | Insrr | insulin receptor-related receptor | 78 | 39462 | 503 |
| 3 | Hdc | histidine decarboxylase | 511 | 198375 | 388 |
| 4 | Chga | chromogranin A | 3944 | 1450581 | 368 |
| 5 | Kcnh6 | potassium voltage-gated channel, subfamily H (eag-related), member 6 | 30 | 9320 | 316 |
| 6 | Insm1 | insulinoma-associated 1 | 26 | 7553 | 293 |
| 7 | Slc18a2 | solute carrier family 18 (vesicular monoamine), member 2 | 283 | 77130 | 276 |
| 8 | Sv2a | Synaptic vesicle glycoprotein 2A | 15 | 4010 | 276 |
| 9 | Syt5 | Synaptotagmin 5 | 28 | 7275 | 255 |
| 10 | Cadps | Calcium dependent secretion activator | 20 | 5062 | 254 |
| 11 | Slc29a4 | solute carrier family 29 (nucleoside transporters), member 4 | 22 | 5364 | 244 |
| 12 | Chgb | chromogranin B | 582 | 140324 | 241 |
| 13 | Ncam1 | neural cell adhesion molecule 1 | 12 | 2882 | 233 |
| 14 | Scn3a | sodium channel, voltage-gated, type III, alpha | 7 | 1610 | 231 |
| 15 | Myt1 | myelin transcription factor 1 | 8 | 1677 | 223 |
| 16 | Slc7a14 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 | 22 | 4777 | 217 |
| 17 | Gck | glucokinase | 10 | 2188 | 214 |
| 18 | Pcsk1 | proprotein convertase subtilisin/kexin type 1 | 70 | 14918 | 214 |
| 19 | Cacna1h | Voltage-dependent T-type calcium channel subunit alpha-1H | 36 | 7234 | 198 |
| 20 | Snap25 | synaptosomal-associated protein 25 | 49 | 9404 | 197 |
| 21 | Slc16a12 | solute carrier family 16 (monocarboxylic acid transporters), member 12 | 9 | 1674 | 195 |
| 22 | Arx | aristaless related homeobox | 11 | 2022 | 188 |
| 23 | Stx1a | syntaxin 1A | 16 | 2973 | 185 |
| 24 | Scg5 | secretogranin 5 | 132 | 23458 | 178 |
| 25 | St18 | suppression of tumorigenicity 18 | 71 | 12049 | 169 |
The variance normalized read counts for each expressed gene in HDC+ and HDC− (NEG) cells. Differential expression was calculated using the ratio of read counts for HDC+ over NEG and ranked by the absolute fold change.
Histidine decarboxylase showed the nearly 400-fold enrichment in HDC+ cells. A number of solute transporters were differentially expressed in HDC+ cells including Slc18a2 (VMAT2), which is commonly expressed in neuroendocrine cells. Several voltage-gated sodium and potassium channels, calcium activated potassium channels, and calcium channels as well as members of the SNARE complex including Syntaxin 1a, Snap25, and synaptotagmin 5 are listed in Table 1. Further analysis showed expression levels that were increased 10- and 532-fold for 15 potassium channels, 8 calcium channels, 5 sodium channels, 28 solute transporter genes, as well as increased expression of genes associated with endocrine cell function including glucokinase (Gck) and prohormone convertase 1 (Pcsk1).
Transcription factors represent another class of genes showing differentially upregulated expression that may drive the differentiation of ECL cells. We queried our dataset for expression data on approximately 1200 transcription factor genes, filtering out factors expressed at relatively low expression levels (<100 mean counts). We identified 33 transcription factors that were upregulated 10 to 564 fold in ECL cells relative to the HDC− control cells, (Fig. 5D). The selected genes included many of the major structural classes of transcription factors associated with development and differentiation, including proteins with homeodomains, winged helix motifs, bHLH-PAS proteins, zinc finger proteins, nuclear receptors, LIM-homeo domain proteins, and basic helix loop helix proteins.
Of the proteins identified, most have not been previously implicated in digestive tract or pancreatic endocrine differentiation. Notably, several transcription factors previously implicated in intestinal endocrine cell differentiation (Neurog3, Isl1, Nkx2-2, Pax4, NeuroD, and Pax6) were absent or expressed at negligible levels in HDC+ cells. The low level of Neurog3 expressed was anticipated, since Neurog3 mRNA is relatively unstable, resulting in transient expression in enteroendocrine precursor cells, prior to the expression of endocrine markers.
ECL cells play an important role in stimulating acid secretion by parietal cells through gastrin stimulated release of histamine. Consistent with this function, the Cckb receptor was enriched and expressed at high levels in HDC+ cells (Fig. 4C). Expression of Gipr (GIP receptor) was 81-fold greater in ECL cells than HDC- control cells, potentially accounting for the effects of GIP on inhibiting acid secretion. We identified 500-fold increased expression of Insrr21, 22, a member of the insulin receptor family, in ECL cells. Insrr has been previously identified in islets and more recently has been shown to function as pH sensing receptor in the kidney.
Several serine proteases associated with connective tissue type mast cells were expressed in HDC+ cells at relatively low levels with limited enrichment over HDC− cells (Table 2). More importantly, HDC+ cells did not express the proteins CD34 and Fcer1a, which are produced in most mast cell types. ECL cells showed moderate to high expression of a number of keratins, tight junction associated proteins, and cell adhesion molecules expressed in epithelial cells (Table 2, upper). GO enrichment analysis indicated that genes related to biological processes involved in mast cell function, and the immune response were significantly overrepresented in HDC− cells compared to HDC+ cells (Suppl. Table 1B) indicating that the gastric mast cells did not sort with HDC+ cells.
Table 2.
Epithelial and mast cell markers in HDC and NEG cells
| Gene | Description | Counts |
||
|---|---|---|---|---|
| NEG | HDC | |||
| Epithelial cell marker | Alcam | activated leukocyte cell adhesion molecule | 3476 | 32167 |
| Dsp | desmoplakin | 29603 | 14144 | |
| Cdh1 | cadherin 1 | 38851 | 13993 | |
| Epcam | epithelial cell adhesion molecule | 5521 | 10251 | |
| Cldn18 | claudin 18 | 27967 | 9145 | |
| Krt7 | keratin 7 | 308 | 8041 | |
| Krt18 | keratin 18 | 25951 | 5760 | |
| Krt8 | predicted gene 5604; keratin 8 | 49614 | 5105 | |
| Krt19 | keratin 19 | 80864 | 3444 | |
| Prom1 | prominin 1 | 4469 | 2621 | |
| Prlr | prolactin receptor | 96 | 1992 | |
| Muc1 | mucin 1, transmembrane | 13430 | 944 | |
| Cldn3 | claudin 3 | 18 | 665 | |
| Plau | plasminogen activator, urokinase | 499 | 21 | |
|
| ||||
| Mast cell genes | Kit | kit oncogene | 774 | 1346 |
| Mcpt2 | mast cell protease 2 | 506 | 1093 | |
| Tpsab1 | tryptase alpha/beta 1 | 537 | 1009 | |
| Mcpt1 | mast cell protease 1 | 535 | 894 | |
| Cpa3 | mast cell carboxypeptidase A3 | 136 | 665 | |
| Mcpt4 | mast cell protease 4 | 24 | 620 | |
| Tpsb2 | tryptase beta 2 | 128 | 603 | |
| Cma1 | chymase 1, mast cell | 30 | 439 | |
| Cd34 | CD34 antigen | 32 | 43 | |
| Fcer1a | Fc receptor, IgE, high affinity I | 10 | 37 | |
| Mcpt8 | mast cell protease 8 | 2 | 13 | |
| Cma2 | chymase 2, mast cell | 1 | 7 | |
| Mcpt9 | mast cell protease 9; mast cell protease-like | 4 | 2 | |
| Il2ra | interleukin 2 receptor, alpha chain | 0 | 0 | |
| Prss34 | protease, serine, 34 | 0 | 0 | |
Variance normalized counts for each expressed gene shown in HDC cells (NEG) and HDC+(HDC) cells.
DISCUSSION
Two recent studies have examined hormone gene expression in enriched enteroendocrine cells isolated by FACS from transgenic reporter mice for CCK23, GIP24, and glucagon24. Both studies revealed expression of multiple hormone gene transcripts including GIP, CCK, secretin, and glucagon transcripts in enriched populations. Microarray expression profiling showed overlapping expression that was dependent on the region of the intestine examined24. In the stomach body, we have identified limited overlapping expression between HDC, ghrelin, and somatostatin cells. However, corpus ECL cells show is little overlapping hormone expression with the intestine enteroendocrine cells.
In addition to enteroendocrine cells, mast cells8, 16 and immature myeloid8, 16 cells secrete serotonin and/or histamine. We unexpectedly found that mucosal serotonin cells in the corpus were mast cells, expressing genes associated with mast cells, but not enteroendocrine cells. Their mast cell origin was subsequently established by the absence of corpus serotonin-expressing cells in c-Kitwsh/wsh mutant mice and further confirmed by bone marrow transplantation. The labeling of serotonin cells with donor cells probably underestimated the contribution of bone marrow as we did not account for post-transplant chimerism or for cells that may turnover very slowly. In the human corpus, serotonin cells are relatively rare and do not stain for c-kit (not shown). Unlike the mouse, mucosal mast cells are extremely rare in the human corpus.
Unlike serotonin cells, our work indicates that ECL cells did not arise from bone marrow. The expressed transcriptome of gastric ECL cells shows enriched expression of genes associated with endocrine differentiation, including hormones, ion channels, solute transporters, and transcription factors. A recent study examined expression of several stomach genes by qPCR in FACS isolated cells from the stomach of HDC-Cre; ROSATom mice25. From enriched expression of Tph1, and tryptase beta2, the authors suggested that gastric HDC cells were mast cells. However, in this study, mast cell markers were only seen in lamina propria and not in the mucosa unlike the present work. The cell population was isolated on the basis of recombination in a Cre reporter by Hdc-Cre rather than by HDC expression (as a reporter) as described here. Since tomato red expression was driven by the ROSA26 locus, rather than by HDC, it is likely that some of the sorted cells arose from cells that transiently expressed HDC in bone marrow. Our RNA-seq studies showed expression of a small number of proteases at relatively low levels in ECL cells with limited enrichment. GO enrichment analysis did not identify overrepresentation of genes associated with mast cell differentiation in HDC+ cells whereas a number of biological processes associated with mast cells and immune function were overrepresented in HDC− cells, suggesting that mast cells in the stomach remained in the HDC− cell population during sorting. Instead, GO analysis identified highly significant enrichment of GO terms associated with endocrine function in HDC+ cell population.
Isolation of highly enriched populations of serotonin cells and ECL cells from the gastric corpus by FACS enabled us to directly study genes expressed in these two previously inaccessible cell types that differentiate independently of NeuroD and possibly Neurog3 unlike other enteroendocrine cells. We identified a number of transcription factors enriched between 10 and 300- fold that may function as potential drivers of HDC cell differentiation. Some have been previously associated with enteroendocrine cell differentiation including Insm126, Arx27, and Fev28 Several others, including, Nkx6-229, Prox130, Rfx631, Myt132, Rfx333, Insm126, Arx27, and St1834 contribute to development of the endocrine pancreas. Over 70% of the enriched transcription factors including Lhx3, Lhx5, Lmx1b, and Klf12, have not been implicated in differentiation of gut or pancreatic endocrine cells. A number of these transcription factors expressed at high levels in ECL cells have been implicated in brain development and neuronal differentiation, including ARX, Myt1, Lhx3, Lhx5, and Lmx1b. Similarities between neurons and digestive tract endocrine cells are widely recognized despite their origins in different germ layers. These similarities included regulated secretion and transcription factors required for their differentiation, e.g. NeuroD, Neurog3. Several transcription factors previously identified in enteroendocrine cells including Pax4, Pax6, Isl1, Nkx2.2, and NeuroD were not expressed in ECL cells.
Surprisingly, transcription factors that are selectively expressed in ECL cells have little overlap with those in intestinal enteroendocrine cells24 (Fig. 5E). Low expression levels of intestinal transcription factors in ECL cells may restrict their differentiation to the stomach expression and account for the relatively limited overlapping expression of intestinal hormones. Likewise, stomach and ECL specific transcription factors not expressed in the intestine may account for the absence of Hdc expression in the intestine.
For several decades, histamine secreted by ECL cells has been identified as the major stimulus of gastric acid secretion by parietal cells35. Gastrin plays a major role in regulating histamine release by binding to its receptor on ECL cells rather than from direct effects on parietal cells. The introduction of histamine 2 receptor antagonists represented a major breakthrough for treating diseases related to acid hypersecretion by blocking the effects of ECL cell secreted histamine. The majority of gastric carcinoid tumors are slow growing neoplasms associated with hypergastrinemia from atrophic gastritis (type1) and gastrinomas (type 2). Unlike midgut neuroendocrine tumors, gastric carcinoids arise from ECL cells and rarely produce serotonin, consistent with our present findings that ECL cells but not serotonin-producing cells in the stomach are of neuroendocrine origin. As expected the present work confirms that the CCKB (gastrin) receptor is highly enriched in ECL cells. The identification of insulin receptor related receptor (INSRR), as a highly expressed gene in ECL cells, may be of considerable interest since it has been shown to be a pH sensing receptor in the kidney that could have a role in regulating gastric acid secretion 21, 22. This latter finding is one example of the potential power of RNA sequencing to define the transcriptome of enteroendocrine cells in far greater detail than previously possible.
Supplementary Material
Acknowledgments
Grant Support: Caring for Carcinoid Foundation (ABL), NIH DK90000
We thank Timothy Wang and Walden Ai for providing the HDC-BAC and cloning oligonucleotides. We acknowledge the assistance of the Transgenic Animal Model, Flow Cytometry, and Deep Sequencing Cores at the University of Massachusetts Medical School.
FUNDING
Supported by National Institutes of Health grants R01DK9000 and an award from the Caring for Carcinoid Foundation (to ABL)
Abbreviations
- EC
enterochromaffin
- ECL
enterochromaffin-like
- Neurog3
Neurogenin3
- bHLH
basic helix loop helix
- BAC
bacterial artificial chromosome
- ChgA
chromogranin A
- HDC
histidine decarboxylase
- CFP
cyan fluorescent protein
- tdTom
tandem repeat Tomato gene
- 7-AAD
7-amino-actinomycin D
- EE
enteroendocrine
- FACS
fluorescent activated cell sorting
- GI
gastrointestinal
- DEG
differential expressed genes
- RT-PCR
reverse transcription polymerase chain reaction
- YFP
yellow fluorescent protein
- VAMP
vesicle-associated membrane protein
Footnotes
Disclosures: None
Author contributions:
HJL Study concept and design, data acquisition, data analysis; BJ data acquisition; data analysis; DA, data analysis and interpretation, statistical analysis. DC data analysis and interpretation, statistical analysis; MGM, GR, data acquisition; ABL study concept and design, data analysis, critical revision of manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schubert ML. Stomach and duodenum. Curr Opin Gastroenterol. 2002;18:637–8. doi: 10.1097/00001574-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature. 2006;444:854–9. doi: 10.1038/nature05484. [DOI] [PubMed] [Google Scholar]
- 3.Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–54. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Jenny M, Uhl C, Roche C, et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO Journal. 2002;21:6338–47. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CS, Perreault N, Brestelli JE, et al. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes and Development. 2002;16:1488–97. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HJ, Kapoor A, Giel-Moloney M, et al. Notch signaling differentially regulates the cell fate of early endocrine precursor cells and their maturing descendants in the mouse pancreas and intestine. Dev Biol. 2012;371:156–69. doi: 10.1016/j.ydbio.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu D, Ellis HM, Lee EC, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5978–83. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang XD, Ai W, Asfaha S, et al. Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nature Medicine. 2011;17:87–95. doi: 10.1038/nm.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Willet SG, Bankaitis ED, et al. Non-parallel recombination limits cre-loxP-based reporters as precise indicators of conditional genetic manipulation. Genesis. 2013;51:436–42. doi: 10.1002/dvg.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain RN, Brunkan CS, Chew CS, et al. Gene expression profiling of gastrin target genes in parietal cells. Physiol Genomics. 2006;24:124–32. doi: 10.1152/physiolgenomics.00133.2005. [DOI] [PubMed] [Google Scholar]
- 14.Prasad P, Yanagihara AA, Small-Howard AL, et al. Secretogranin III directs secretory vesicle biogenesis in mast cells in a manner dependent upon interaction with chromogranin A. Journal of Immunology. 2008;181(7):5024–34. doi: 10.4049/jimmunol.181.7.5024. [DOI] [PubMed] [Google Scholar]
- 15.Marathias K, Lambracht-Hall M, Savala J, et al. Endogenous regulation of rat brain mast cell serotonin release. International Archives of Allergy and Applied Immunology. 1991;95:332–40. doi: 10.1159/000235470. [DOI] [PubMed] [Google Scholar]
- 16.Ringvall M, Ronnberg E, Wernersson S, et al. Serotonin and histamine storage in mast cell secretory granules is dependent on serglycin proteoglycan. Journal of Allergy and Clinical Immunology. 2008;121:1020–6. doi: 10.1016/j.jaci.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 17.Kushnir-Sukhov NM, Brown JM, Wu Y, et al. Human mast cells are capable of serotonin synthesis and release. Journal of Allergy and Clinical Immunology. 2007;119:498–9. doi: 10.1016/j.jaci.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Tono T, Tsujimura T, Koshimizu U, et al. c-kit Gene was not transcribed in cultured mast cells of mast cell-deficient Wsh/Wsh mice that have a normal number of erythrocytes and a normal c-kit coding region. Blood. 1992;80:1448–53. [PubMed] [Google Scholar]
- 19.Duttlinger R, Manova K, Chu TY, et al. W-sash affects positive and negative elements controlling c-kit expression: ectopic c-kit expression at sites of kit-ligand expression affects melanogenesis. Development. 1993;118:705–17. doi: 10.1242/dev.118.3.705. [DOI] [PubMed] [Google Scholar]
- 20.Chen CC, Grimbaldeston MA, Tsai M, et al. Identification of mast cell progenitors in adult mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11408–13. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deyev IE, Sohet F, Vassilenko KP, et al. Insulin receptor-related receptor as an extracellular alkali sensor. Cell Metab. 2011;13:679–89. doi: 10.1016/j.cmet.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raile K, Klammt J, Laue S, et al. Glucose concentration and AMP-dependent kinase activation regulate expression of insulin receptor family members in rat islets and INS-1E beta cells. Diabetologia. 2005;48(9):1798–809. doi: 10.1007/s00125-005-1860-x. [DOI] [PubMed] [Google Scholar]
- 23.Egerod KL, Engelstoft MS, Grunddal KV, et al. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153:5782–95. doi: 10.1210/en.2012-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib AM, Richards P, Cairns LS, et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153:3054–65. doi: 10.1210/en.2011-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker AK, Park WM, Chuang JC, et al. Characterization of gastric and neuronal histaminergic populations using a transgenic mouse model. PLoS One. 2013;8:e60276. doi: 10.1371/journal.pone.0060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gierl MS, Karoulias N, Wende H, et al. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 2006;20:2465–78. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beucher A, Gjernes E, Collin C, et al. The homeodomain-containing transcription factors Arx and Pax4 control enteroendocrine subtype specification in mice. PLoS One. 2012;7:e36449. doi: 10.1371/journal.pone.0036449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YC, Zuraek MB, Kosaka Y, et al. The ETS oncogene family transcription factor FEV identifies serotonin-producing cells in normal and neoplastic small intestine. Endocr Relat Cancer. 2010;17:283–91. doi: 10.1677/ERC-09-0243. [DOI] [PubMed] [Google Scholar]
- 29.Sander M, Sussel L, Conners J, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2. 2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–40. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Kilic G, Aydin M, et al. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev Biol. 2005;286:182–94. doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Soyer J, Flasse L, Raffelsberger W, et al. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137:203–12. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Wang S, Zhang J, et al. The fringe molecules induce endocrine differentiation in embryonic endoderm by activating cMyt1/cMyt3. Dev Biol. 2006;297:340–9. doi: 10.1016/j.ydbio.2006.04.456. [DOI] [PubMed] [Google Scholar]
- 33.Ait-Lounis A, Bonal C, Seguin-Estevez Q, et al. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010;59:1674–85. doi: 10.2337/db09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tennant BR, Islam R, Kramer MM, et al. The transcription factor Myt3 acts as a pro-survival factor in beta-cells. PLoS One. 2012;7:e51501. doi: 10.1371/journal.pone.0051501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schubert ML, Peura DA. Control of gastric acid secretion in health and disease. Gastroenterology. 2008;134:1842–60. doi: 10.1053/j.gastro.2008.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.