Abstract
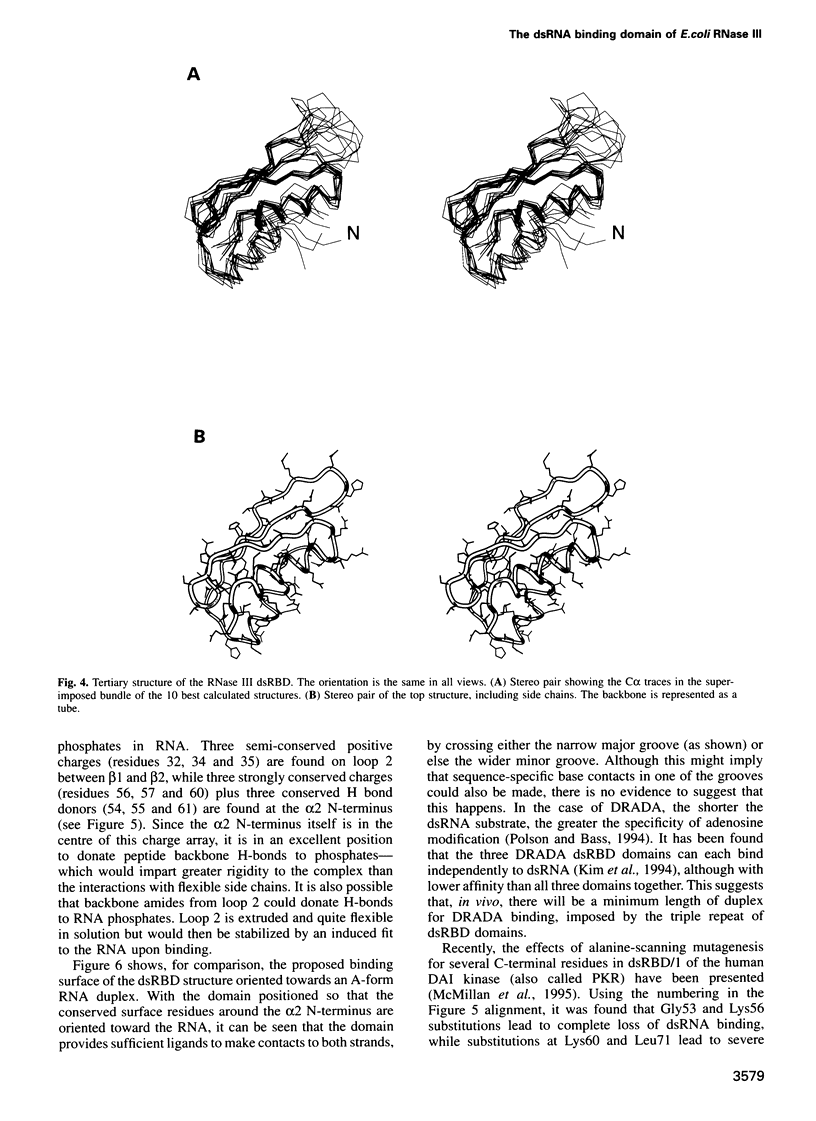
The double-stranded RNA binding domain (dsRBD) is a approximately 70 residue motif found in a variety of modular proteins exhibiting diverse functions, yet always in association with dsRNA. We report here the structure of the dsRBD from RNase III, an enzyme present in most, perhaps all, living cells. It is involved in processing transcripts, such as rRNA precursors, by cleavage at short hairpin sequences. The RNase III protein consists of two modules, a approximately 150 residue N-terminal catalytic domain and a approximately 70 residue C-terminal recognition module, homologous with other dsRBDs. The structure of the dsRBD expressed in Escherichia coli has been investigated by homonuclear NMR techniques and solved with the aid of a novel calculation strategy. It was found to have an alpha-beta-beta-beta-alpha topology in which a three-stranded anti-parallel beta-sheet packs on one side against the two helices. Examination of 44 aligned dsRBD sequences reveals several conserved, positively charged residues. These residues map to the N-terminus of the second helix and a nearby loop, leading to a model for the possible contacts between the domain and dsRNA.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res. 1993 Jul 1;21(13):3093–3096. doi: 10.1093/nar/21.13.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., Hurst S. R., Singer J. D. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr Biol. 1994 Apr 1;4(4):301–314. doi: 10.1016/s0960-9822(00)00069-5. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar S., Krainer A. R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993 Dec 25;21(25):5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Grünert S., Murzin A. G., Proctor M., St Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995 Jul 17;14(14):3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglone Morelli M. A., Stier G., Gibson T., Joseph C., Musco G., Pastore A., Travè G. The KH module has an alpha beta fold. FEBS Lett. 1995 Jan 23;358(2):193–198. doi: 10.1016/0014-5793(94)01422-w. [DOI] [PubMed] [Google Scholar]
- Chelladurai B. S., Li H., Nicholson A. W. A conserved sequence element in ribonuclease III processing signals is not required for accurate in vitro enzymatic cleavage. Nucleic Acids Res. 1991 Apr 25;19(8):1759–1766. doi: 10.1093/nar/19.8.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkers P. J., Nilges M., Folmer R. H., Konings R. N., Hilbers C. W. The solution structure of the Tyr41-->His mutant of the single-stranded DNA binding protein encoded by gene V of the filamentous bacteriophage M13. J Mol Biol. 1994 Feb 11;236(1):229–246. doi: 10.1006/jmbi.1994.1132. [DOI] [PubMed] [Google Scholar]
- Ghisolfi L., Kharrat A., Joseph G., Amalric F., Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992 Oct 15;209(2):541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Hyvönen M., Musacchio A., Saraste M., Birney E. PH domain: the first anniversary. Trends Biochem Sci. 1994 Sep;19(9):349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Thompson J. D. Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: implications for monomeric RNA helicases. Nucleic Acids Res. 1994 Jul 11;22(13):2552–2556. doi: 10.1093/nar/22.13.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. J., Thompson J. D., Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993 Jun 21;324(3):361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Green S. R., Mathews M. B. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992 Dec;6(12B):2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E., Carpousis A. J., Régnier P. Nucleolytic inactivation and degradation of the RNase III processed pnp message encoding polynucleotide phosphorylase of Escherichia coli. J Mol Biol. 1994 Jun 17;239(4):439–454. doi: 10.1006/jmbi.1994.1387. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol W. G., Halie L. M., Sander C. Dipoles of the alpha-helix and beta-sheet: their role in protein folding. Nature. 1981 Dec 10;294(5841):532–536. doi: 10.1038/294532a0. [DOI] [PubMed] [Google Scholar]
- Kao P. N., Chen L., Brock G., Ng J., Kenny J., Smith A. J., Corthésy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994 Aug 12;269(32):20691–20699. [PubMed] [Google Scholar]
- Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992 Jul;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. G., Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993 Aug 5;268(22):16822–16830. [PubMed] [Google Scholar]
- Lee W., Harvey T. S., Yin Y., Yau P., Litchfield D., Arrowsmith C. H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994 Dec;1(12):877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- Levy R. M., Bassolino D. A., Kitchen D. B., Pardi A. Solution structures of proteins from NMR data and modeling: alternative folds for neutrophil peptide 5. Biochemistry. 1989 Nov 28;28(24):9361–9372. doi: 10.1021/bi00450a017. [DOI] [PubMed] [Google Scholar]
- Macias M. J., Musacchio A., Ponstingl H., Nilges M., Saraste M., Oschkinat H. Structure of the pleckstrin homology domain from beta-spectrin. Nature. 1994 Jun 23;369(6482):675–677. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- McMillan N. A., Carpick B. W., Hollis B., Toone W. M., Zamanian-Daryoush M., Williams B. R. Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J Biol Chem. 1995 Feb 10;270(6):2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- Meadows R. P., Olejniczak E. T., Fesik S. W. A computer-based protocol for semiautomated assignments and 3D structure determination of proteins. J Biomol NMR. 1994 Jan;4(1):79–96. doi: 10.1007/BF00178337. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilges M. A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins. 1993 Nov;17(3):297–309. doi: 10.1002/prot.340170307. [DOI] [PubMed] [Google Scholar]
- Nilges M. Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J Mol Biol. 1995 Feb 3;245(5):645–660. doi: 10.1006/jmbi.1994.0053. [DOI] [PubMed] [Google Scholar]
- Nilges M., Gronenborn A. M., Brünger A. T., Clore G. M. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 1988 Apr;2(1):27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- O'Connell M. A., Krause S., Higuchi M., Hsuan J. J., Totty N. F., Jenny A., Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995 Mar;15(3):1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C., Ito N., Evans P. R., Teo C. H., Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994 Dec 1;372(6505):432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Pastore A., Saudek V., Ramponi G., Williams R. J. Three-dimensional structure of acylphosphatase. Refinement and structure analysis. J Mol Biol. 1992 Mar 20;224(2):427–440. doi: 10.1016/0022-2836(92)91005-a. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992 Nov;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Polson A. G., Bass B. L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994 Dec 1;13(23):5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V., White S. W. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992 Aug 27;358(6389):768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Fuchs R., Higgins D. G., Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1993 Jul 1;21(13):2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Saudek V., Pastore A., Morelli M. A., Frank R., Gausepohl H., Gibson T. The solution structure of a leucine-zipper motif peptide. Protein Eng. 1991 Jun;4(5):519–529. doi: 10.1093/protein/4.5.519. [DOI] [PubMed] [Google Scholar]
- Schweisguth D. C., Chelladurai B. S., Nicholson A. W., Moore P. B. Structural characterization of a ribonuclease III processing signal. Nucleic Acids Res. 1994 Feb 25;22(4):604–612. doi: 10.1093/nar/22.4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993 Mar 11;21(5):1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl M. J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993 Dec;17(4):355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown N. H., Gall J. G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput Appl Biosci. 1994 Feb;10(1):19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- Weber P. L., Morrison R., Hare D. Determining stereo-specific 1H nuclear magnetic resonance assignments from distance geometry calculations. J Mol Biol. 1988 Nov 20;204(2):483–487. doi: 10.1016/0022-2836(88)90589-x. [DOI] [PubMed] [Google Scholar]
- Wilson R., Ainscough R., Anderson K., Baynes C., Berks M., Bonfield J., Burton J., Connell M., Copsey T., Cooper J. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994 Mar 3;368(6466):32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- van der Oost J., Lappalainen P., Musacchio A., Warne A., Lemieux L., Rumbley J., Gennis R. B., Aasa R., Pascher T., Malmström B. G. Restoration of a lost metal-binding site: construction of two different copper sites into a subunit of the E. coli cytochrome o quinol oxidase complex. EMBO J. 1992 Sep;11(9):3209–3217. doi: 10.1002/j.1460-2075.1992.tb05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]