Abstract
Developmental defects and disruption of molecular pathways of the cardiac conduction system (CCS) can cause life-threatening cardiac arrhythmias. Despite decades of effort, knowledge about the development and molecular control of the CCS remains primitive. Mouse genetics, complementary to other approaches such as human genetics, has become a key tool for exploring the developmental processes of various organs and associated diseases. Genetic analysis using mouse models will likely provide great insights about the development of the CCS, which can facilitate the development of novel therapeutic strategies to treat arrhythmias. To enable genetic studies of the CCS, CCS-associated Cre mouse models are essential. However, existing mouse models with Cre activity reported in the CCS have various limitations such as Cre leak, haploinsufficiency, and inadequate specificity of the Cre activity. To circumvent those limitations, we successfully generated Hcn4-CreERT2 BAC transgenic mice using BAC recombineering in which Cre activity was specifically detected in the entire CCS after tamoxifen induction. Our Hcn4-CreERT2 BAC transgenic line will be an invaluable genetic tool with which to dissect the developmental control of CCS and arrhythmias.
Keywords: Cardiac conduction system, inducible Cre line, Hcn4-CreERT2, BAC
Heart growth and its proper function depend on the coordinated development of various cardiac cell types (Garg, 2006; McCulley and Black, 2012; Srivastava, 2006). The early heart tube contracts peristaltically to pump blood to support embryo growth. As it develops further, the heart undergoes growth and chamber septation and, eventually remodels into a four-chambered structure. During these developmental processes, a tiny pool of specialized cells, constituting the cardiac conduction system (CCS), orchestrates coordinated, sequential cardiac contraction (Christoffels et al., 2010; Mikawa and Hurtado, 2007). The CCS consists of the SAN (sinoatrial node), AVN (atrioventricular node), His bundle, bundle branches, and Purkinje fibers. It has been established that the His-Purkinje system has a myogenic origin, but how it develops and assembles into a fast CCS is debated (Christoffels and Moorman, 2009; Gourdie et al., 1995). Recent retrospective clonal analysis support the notion that common progenitors give rise to His-Purkinje cells and contractile cardiomyocytes, and these lineage-restricted cells then proliferate (Miquerol et al., 2010). Although exciting progress has been made, numerous questions remain to be answered. For example, the cell lineage of the SAN is not certain (Bressan et al., 2013; Liang et al., 2013).
Coordinated cardiac contraction is vital for the heart to pump blood efficiently. In the adult heart, the SAN initiates the action potential, which travels through the atria to the AVN, where the electrical signal is delayed. The electrical signal then rapidly travels to the His bundle, bundle branches, and Purkinje fibers, eventually traveling throughout the ventricles to trigger ventricular contraction.
Cardiac arrhythmias can be life-threatening. Developmental abnormalities of CCS lineages may be associated with arrhythmogenic regions in adult hearts (Jongbloed et al., 2012). Disorders of the CCS include sick sinus syndrome, heart block, AV nodal reentry (Park and Fishman, 2011). Ventricular arrhythmias such as idiopathic fascicular tachycardia and arrhythmias in patients after myocardial infarction can arise in the ventricular conduction system (Scheinman, 2009). While it is well established that dysregulation of ion channels or their mutations can cause arrhythmias, how developmental defects and disruptions of molecular control of CCS cause arrhythmias is not clear.
Although various components of the CCS were discovered more than 100 years, our understanding of the developmental and molecular control of the CCS remains limited (Christoffels and Moorman, 2009; Munshi, 2012). Several transcription factors such as Tbx3, Tbx5, Tbx18, Id2, Irx3, Nkx2.5, Shox2, and Notch have been identified as playing important roles in the CCS (Blaschke et al., 2007; Espinoza-Lewis et al., 2009; Frank et al., 2012; Hoogaars et al., 2007; Moskowitz et al., 2007; Moskowitz et al., 2004; Rentschler et al., 2011; Wiese et al., 2009; Zhang et al., 2011). A complete delineation of the developmental control of CCS relies on specific genetic manipulations such as loss-of-function or gain-of-function studies. Over the past several years, several Cre lines with Cre activity in the CCS have been reported (Arnolds and Moskowitz, 2011; Beyer et al., 2011; Hoesl et al., 2008; Liang et al., 2013; Sun et al., 2013). However, these lines have several limitations. First, in all of these lines except for the minK-CreERT2 line, a knock-in strategy was used. Given that Hcn4, Cx40, and Shox2 play key roles in CCS, haploinsufficiency is a concern when using these knock-in mice to delete important genes in the CCS as noted previously (Liang et al., 2013). In this situation the haploinsufficiency can complicate the interpretation of genetic deletion in CCS. Second, Cre activity is not specifically in the CCS but in other cardiac cells as well. For example, Cx40 encodes a gap junction protein responsible for the rapid propagation of cardiac action potential in the His-Purkinje system and for cell-cell communication in arterial endothelial cells. As such, Cre activity is seen in His-Purkinje system and arterial endothelial cells in Cx40-CreERT2 mice. Third, leakage of Cre activity limits the usefulness of the minK-CreERT2 line for certain applications such as lineage tracing. Finally, Cre activity in Cx40-CreERT2, minK-CreERT2, and Shox2-Cre is only seen in part of the CCS. A comparison between these Cre lines is summarized in the Table 1.
Table 1.
Comparison of various Cre and inducible Cre lines with reported Cre activity in the CCS.
| Cre lines | Cre activity |
KI | BAC | Leaky | other cardiac expression |
reference | |||
|---|---|---|---|---|---|---|---|---|---|
| SAN | AVN | His/BB | Purkinje | ||||||
| Hcn4-CreERT2 | + | + | + | + | yes | no | no | no | Hoesl et al., 2008 |
| Hcn4-CreERT2 | + | + | + | + | yes | no | no | no | Liang et al. 2013 |
| Cx40-CreERT2 | − | − | + | + | yes | no | no | yes | Beyer et al., 2011 |
| minK-CreERT2 | − | + | + | + | no | yes | yes | no | Arnolds et al., 2011 |
| Shox2-Cre | + | n/a | +/− | − | yes | no | − | n/a | Sun et al., 2013 |
| Hcn4-CreERT2 | + | + | + | + | no | yes | no | no | This report |
SAN: sinoatrial node; AVN: atrioventricular node; BB: bundle branches; KI: knockin
To elucidate developmental control of the CCS, we sought to generate a CCS-specific, inducible Cre line without those limitations. For this purpose, we chose Hcn4 as a marker of the CCS. Hcn4 belongs to a family of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels that consists of Hcn1-4. Hcn4 is specifically expressed in the entire CCS in all species examined so far. The voltage-gated ion channel Hcn4 mediates the inward sodium-potassium current and is responsible for initiating the diastolic depolarization of SAN cells (DiFrancesco, 2010). Human genetic studies showed that mutations in HCN4 cause bradycardia and sick sinus syndrome (DiFrancesco, 2010; Herrmann et al., 2007). To avoid haploinsufficiency, we chose a transgenic strategy using a bacterial artificial chromosome (BAC) as BACs are likely to contain all of the regulatory elements necessary to confer endogenous expression in vivo (Gong et al., 2003). A CreERT2 cassette was selected to control Cre recombinase activity in a tempo-spatial fashion. CreERT2 encodes a Cre recombinase fused to a mutant estrogen ligand-binding domain (ERT2). In the presence of the estrogen receptor antagonist tamoxifen, CreERT2 rapidly relocates into the nucleus to excise LoxP-flanked DNA regions.
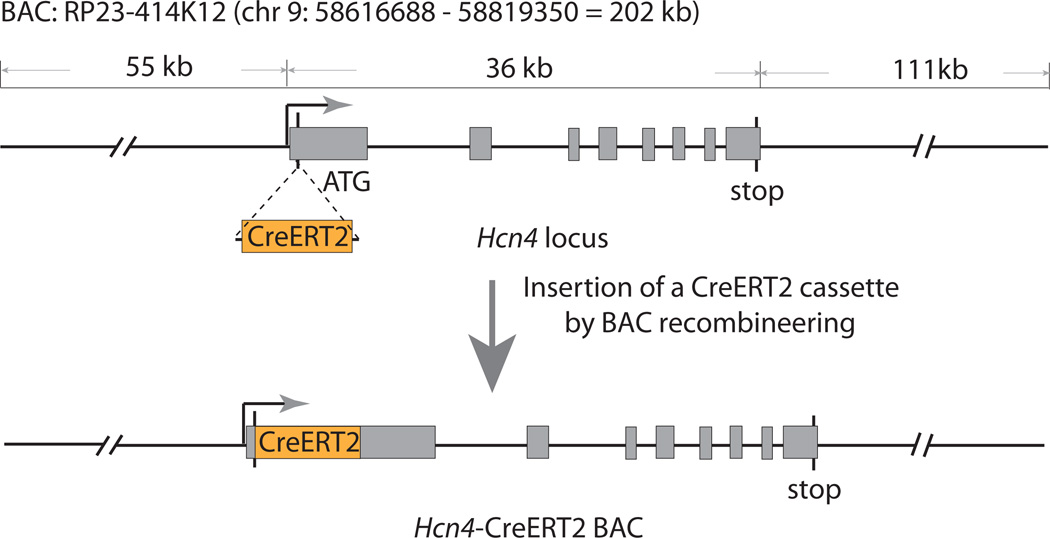
To develop the Hcn4-CreERT2 BAC transgenic mice, a 200-kb BAC containing Hcn4 locus (RP23-414K12) was chosen to direct the expression of CreERT2 (FIG. 1). This BAC does not contain other protein-coding genes or microRNAs, which excludes the over-expression of other genes in the resulting transgenic mice. A CreERT2 cassette was inserted into the Hcn4 locus in the BAC by BAC recombineering (FIG. 1) (Warming et al., 2005). The successfully engineered BAC was purified and used for pronuclear injection of fertilized eggs. Two independent founders were generated from the injection. Both transgenic founders showed essentially identical Cre activity after tamoxifen induction so we selected one of the lines for the detailed analysis described below.
FIG. 1.
Generation of the Hcn4-CreERT2 BAC by recombineering. A CreERT2 cassette with a poly(A) tail was specifically inserted immediately after first ATG of Hcn4 locus in BAC using an homologous recombination-based technique. The correct insertion and integrity of modified BAC was verified by Sanger sequencing before pro-nuclear injection.
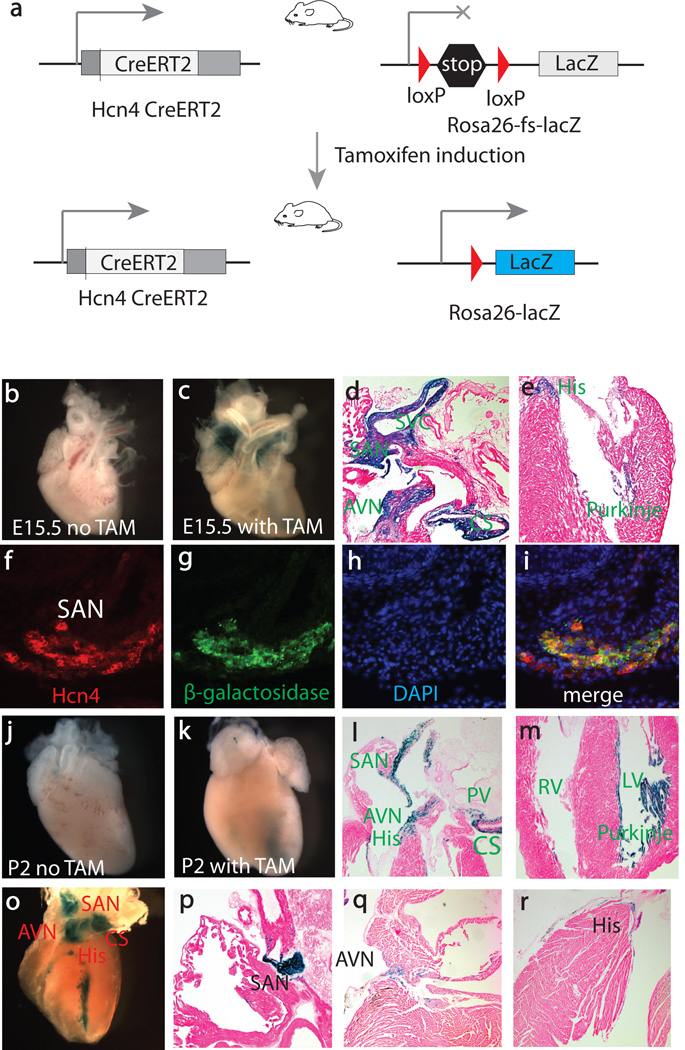
To evaluate the inducible properties of the transgenic Hcn4-CreERT2 allele, we crossed our line with the R26R reporter line (FIG. 2a). The R26R reporter mice have been widely used for testing Cre activity and lineage tracing, as deletion of a floxed stop cassette leads to constitutive expression of β-galactosidase (Soriano, 1999). In the Hcn4-CreERT2:R26R mice, no β-galactosidase activity was seen without tamoxifen induction at any stages (FIG. 2b & 2j). To examine the Cre activity of our Hcn4-CreERT2 transgenic line, tamoxifen was administrated by intraperitoneal (IP) injection at E12.5, and embryos were stained with X-gal at E15.5 (FIG. 2c–e). X-gal-stained cells were detected in the SAN, AVN, His bundle, bundle branches, and Purkinje fibers in the left ventricle. Only a few Purkinje fibers in the right ventricle were stained blue, consistent with the finding that the majority of right ventricular conduction system does not arise from Hcn4 progenitor cells but rather from the second heart field (SHF) (Liang et al., 2013). In addition to the CCS, X-gal-stained cells were seen in the right superior vena cava and coronary sinus but not in other parts of the embryos (FIG. 2d and Supplemental FIG. 1).
FIG. 2.
Tamoxifen-induced Cre activity in the entire CCS. (a) Breeding scheme to test functionality of the Hcn4-CreERT2 BAC transgenic line. (b–e) Cre activity after tamoxifen induction at E12.5. (f–i). Double-label fluorescent immnohistochemistry at E15.5 to show expression of Hcn4 and β-galactosidase. Hcn4 (f, red), β-galactosidase (g, green), DAPI (h, blue), Three imaged are merged in (i). (j–m) Cre activity after tamoxifen induction at perinatal stage. (o–r) Cre activity was detected in the entire CCS after tamoxifen induction in adults. TAM, tamoxifen; SVC, superior vena cava; SAN, sinoatrial node; AVN, atrioventricular node; PV, pulmonary vein; CS, coronary sinus, RV, right ventricle; LV, left ventricle.
To confirm that the blue cells stained by X-gal were CCS cells, we evaluated if the ones in the developing SAN at E15.5 co-expressed β-galactosidase and Hcn4. Double-label fluorescent immunohistochemistry was performed using antibodies against β-galactosidase and Hcn4 on frozen sections. As shown in FIG. 2f–I, β-galactosidase and Hcn4 were completely co-localized in the SAN, documenting that the β-galactosidase-expressing cells were bona fide CCS cells.
The CCS matures after birth, a process that is completed by the age of 1 month in mice (Moskowitz et al., 2004). Having shown specific induction of Cre activity in the developing heart at mid-gestation, we wanted to determine if this Cre line worked at the neonatal stage and in adults as well. Tamoxifen was administrated at E16.5, and X-gal staining was performed at P2 (FIG. 2j, k). After induction, X-gal-stained cells were detected in the SAN, AVN, His bundle, bundle branches, and Purkinje fibers in the left ventricle. A few Purkinje fibers in the right ventricle were also stained (FIG. 3l,m). These lineage tracing data are similar to those with induction at E12.5. In addition, X-gal-stained cells were seen in the right superior vena cava, pulmonary vein, coronary sinus, and atrioventricular ring bundles around mitral valve and tricuspid valve (FIG. 2l & m). These regions are closely related to CCS lineage development and are arrhythmogenic foci responsible for atrial and ventricular arrhythmia (Jongbloed et al., 2012; Liang et al., 2013; Yamamoto et al., 2006). No blue cells were seen in other parts of the heart except for a few cells in the right side of interatrial septum (Supplemental FIG. 2). These cells may represent the internodal conduction tracts that are related to the development of the left venous valves and septum spurium (Blom et al., 1999; Jongbloed et al., 2012). A detailed characterization of these blue cells using our Hcn4-CreERT2 model may provide novel insights on how these regions causes arrhythmia in various heart diseases. To evaluate the Cre activity in adult mice, tamoxifen was administrated by IP injection at 1-month and the Cre activity was analyzed. X-gal staining was specifically detected in the entire CCS (FIG. 2o–r). Similar to the observation in the embryos, no X-gal-stained cells were seen in other organs (Supplemental FIG. 3).
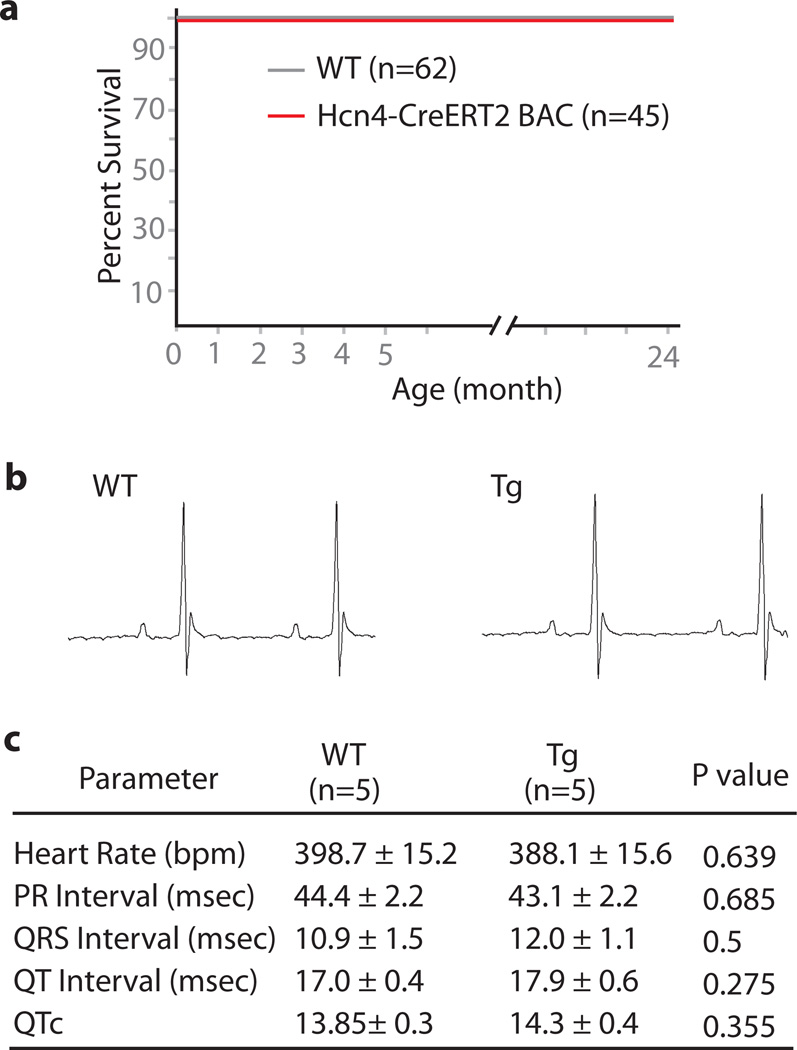
FIG. 3.
The Hcn4-CreERT2 animal homozygous for the transgene insertion showed normal life span (a) and normal EKG parameter (b & c). Representative traces of EKG in lead II was shown (b).
Recent studies have found that Hcn4 is transiently expressed in the progenitors of first heart field (FHF), and that Hcn4+ progenitor cells give rise to the left ventricle, part of both atria, and almost all components of the CCS (Liang et al., 2013; Spater et al., 2013). Tamoxifen induction at E7.5 in Hcn4-CreERT2 transgenic mice confirmed these findings (Supplemental FIG. 4), suggesting that our Hcn4-CreERT2 model can also be used to study development of FHF as well.
Because our model is likely to be widely used to conditionally inactivate genes in the CCS, it is important to explore if the transgenic line is normal. The Hcn4-CreERT2 mice heterozygous as well as homozygous for the transgene insertion were viable and fertile. We have not seen any difference in terms of life span or breeding properties between homozygous transgenic mice and wild-type controls (FIG.3a). Surface electrocardiogram (EKG) revealed no discernible difference between transgenic mice and wild-type littermate controls (FIG. 3b & c). In conclusion, these transgenic mice were indistinguishable from wild-type mice.
Taken together, we have generated a new transgenic mouse line, Hcn4-CreERT2, which exhibits Cre activity specifically in the entire CCS under tight control with tamoxifen.
This transgene can permit conditional gene deletion in the CCS during development and in adults, and it can also be harnessed as an important tool to model CCS diseases. As Hcn4 is transiently expressed in the FHF, this line can be a novel genetic tool to study early heart development as well.
MATERIALS AND METHODS
Mouse Strains
R26R reporter mice were ordered from Jackson Laboratories. All experiments with animals were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai.
Generation of the Hcn4-CreET2 BAC transgenic Line
A BAC was purchased from the BACPAC Resources Center at Children's Hospital Oakland Research Institute in Oakland, California. A 2.0-kb CreERT2 cassette was inserted immediately after the first ATG of the Hcn4 gene in BAC using recombineering. The BAC ends and the flanking regions of CreERT2 in the Hcn4 locus were confirmed by sequencing. BAC DNA was purified using NucleoBond BAC-100 (Clontech). The PCR primers used to genotype the Hcn4-CreERT2 transgenic mice amplifying a 401-bp sequence are:
CreERT2-F: GTGCCTGGCTAGAGATCCTG;
CreERT2-R: GATGTGGGAGAGGATGAGGA
We identified putative homozygous transgenic mice using quantitative PCR with primers that detect a 179-bp fragment from the CreERT2 cassette.
CreERT2-F: GACAGGAACCAGGGAAAATG
CreERT2-R: TCCAGAGACTTCAGGGTGCT
Putative homozygous transgenic mice were crossed with wild-type mice. If all of the pups born from these crosses were heterozygous, then we concluded that those transgenic mice were homozygous. The Hcn4-CreERT2 BAC transgenic line will be available to the research community on request.
Tamoxifen Administration
Tamoxifen (Sigma T5648) was dissolved in 100% ethanol at a concentration of 200 mg/ml, and then diluted in sesame oil (Sigma S3547) to a concentration of 10 mg/ml.
For tamoxifen induction in embryos, pregnant dams were injected with 2 mg/25 g body weight by intraperitoneal injection once. For tamoxifen induction in adults, adult mice were injected intraperitoneally at the amount of 1 mg tamoxifen/25 g body weight for 3 consecutive days and sacrificed three days after the final injection to analyze recombination.
X-Gal staining
For whole mount staining, the hearts were dissected out in ice-cold PBS and fixed with 4% paraformaldehyde in PBS at 4 °C for 1 h, then rinsed 3 × 30 min at room temperature in β-galactosidase rinse buffer (0.2 M sodium phosphate, pH7.3, 2 mM magnesium chloride, 0.02% NP40, 0.01% sodium deoxycholate). The hearts were stained in X-gal staining solution (4 mM ferrocyanide, 4 mM ferricyanide, 2 mM MgCl2, 1 mg/ml X-gal) at room temperature overnight and postfixed in 4% paraformaldehyde in PBS. To examine the X-gal-stained cells in sections, tissues were cryosectioned at a thickness of 10 µm, and fixed for 10 min at room temperature in 4% paraformaldehyde in PBS. The slides were rinsed in PBS and incubated in X-gal staining solutions at 37 °C overnight. After staining, the slides were rinsed briefly in PBS and then stained with Eosin Y.
Immunohistochemistry
Immunohistochemistry was performed on cryosections. The hearts were infused in 0.5 M sucrose for 2–3 h, and then in OCT frozen in dry ice. Samples were sectioned at the thickness of 10 µm. Immediately before performing immunohistochemistry, the sections were immersed in methanol at −20 °C for 5 min and washed three times with PBS for 5min. The sections were blocked with 1× Animal-Free Blocker (SP-5030, Vector lab) for 1 h. Hcn4 antibody (ab32675, Abcam, 1:200 dilution) and anti-β-galactosidase antibody (ab9361, Abcam, 1:200 dilution) were applied overnight at 4 °C. The slides were washed in PBS and then were incubated in secondary antibodies (Alexa Fluor® 488 Donkey anti-Chicken, Jackson Immuno, 1:3000 dilution, Alexa Fluor® 594 Donkey Anti-rat, Invitrogen, 1:3000) for 1 h at room temperature. The slides were mounted in VECTASHIELD Mounting Medium with DAPI.
Surface EKG
Cardiac electrophysiological function was assessed with surface electrocardiography as previously described . Data analysis was performed using the Chart5Pro (v 5.4.2, AD Instruments).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Bruce Gelb for comments on the manuscript. Y.Z. was supported by an SDG grant (AHA), a Basil O'Connor Starter Scholar Research Award (March of Dimes Foundation), and NIH awards (1R01HL107376 & 1K02HL103597).
LITERATURE CITED
- Arnolds DE, Moskowitz IP. Inducible recombination in the cardiac conduction system of minK: CreERT(2) BAC transgenic mice. Genesis. 2011;49:878–884. doi: 10.1002/dvg.20759. [DOI] [PubMed] [Google Scholar]
- Beyer S, Kelly RG, Miquerol L. Inducible Cx40-Cre expression in the cardiac conduction system and arterial endothelial cells. Genesis. 2011;49:83–91. doi: 10.1002/dvg.20687. [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Blom NA, Gittenberger-de Groot AC, DeRuiter MC, Poelmann RE, Mentink MM, Ottenkamp J. Development of the cardiac conduction tissue in human embryos using HNK-1 antigen expression: possible relevance for understanding of abnormal atrial automaticity. Circulation. 1999;99:800–806. doi: 10.1161/01.cir.99.6.800. [DOI] [PubMed] [Google Scholar]
- Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Moorman AF. Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others? Circ Arrhythm Electrophysiol. 2009;2:195–207. doi: 10.1161/CIRCEP.108.829341. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Smits GJ, Kispert A, Moorman AF. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- DiFrancesco D. The role of the funny current in pacemaker activity. Circ Res. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci U S A. 2012;109:E154–163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:1141–1148. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gourdie RG, Mima T, Thompson RP, Mikawa T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development. 1995;121:1423–1431. doi: 10.1242/dev.121.5.1423. [DOI] [PubMed] [Google Scholar]
- Herrmann S, Stieber J, Ludwig A. Pathophysiology of HCN channels. Pflugers Arch. 2007;454:517–522. doi: 10.1007/s00424-007-0224-4. [DOI] [PubMed] [Google Scholar]
- Hoesl E, Stieber J, Herrmann S, Feil S, Tybl E, Hofmann F, Feil R, Ludwig A. Tamoxifen-inducible gene deletion in the cardiac conduction system. J Mol Cell Cardiol. 2008;45:62–69. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed MR, Vicente Steijn R, Hahurij ND, Kelder TP, Schalij MJ, Gittenberger-de Groot AC, Blom NA. Normal and abnormal development of the cardiac conduction system; implications for conduction and rhythm disorders in the child and adult. Differentiation. 2012;84:131–148. doi: 10.1016/j.diff.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Hurtado R. Development of the cardiac conduction system. Semin Cell Dev Biol. 2007;18:90–100. doi: 10.1016/j.semcdb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Moreno-Rascon N, Beyer S, Dupays L, Meilhac SM, Buckingham ME, Franco D, Kelly RG. Biphasic development of the mammalian ventricular conduction system. Circ Res. 2010;107:153–161. doi: 10.1161/CIRCRESAHA.110.218156. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, Roden D, Berul CI, Seidman CE, Seidman JG. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- Munshi NV. Gene regulatory networks in cardiac conduction system development. Circ Res. 2012;110:1525–1537. doi: 10.1161/CIRCRESAHA.111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Fishman GI. The cardiac conduction system. Circulation. 2011;123:904–915. doi: 10.1161/CIRCULATIONAHA.110.942284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, Morley GE, Patel VV, Epstein JA. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–533. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinman MM. Role of the His-Purkinje system in the genesis of cardiac arrhythmia. Heart Rhythm. 2009;6:1050–1058. doi: 10.1016/j.hrthm.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spater D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Sun C, Zhang T, Liu C, Gu S, Chen Y. Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis. 2013;51:515–522. doi: 10.1002/dvg.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Dobrzynski H, Tellez J, Niwa R, Billeter R, Honjo H, Kodama I, Boyett MR. Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc Res. 2006;72:271–281. doi: 10.1016/j.cardiores.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R, Delgado-Olguin P, Davis M, Chi NC, Puviindran V, Gaborit N, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci U S A. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.