Abstract
Stem cell fate and function are dynamically modulated by the interdependent relationships between biochemical and biophysical signals constituting the local 3D microenvironment. While approaches to recapitulate the stem cell niche have been explored for directing stem cell differentiation, a quantitative relationship between embryonic stem cell (ESC) morphogenesis and intrinsic biophysical cues within three-dimensional microtissues has not been established. In this study, we demonstrate that mesenchymal embryonic microtissues induced by BMP4 exhibited increased stiffness and viscosity accompanying differentiation, with cytoskeletal tension significantly contributing to multicellular stiffness. Perturbation of the cytoskeleton during ESC differentiation led to modulation of the biomechanical and gene expression profiles, with the resulting cell phenotype and biophysical properties being highly correlated by multivariate analyses. Together, this study elucidates the dynamics of biophysical and biochemical signatures within embryonic microenvironments, with broad implications for monitoring tissue dynamics, modeling pathophysiological and embryonic morphogenesis and directing stem cell patterning and differentiation.
The biochemical composition and physical structure of tissue microenvironments are critical regulators of stem cell fate and function1,2. In vivo, native stem cell niches maintain the self-renewing state through short range soluble signaling3 and extracellular signals4, such as direct anchorage via intercellular and/or matrix adhesions. Conversely, dysregulation of niche elements leads to differentiation and migration of resident stem cell populations5. Therefore, the synergy of biophysical and biochemical signals and the complexity of the local microenvironment have been increasingly recognized as critical mediators of the delicate balance between self-renewal and differentiation. However, while engineering approaches often aim to dissect the relative influence of individual stimuli, the signals regulating stem cell differentiation are inherently interrelated, motivating the multiparametric analysis of stem cell structure and phenotype accompanying dynamic cell fate decisions.
The pre-implantation embryo is one of the most dynamic tissue microenvironments, whereby the patterning of structurally and functionally distinct tissue structures, such as liver, skin, and heart arise from physically adjacent, yet precise spatially defined, regions during development6. In particular, embryonic gastrulation mediates the first somatic cell fate decisions, and results in spatially distinct localization of the three germ lineages – endoderm, ectoderm, and mesoderm. During gastrulation, the cells of the epiblast that will comprise the mesoderm lineage undergo a rapid and concerted morphogenesis7,8 through which the tightly adherent, epithelial cells become less adhesive mesenchymal cells, in a process known as the epithelial-to-mesenchymal transition (EMT)9. The dynamics and patterning of EMT are tightly regulated by the biophysical and biochemical tissue microenvironment, through signals that maintain or abrogate the apical-basal polarity of epithelial cells. While the role of biochemical signals are well established in the induction of EMT, the resulting changes in cell shape that occur during gastrulation are a fundamentally biomechanical process, through which forces arising as a result of adhesion remodeling are transmitted via changes in cytoskeletal tension10,11,12. Biophysical signals also act in conjunction with biochemical cues to mediate different stages of embryonic morphogenesis, including mesoderm intercalation13, dorsal elongation14, and neural tube closure15. Moreover, mechanical cues alter the responsiveness of cells to chemical signals in order to mediate proliferation and dictate organ size16, thereby demonstrating the complexity of deciphering signaling mechanisms regulating tissue patterning.
While the phenotypic17 and mechanical18 changes arising during embryonic development have been characterized through orthogonal approaches in various species6,19,20, several questions remain regarding the role of cellular biomechanics in determining cell fate, particularly during mammalian embryogenesis. The differentiation of pluripotent embryonic stem cells (ESCs), the in vitro analog to the inner cell mass of pre-implantation embryos, parallels many of the in vivo cell fate transitions, reflecting the utility of ESCs as a model for systematically studying mammalian embryonic development21. ESC differentiation approaches have aimed to recapitulate the biochemical and/or biophysical milieu of the gastrulating embryo, including the perturbation of developmentally relevant signaling pathways22 and the manipulation of the composition and mechanical properties of adherent substrates23,24. However, when assembled as three-dimensional aggregates, ESCs undergo morphogenic events, including the deposition of extracellular mediators of EMT, such as hyaluronan and versican25, which initiates dynamic, developmentally relevant processes, thereby offering unique opportunities to study biomechanics in parallel with changes in pluripotent cell fate and morphogenesis.
The objective of this study, therefore, was to define the intrinsic cellular biophysical characteristics and phenotypic changes that arise simultaneously during cell fate specification and morphogenesis of three-dimensional pluripotent stem cell microenvironments. A multifaceted approach was employed to assess the dynamics of morphogenesis and differentiation, characterize the biomechanical properties of 3D microtissues, and determine correlative associations between gene expression and mechanical signatures via multivariate data modeling. Overall, this study demonstrates that morphogenesis arises via EMT during mesoderm differentiation of pluripotent microtissues, resulting in dynamic temporal changes in phenotype that are highly correlated with biomechanical properties. Ultimately, understanding the biophysical changes accompanying epithelial-to-mesenchymal transition of ESCs has broad implications to reveal physical mechanisms underlying mammalian embryonic development, tissue homoeostasis, pathological remodeling and cancer metastasis.
Results
BMP4 induces mesoderm differentiation in ESC microtissues
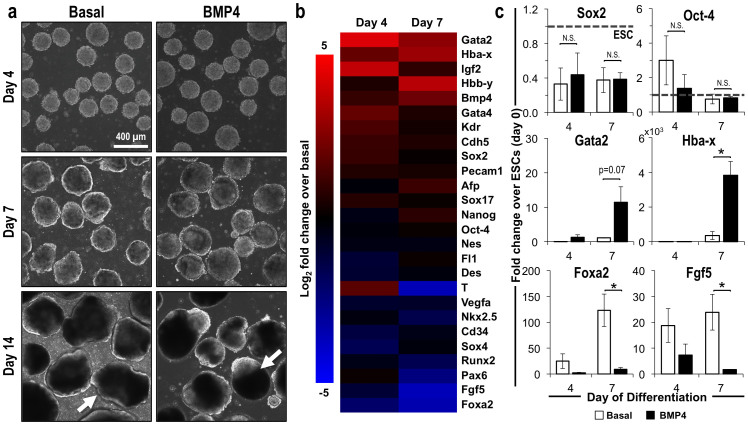
Homogeneous populations of EBs were formed via forced aggregation and maintained in controlled hydrodynamic suspension culture, as described previously26, in order to systematically examine three-dimensional stem cell morphogenesis. As expected, supplementation with BMP4 (10 ng/mL) altered the EB morphology (Fig. 1a), as evidenced by changes in the EB circularity and optical density (arrows), and overall gene expression profile (Fig. 1b), resulting in the increased expression of genes related to mesoderm lineages (Supplementary Table S1) at days 4 and 7 of differentiation compared to those maintained in basal, serum-free culture conditions. Although decreases in the pluripotency markers Sox2 and Oct-4 did not differ significantly between the two culture conditions, markers of hematopoietic mesoderm (Gata2, Hba-x) were increased by BMP4 treatment (Gata2 p = 0.07; Hba-x p = 0.03), and genes related to endoderm (Foxa2) and ectoderm (Fgf5) were significantly decreased (Foxa2 p = 0.04; Fgf5 p = 0.004), thereby illustrating the morphogenic influence of BMP4 in directing mesoderm differentiation in 3D cultures, analogous to previous reports using monolayer differentiation techniques27,28.
Figure 1. Induction of mesoderm differentiation within EBs via BMP4 treatment.
(a) Phase contrast images demonstrate similar gross morphology of EBs cultured in basal, serum-free media or media supplemented with BMP4 after 4 days of differentiation, with differences in circularity (basal) and optical density (BMP4), particularly apparent after 14 days of differentiation (arrows). (b) Gene expression profiles also differed between EBs cultured in different soluble culture environments after 4 and 7 days of differentiation, (c) resulting in similar levels of pluripotency factors, with divergent expression of markers for hematopoietic mesoderm (Gata2, Hba-x), endoderm (Foxa2), and ectoderm (Fgf5). Scale bar = 200 μm; n = 3; * = p < 0.05 using two-way ANOVA and post-hoc Tukey analysis.
ESCs undergo mesenchymal morphogenesis during 3D mesoderm differentiation
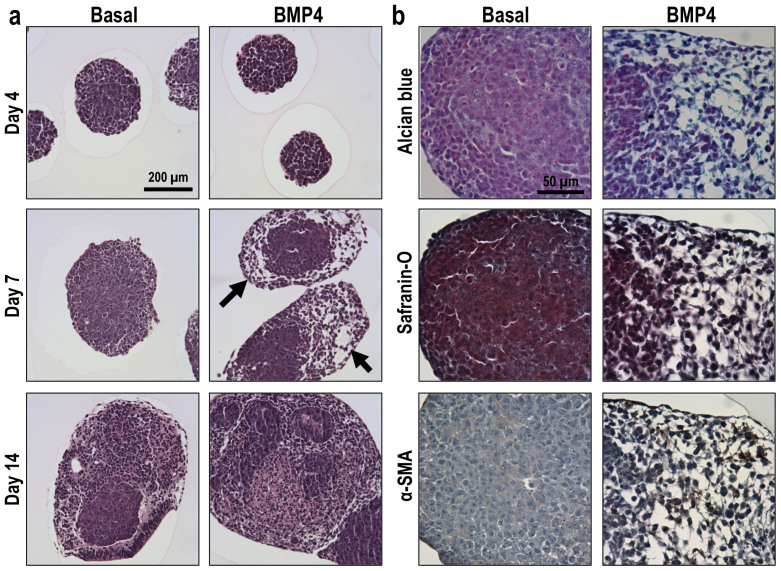
Consistent with the gross changes in EB morphology, distinct differences in cellular morphology and organization within EBs were observed during the course of differentiation, in a soluble factor-dependent manner (Fig. 2a). While EBs within both conditions exhibited similarly homogenous populations of cells with epithelial morphologies initially (day 4), biochemical-mediated differentiation led to changes in cellular organization and morphogenesis, with basal EBs continuing to exhibit tightly packed, epithelial cell clusters, in contrast to BMP4-treated EBs, in which populations of elongated, mesenchymal-like cells were apparent at day 7 of differentiation. Interestingly, the mesenchymal morphology observed at day 7 was no longer apparent by 14 days, indicating a transition phase accompanying the specific stage of mesoderm differentiation. Consistent with the GAG-rich pericellular matrix of undifferentiated ESCs, the EBs maintained in basal conditions exhibited positive safranin-O staining. In addition to the morphological changes, BMP4-treated EBs exhibited a glycosaminoglycan (GAG) rich matrix at day 7, with spatial variation in composition, as evidenced by positive alcian blue (pH 2.5) and negative safranin-O staining within the interstitial space between the regions of mesenchymal cells (Fig. 2b). The putative mesenchymal cells also expressed α-smooth muscle actin (α-SMA), an established marker of EMT29. Taken together, EB mesoderm differentiation induced by BMP4 treatment results in EMT morphogenesis to yield characteristic mesenchymal cell populations.
Figure 2. Mesenchymal morphogenesis of ESCs within EBs.
(a) EBs exhibited dynamic remodeling of the microenvironment through 14 days of differentiation, with distinct differences apparent between EBs (H&E staining) in different soluble media formulations. (b) After 7 days of differentiation, mesenchymal-like regions (arrows) within EBs exhibited positive staining for alcian blue (blue) and α-SMA (brown/black), whereas safranin-O (red/brown) stained positively within epithelial-like regions and was largely absent within the mesenchymal-like regions of BMP4-treated EBs. Scale bars (A) = 200 μm, (B) = 50 μm; n = 3; Nuclei counterstained with: fast green (safranin-O), nuclear fast red (alcian blue), and hematoxylin (α-SMA).
Biophysical microtissue properties are dynamically modulated during ESC differentiation
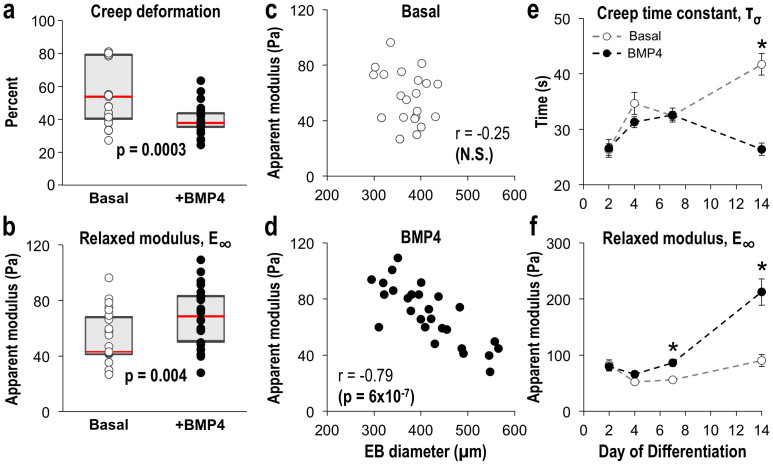
To characterize the physical remodeling of the EB microenvironment during ESC differentiation, biomechanical microtissue properties were measured via micron-scale parallel plate compression (Supplementary Fig. S1a). Descriptive physical characteristics, including moduli, were calculated by measuring the viscoelastic creep (change in deformation under constant force), which was fitted to a linear viscoelastic model (average R2 = 0.97; Supplementary Fig. S1b,c). EBs treated with BMP4 exhibited increased viscous resistance to deformation (p = 0.0003), as evidenced by decreased creep compared to those differentiated in basal conditions at day 7 of differentiation (Fig. 3a). BMP4-treated EBs were 1.5-fold stiffer than basal EBs after 7 days of differentiation (p = 0.004; Fig. 3b). Therefore, morphologically distinct EBs also exhibited biomechanical differences, both in terms of viscoelasticity and microtissue stiffness. Interestingly, the size distribution of EBs was altered upon BMP4 treatment, which led to increased dispersity after 7 days of differentiation (median absolute deviation, MAD = 31 μm for basal EBs; MAD = 62 μm for BMP4 EBs). While the stiffness and size of basal EBs were not strongly correlated (Fig. 3c), those differentiated with BMP4 demonstrated an inverse relationship (p < 0.001), with smaller BMP4-treated EBs exhibiting increased stiffness (Fig. 3d), thereby suggesting that smaller EBs may undergo more robust mesenchymal responses to BMP4, potentially due to transport-related phenomena30. In addition to the increased stiffness of the BMP4-treated population of EBs, the correlation between aggregate size and stiffness reflects the physical relationship between morphogenesis and biomechanical properties, thereby providing a novel quantitative metric to assess the state of EB differentiation.
Figure 3. Dynamic modulation of EB mechanical properties.
EBs exhibited differences in (a) viscoelastic creep and (b) modulus after 7 days of differentiation, including (c,d) a significant correlation between modulus and EB diameter within BMP4-treated EBs. In addition, (e) modulus and (f) viscoelastic time constant characteristics were modulated as a function of time, with differences between culture conditions after 14 days of differentiation. (a,b) gray shaded regions represent the upper and lower quartiles while the red center line denotes the median of the populations. (a–d) n = 27 EBs from 5 independent experiments; (e), (f) n = 6 EBs; * = p < 0.05 using two-way ANOVA and post-hoc Tukey analysis.
Moreover, the biomechanical properties of EBs were dynamically modulated during the course of differentiation, whereby significant differences in the stiffness and viscosity were measured over time, both within and between individual biochemical differentiation conditions (Fig. 3e,f and Supplementary Fig. S2). Specifically, basal conditions exhibited decreased stiffness between days 2 and 4 of differentiation (p = 0.04), whereas BMP4-treated EBs exhibited similar stiffness throughout the first 7 days of differentiation; the stiffness of basal and BMP4-treated EBs was significantly increased at day 14 compared to day 4 (basal p = 0.01; BMP4 p < 0.001), thus indicating an overall increase in stiffness during differentiation. Both conditions also exhibited similar initial viscosity dynamics, whereby the viscous response decreased (i.e. increasing deformation time, or creep time constant) during the initial 7 days of differentiation (basal p = 0.04; BMP4 p = 0.04); however, the viscosity of EBs from basal conditions continued to decrease through day 14 of differentiation, whereas the viscosity of BMP4-treated EBs increased between days 7 and 14 of differentiation, resulting in significantly increased viscosity of BMP4-treated EBs compared to basal EBs (p < 0.001). Overall, the biomechanical properties of multicellular ESC aggregates change dynamically during differentiation, with BMP4-treated EBs stiffening and exhibiting increased viscosity as cells commit toward mesodermal lineages and undergo EMT.
Cytoskeletal tension regulates biophysical microtissue environments
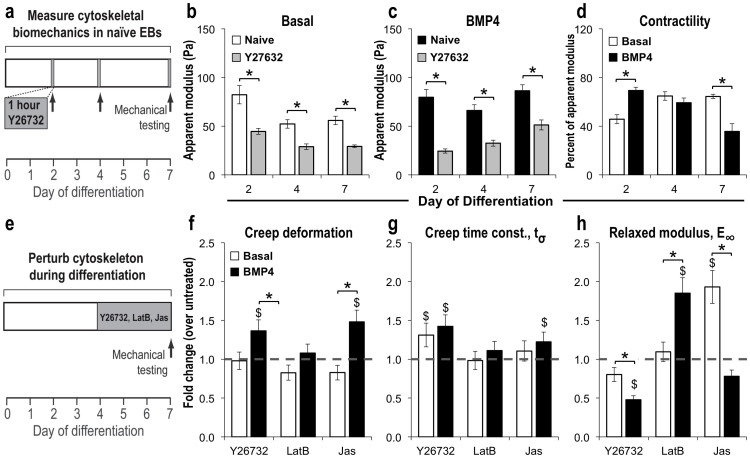
The actomyosin cytoskeleton contributes to the primary structural mechanics of cells and is particularly important in mediating cell shape, force generation, and stiffness in embryonic tissues31. Therefore, we hypothesized that the biophysical and phenotypic changes arising during three-dimensional pluripotent stem cell morphogenesis and differentiation were mediated by cytoskeletal tension. The actin filament organization of cells comprising basal serum-free EBs retained a largely cortical structure at all stages of differentiation, consistent with an epithelial cell phenotype; however, BMP4-treated EBs exhibited decreased assembly of F-actin, with punctuated cortical structures after 4 days of differentiation (Supplementary Fig. S3). Moreover, consistent with the mesenchymal morphology, cells within BMP4-treated EBs exhibited characteristic elongated cellular morphologies and filamentous cytoskeletal architecture by day 7 of differentiation, which implicates the cytoskeleton in mediating microtissue stiffness during EB morphogenesis. Disruption of the Rho associated protein kinase (ROCK) pathway with a small molecule inhibitor (Y27632; 10 μM; 1 hour)32 (Fig. 4a) acutely decreased the stiffness of EBs from both differentiation conditions at all time points examined (p < 0.03 for all conditions), indicating that the actomyosin cytoskeleton significantly contributes to the stiffness of EBs (~50%) (Fig. 4 b,c). In addition, despite similar EB stiffness, the ROCK-mediated contractility of BMP4-treated was increased compared to basal EBs after 2 days of differentiation (p < 0.001), demonstrating the emergence of unique biophysical traits at early stages of differentiation, even before morphological or phenotypic changes could be observed (Fig. 4d). Moreover, although the magnitude of stiffness increased during differentiation, the relative contribution of cytoskeletal tension to the overall modulus of BMP4-treated EBs decreased, indicating an increasing role for other structural mechanisms (e.g. nuclear, extracellular) at later stages of mesenchymal morphogenesis. Altogether, cytoskeletal tension significantly contributes to ESC microtissue mechanics and the actomyosin cytoskeleton is remodeled during mesenchymal morphogenesis.
Figure 4. Influence of cytoskeletal tension on EB mechanical properties.
(a) Treatment of EBs with the ROCK inhibitor Y27632 for 1 hour significantly decreased the modulus of (b) basal and (c) BMP4-treated EBs and (d) demonstrated differences in the relative contribution to the overall naïve EB modulus at 2 and 7 days of differentiation. (e) In addition, EBs were differentiated in the presence of cytoskeletal agonists and antagonists, and exhibited changes in the (f) viscoelastic creep and (g) time constant responses, as well as (h) modulus after 7 days of differentiation. n = 6 EBs; * = p < 0.05 compared to basal; $ = p < 0.05 compared to untreated, using two-way ANOVA and post-hoc Tukey analysis.
While the cytoskeletal structure directly mediates tissue stiffness, the dynamics of actin polymerization have also been implicated in morphogenic processes, such as EMT33. Therefore, the actin polymerization was perturbed via treatment with an agonist (Jasplakinoide, Jas; 50 nM)34 or antagonist (latrunculin B, LatB; 100 nM)35 in addition to perturbation of ROCK-mediated contractility with Y27632 during the initial period of ESC morphogenesis (days 4–7) in order to assess the impact on biomechanics, morphogenesis, and cell phenotype (Fig. 4e). BMP4-treated EBs exhibited decreased viscosity in response to Jas and Y27632, compared to untreated (Jas p < 0.001; Y27632 p = 0.01) EBs (Fig. 4f); in contrast, the viscous response of basal EBs remained unchanged across all conditions. Despite opposite mechanisms of action, the decreased viscosity in response to both Jas (agonist) and Y27632 (antagonist) demonstrates additional time-dependent biophysical properties, which may arise from feedback or compensatory changes arising from dynamic changes in cellular organization and phenotype, as well as remodeling of adhesions, or modulation of the spatial distribution or relative contributions of intracellular structural elements, including actin, myosin, and intermediate filaments, in response to cytoskeletal perturbations. The kinetics of the viscous response, reflected by the creep time constant, were increased by treatment with Y27632 in BMP-4 and basal conditions (Fig. 4g), suggesting that inhibiting ROCK-mediated contractility decreased aggregate viscosity. In addition, Jas increased the stiffness of EBs cultured in basal, serum-free media compared to untreated EBs, consistent with its known role stimulating actin polymerization (p < 0.001); however, the stiffness of BMP4-treated EBs was not affected by Jas (Fig. 4h). In contrast, Y27632 significantly decreased the stiffness of BMP4 EBs compared to untreated conditions (p < 0.001), but did not affect EBs differentiated in basal conditions. Interestingly, although LatB has been reported to decrease actin polymerization, BMP4-treated EBs were significantly stiffer when cultured in LatB compared to untreated EBs (p < 0.001). Overall, these studies indicate that the biomechanical properties of pluripotent microtissues undergoing differentiation and morphogenesis are significantly altered by the dynamics of actin polymerization contributing to cytoskeletal tension.
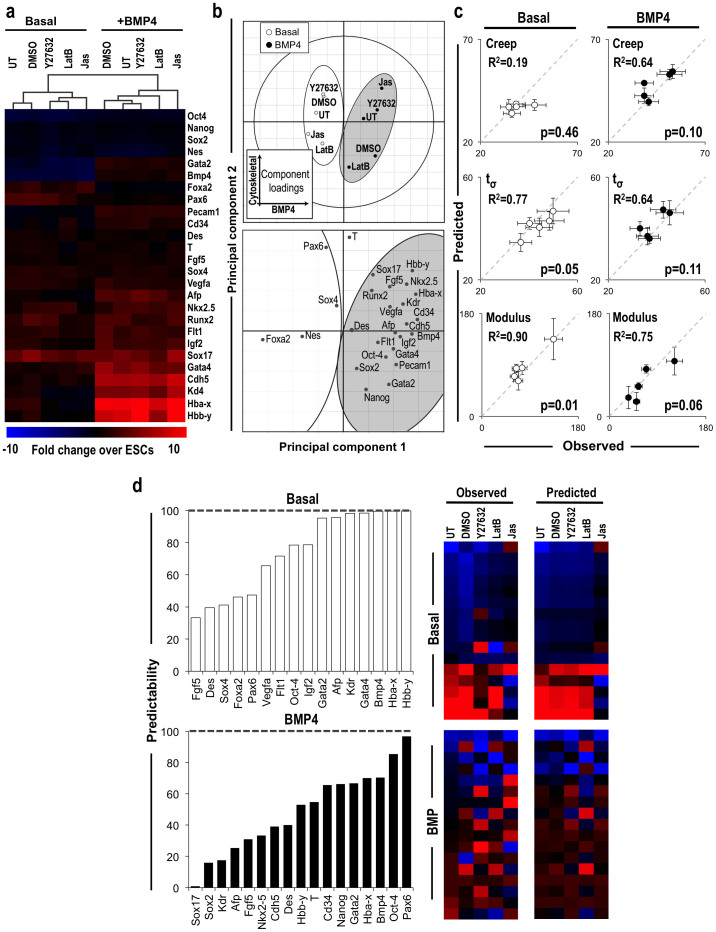
Biomechanics predict microtissue phenotypic profiles
The impact of biomechanical forces on stem cell phenotype has been increasingly appreciated as an exogenous means to direct cell fate24,36,37; however, a quantitative relationship between ESC phenotype and biophysical properties of three-dimensional microtissues has not been established. Therefore, the morphology and phenotype of ESCs were assessed in parallel with the changes in microtissue stiffness upon treatment with small molecules. The cytoskeletal agonist and antagonists did not induce marked morphological changes, based on gross histology (Supplementary Fig. S4); however, subtle differences were noted, as LatB-treated BMP4 EBs exhibited more epithelial-like cells, similar to basal EBs (Supplementary Fig. S4s), whereas Jas and Y27632 treatment induced more mesenchymal morphogenesis within BMP4 EBs (Supplementary Fig. S4q,r). In addition, treatment with the small molecules altered the gene expression profiles of both basal and BMP4 EB populations after 7 days of differentiation; hierarchical clustering indicated marked phenotypic differences for Jas treated EBs from both conditions relative to untreated EBs (Fig. 5a). Similarly, a PLS model (R2Y = 0.70, Q2 = 0.10; 3 significant PCs) illustrated the variance in gene expression signatures due to BMP4 treatment and cytoskeletal perturbations, which were distinctly separated along the first and second principal components, respectively (Fig. 5b); although the biochemical morphogen treatment in the presence or absence of BMP4 dominated the phenotypic gene expression, subtle variations in the phenotypic profile were induced by cytoskeletal perturbations, thereby indicating the utility of PLS modeling for capturing variations not detectable through traditional analytical methods, such as hierarchical clustering. Consistent with the histological observations, LatB-treated EBs exhibited phenotypic profiles indicative of decreased EMT, similar to basal EBs, whereas Jas-treated BMP4 EBs exhibited increased mesoderm differentiation and mesenchymal morphogenesis, as evidenced by the position along the first principal component. Increased expression of pluripotency genes such as Nanog, Sox-2, and Oct-4, indicated that LatB treatment delayed the differentiation kinetics of ESCs for both culture media conditions. Interestingly, the same mesendoderm genes (Hbb-y, Sox17, Nkx2.5, Fgf5) increased by Jas treatment of BMP4 EBs were decreased by Jas in basal EBs, thus demonstrating that actin polymerization was not sufficient to induce a mesendoderm gene expression profile within basal EBs (Fig. 5b).
Figure 5. Prediction of microtissue stiffness and ESC phenotypes by multivariate modeling.
(a) PCR array analysis of gene expression suggested differences in cell fate after 7 days of differentiation due to small molecule-mediated perturbation of the cytoskeleton from days 4–7. (b) In addition, the score plot demonstrates distinctions between culture conditions across the first and second principal components, based upon gene expression profiles, shown in the loadings plot. (c) Separate PLS with gene expression as inputs and mechanical properties as outputs demonstrated a high degree of correlation between mechanical properties and phenotypic traits. (d) In addition, PLS models based upon mechanical characteristics demonstrated different degrees of correlation for the associated gene expression profiles when trained with individual or full biomechanical profiles.
Based on the biochemical-mediated variability between experimental groups captured by the first principal component, separate PLS models were constructed based upon the gene expression (input, X) and mechanical properties (output, Y) to distinguish the cellular responses to cytoskeletal perturbations for EBs differentiated in basal, serum-free media (R2Y = 0.70, Q2 = 0.10; 1 significant PC) and in the presence of BMP4 (R2Y = 0.65, Q2 = 0.30; 1 significant PC) (Fig. 5c). The models reflect changes in biomechanical properties as a function of the gene expression profile, thus enabling a quantitative relationship between biophysical microtissue environments and ESC differentiated phenotypes. Overall, the gene expression profile was highly correlated with several of the mechanical characteristics; however, the strength of correlations varied between individual biomechanical parameters and across differentiation conditions. The model theoretically predicted the EB stiffness, with R2 coefficients of 0.90 and 0.75 in the basal and BMP4 conditions, respectively. In contrast, the viscous creep response of EBs did not correlate as highly with the phenotype of EBs in either condition, indicating that the changes in individual mechanical properties, such as viscosity, could not be fully explained through phenotypic changes and may also be influenced by extracellular factors, such as ECM deposition. To illustrate the complex relationship between biophysical characteristics and ESC phenotype, the model was instead constructed using the biomechanical parameters to predict gene expression profiles (Fig. 5d). Overall, the biomechanical profile significantly (background Q2Y < 0.05) predicted the responses of 61.5 and 65.4% of the genes, respectively, which established subsets of genes responsive to biophysical perturbations. Moreover, the upper quartile of genes (Basal: Pax-6, Oct-4, Bmp4, Hba-x; BMP4: Hba-x, Hbb-y, Bmp4, Gata4) within each condition were predicted by the models with > 70% accuracy. Interestingly, Bmp4 and Hba-x, which were highly predicted by both models, exhibited divergent responses to small molecule treatments within basal- and BMP4-treated EBs, thereby demonstrating the capacity for multivariate analytics to identify putative markers highly correlated with biomechanics across different environmental conditions. In addition, the genes most highly predicted in basal EBs included those that exhibited decreased (Hba-x, Hbb-y), increased (Gata4), and unchanged (Bmp4) expression patterns in response to cytoskeletal perturbations, indicating that the mechanical model captured an array of different phenotypic responses. In contrast, while the mechanical model largely recapitulated the phenotypic profile of BMP4-treated EBs, several individual genes were not as highly correlated, which may be due to diverse responses arising within less uniform microtissues. The data together indicate that distinct phenotype profiles could be accurately described by multivariate analyses based upon multiple viscoelastic parameters. In summary, these results collectively suggest that ESC differentiated phenotypes are directly related through complex, multiparametric changes in the stiffness and viscoelastic characteristics of EBs.
Discussion
This study elucidates intrinsic biophysical mechanisms mediating three-dimensional pluripotent morphogenesis, which can enable the development of novel approaches to direct the differentiation and patterning of functional ESC-derived microtissues for modeling mammalian pathophysiology and embryogenesis38, pharmacological screening39, and facilitating in vivo tissue repair40. We demonstrate that BMP4-mediated mesoderm differentiation of ESCs recapitulates aspects of embryonic development, particularly in terms of morphogenesis leading to regions of mesenchymal-like cells, analogous to primitive streak formation during gastrulation7,41. Moreover, phenotypic and morphogenic changes were accompanied by remodeling of cytoskeletal elements and modulation of biophysical properties during three-dimensional ESC differentiation, with the phenotypic and mechanical traits correlated by multivariate modeling.
The improved understanding of three-dimensional ESC biophysical and phenotypic dynamics establishes distinctions between embryonic and adult tissue models and provides important principles for the design of relevant microenvironments to direct early cell fate decisions. The biophysical observations noted throughout this study are consistent with the dramatic differences in tissue structure and cellular composition between adult- and embryonic-derived tissues17. Several previous studies have engineered the mechanical properties of stem cell environments through the development of substrates with elasticity comparable to the range of adult tissues (1 kPa–100 kPa)24,42, which is 104 Pa less than rigid standard polystyrene culture dishes. In contrast, the stiffness of morphogenic 3D ESC multicellular aggregates is substantially less than that of native tissues (E∞ < 0.1 kPa). Similar to embryonic tissues43, ESCs undergo rapid growth coincident with morphogenesis, leading to high nuclear density of EBs evidenced by histological analysis (Fig. 2a,b), which is distinct from the structure of adult tissues. In addition, previous characterization of the early EB microenvironment indicated minimal ECM deposition, largely devoid of collagen, during the first 4–7 days of differentiation44; thus, the principal structural attributes of stem cell aggregates are initially constituted by the cells and their cytoskeletal filaments, as well as by the intercellular adhesions (i.e. E-cadherin) supporting EB formation and maintaining multicellular organization. Consistent with our observations, a recent report describing a novel method to measure local forces within cellular aggregates attributed local stresses to myosin activity45. Moreover, the stiffness of embryonic tissue explants has been measured to be in the range of 10–100 Pa14, consistent with our reported data, thereby highlighting the unique physical traits of embryonic microenvironments. This study establishes a biophysical model of multicellular embryo-like tissues in which cytoskeletal tension is transmitted between cells via intercellular adhesions and is dynamically modulated during ESC differentiation and morphogenesis.
In addition to the unique composition of embryonic-derived tissue microenvironments, pluripotent stem cells also exhibit distinctive biophysical characteristics, in contrast to their differentiated progeny. For example, hESCs exhibited a >3 fold increase in modulus upon differentiation toward chondrogenic lineages46 and embryonic tissues stiffen during gastrulation, leading to a 10-fold increase in mesoderm stiffness compared to endoderm14. Similarly, the data presented in our study collectively suggest that stiffness and viscosity increase concomitant with three-dimensional mesoderm differentiation and mesenchymal morphogenesis. The mechanism for increased stiffness upon differentiation has been suggested to result in part from cytoskeletal-mediated changes in structure46,47. In addition, recent studies have demonstrated the increasing stiffness of nuclear structures during mesenchymal stem cell differentiation48, with the biophysical characteristics attributed to lamin-A expression and chromatin structure49, which could account for the increasing contribution of factors independent of ROCK-mediated contractility at later stages of differentiation (Fig. 4d). Morphogenic events responsible for tissue-scale patterning, including local cell proliferation50 and EMT33, occur in regions of increased tension, indicating a role for biophysical regulation of stem cell differentiation and morphogenesis. In this study, perturbation of intracellular cytoskeletal elements during mesenchymal morphogenesis elucidated the relative contribution to biophysical tissue properties; in ESC microenvironments, the actomyosin cytoskeleton significantly contributed (>50% of modulus) to the microtissue stiffness during all stages of differentiation (Fig. 4a–c). Interestingly, early changes in stiffness and cytoskeletal organization have been used to successfully predict the differentiated phenotypes of adipose stem cells (ASCs) and MSCs, respectively51,52. Similarly, differences in cytoskeletal tension between basal and BMP4-treated EBs were detected as early as day 2 of differentiation, suggesting that dynamic cytoskeletal changes occur prior to phenotypic differences commonly assessed by standard molecular, biochemical or histological approaches. In addition, the assessment of biophysical microtissue characteristics provides a direct measurement of the dynamics of active processes mediating morphogenesis, thereby providing metrics to monitor processes such as cell sorting and patterning, analogous to similar events during embryogenesis11,12. Collectively, the data presented herein suggest that stem cell differentiation is reflected by concomitant changes in biophysical properties, thus establishing a novel, non-destructive quantitative metric to assess microtissue morphogenesis and the homogeneity/heterogeneity of multicellular aggregate populations.
Biomechanical regulation of stem cell fate has become a prominent research focus as an approach to engineer stem cell niches23. Manipulation of extracellular mechanical properties has been explored to direct stem cell differentiation, with changes in morphology induced by modulation of cell spreading37 or substrate stiffness24 linked to altered MSC differentiation trajectories. However, monitoring and controlling microtissue biomechanics is particularly challenging due to the limitations associated with manipulating or presenting three-dimensional biophysical cues53 and an incomplete understanding of the physical mechanisms governing 3D stem cell differentiation and patterning. Large-scale bioinformatics techniques based on gene expression signatures are commonly used to define stem cell states54, but such approaches rely on data generated from a single, often destructive, analytical technique. The biomechanical assay demonstrated herein establishes an orthogonal, non-destructive metric for describing three-dimensional stem cell aggregate characteristics. Multivariate modeling (Fig. 5) established a quantitative relationship between gene expression and biomechanical properties of stem cell aggregates differentiated in various biochemical contexts, including those directed toward ectoderm (basal) and mesoderm lineages (BMP4), thereby providing proof-of-principle for the utility of biomechanical metrics to monitor stem cell fate by using non-destructive techniques to measure intact aggregate properties. Therefore, this study presents a novel combination of biophysical metrics to assess the 3D differentiation of ESCs, which highlights opportunities to study the biophysical characteristics of divergent differentiation trajectories, cell phenotypes, and morphogenesis arising in stem cell-derived tissues. Together, the data presented suggest that the phenotype of multicellular mesodermal microtissues can be described by both molecular (gene/protein) and/or biophysical signature55, which together define the morphogenic state of three-dimensional pluripotent stem cell aggregates undergoing differentiation. Ultimately, the dynamic changes in embryonic tissue microenvironments illustrates important principles for the design of strategies to direct differentiation and patterning of complex stem cell-derived tissues amenable to modeling embryogenesis, screening pharmacological compounds, and developing molecular therapeutics.
Methods
Embryonic stem cell expansion
Pluripotent murine ESCs (D3 cell line) were expanded on 0.1% gelatin coated polystyrene tissue culture treated dishes in Dulbecco's Modified Eagle's Medium (DMEM) containing 15% fetal bovine serum (FBS) and supplemented with penicillin (100 U/mL), streptomycin (100 mg), amphotericin (0.25 mg/mL), L-glutamine (2 mM), MEM non-essential amino acid solution (1×), 2-mercaptoethanol (0.1 mM), and leukemia inhibitory factor (LIF; 103 U/mL). Media was exchanged every other day and ESCs were passaged prior to 70% confluence.
Embryoid body formation and culture
A single cell suspension of ESCs was obtained by treatment of monolayer cultures with 0.05% trypsin/0.53 mM EDTA solution. EBs were formed by centrifugation (200 rcf) of ESCs into 400 μm diameter polydimethylsiloxane (PDMS) microwells (Aggrewell;™ Stem Cell Technologies), with approximately 1000 cells per well. After 20 hours of microwell formation in serum containing ESC media without LIF, EBs were transferred to suspension culture (~2000 EBs per dish) and maintained on a rotary orbital shaker platform at 65 rpm, as described previously26. EBs were differentiated in basal serum-free media (N2B27) composed of DMEM/F12 supplemented with N2 (5 μg/ml insulin, 100 μg/ml transferrin, 6 ng/ml progesterone, 16 μg/ml putrescine, 30 nM sodium selenite) and 50 μg/mL bovine serum albumin (BSA), combined 1:1 with Neurobasal™ media supplemented with B2756. Antibiotics and L-glutamine were supplemented at the same concentrations as the undifferentiated ESC media. 90% of EB media, including growth factor supplements, was exchanged every other day following gravity-induced sedimentation of EBs. Mesoderm induction was accomplished, as detailed in the results, by supplementation with 10 ng/mL BMP-4 (R&D Technologies)57. Perturbation of the cytoskeleton was accomplished by a 1-hour treatment with an inhibitor of the Rho-associated protein kinase (ROCK) pathway (Y27632)32, using the standard concentration established for ESC culture (10 μM)58 or by supplementation of the media between days 4–7 of differentiation with Y27632, Jasplakinolide (Jas, 50 nM)34, or Latrunculin B (LatB, 100 nM)35. In all cases, the concentration range for inhibitors was established based upon published measurements of ED50 concentrations35, as well as concentrations employed for extended culture durations (24+ hours)34. Moreover, concentrations were screened to ensure that small molecule treatments did not lead to significant loss of ESC viability and supported EB maintenance. DMSO (<0.1%) was included as a vehicle control.
Parallel plate mechanical compression
The bulk mechanical properties of microtissues were measured using a micron-scale mechanical testing system (Microsquisher; CellScale), which calculates force via measurement of beam deflection in response to user-defined displacements (Supplementary Fig. S1a). All samples were tested in a PBS fluid bath (pH 7.4, containing 0.90 mM Ca2+ and 0.49 mM Mg2+). The cantilever beams were composed of Tungsten (modulus = 411 GPa), and beams of diameters from 76.2 μm to 152.4 μm were employed, depending on the stiffness and sensitivity required to measure different aggregates.
To determine the bulk physical properties of EBs, a viscoelastic creep method was employed, which exerts a constant force while measuring the changes in displacement over time (Supplementary Fig. S1b). The magnitude of force was chosen as the average force corresponding to approximately 40% strain, determined based upon constant strain rate analysis of stress versus strain in n = 3 samples. The time to relaxation was determined to be less than two minutes, which was established based upon standard durations from published literature46,59 and from empirical testing, which established the steady-state phase based upon the constraint of less than 1% change in deformation over 1 second intervals. The creep parameter was calculated as the percent increase in displacement during constant force interval (Supplementary Fig. S1c). Relevant physical properties, including moduli (instantaneous modulus, Eo; relaxed modulus Einf), time constants (creep time constant, τσ; stress relaxation time constant, τε), and apparent viscosity (μ) were calculated based upon a linear viscoelastic model of creep displacement (u) behavior over time, described in equations (1–3), where Do is the initial EB diameter, the stress (σ) is calculated as the constant force normalized to relaxed cross sectional EB area, and H(t) is a unit step function59.
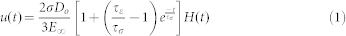 |
 |
 |
Quantitative real time PCR
RNA was extracted from EBs using the RNeasy Mini kit (Qiagen Inc, Valencia, CA) and cDNA was synthesized using the RT2 First Strand Kit (SABiosciences, Frederick, MD) with 900 ng of total RNA. To assess gene expression, a custom RT2Profiler™ PCR Array (SABiosciences) was used in conjunction with a MyIQ iCycler (Bio-rad) and iQ SYBR Green Supermix (Bio-rad). The array consists of 26 genes relevant to pluripotency and differentiation toward the three germ lineages, as well as 3 housekeeping genes (Actb, GAPDH, Hsp90ab1) and PCR efficiency controls (Supplementary Table S1). Alternatively, quantitative real time PCR was performed using primers for the hyaluronan synthase isoforms (HAS-1, HAS-2, and HAS-3), which were designed using Beacon Designer software25. PCR data were normalized to the geometric average of the three housekeeping genes and fold changes in expression were calculated from normalized Ct values, relative to expression of ESCs60. The Genesis software package was used to generate heatmap visualizations of the gene expression data and to calculate the two-dimensional hierarchical clustering, based on Euclidean distance and average linkage clustering.
Histology and immunohistochemistry
EBs sampled from rotary orbital culture were rinsed in PBS and fixed with 10% formalin (4% paraformaldehyde) for 45 minutes with rotation at room temperature. Fixed EBs were embedded within Histogel (Richard Allen Scientific), and processed through a series of ethanol and xylene rinses prior to paraffin embedding. Paraffin embedded EBs were sectioned with a thickness of 5 μm using a rotary microtome (Microtom HM310) and mounted on charged slides (SuperFrost Plus, Fisher Scientific). Sections were deparaffinized prior to staining via ethanol and xylene rinses and subsequently stained with hematoxylin and eosin, alcian blue (pH 2.5; nuclear fast red counterstain) or safranin-O (fast green counterstain).
Alternatively, for immunohistochemistry, slides were blocked in 1.5% normal goat serum (NGS) for 1 hour at room temperature, incubated in mouse monoclonal anti-human smooth muscle actin (α-SMA) primary antibody (Dako; clone 1A4) for 1 hour, and subsequently incubated in biotinylated donkey anti-mouse IgG (1:400, Vector Labs). Slides were then incubated with avidin and biotin-based horseradish peroxidase, based on manufacturer instructions (Elite Vectastain Kit, Vector Labs) and developed using a 3, 3′-diaminobenzidine (DAB) substrate (Vector Labs). Nuclei were counterstained with hematoxylin. After staining, slides were cover slipped and imaged using a Nikon 80i upright microscope. Sections of mouse intestine, cartilage, and heart, were included as positive controls for alcian blue, safranin-O, and α-SMA staining, respectively.
Multivariate partial least squares (PLS) regression modeling
A data matrix (MxN) was constructed with M culture conditions (Jas, LatB, etc), and N experimental parameters (Oct-4, Nanog, modulus, etc), where each column of the independent X (input) matrix represents a unique gene expression signal and the dependent variables in the Y (output) matrix represent mechanical properties. All data were mean-centered and scaled to unit variance and SIMCA-P (Umetrics) was employed to construct the PLSR model using a nonlinear iterative partial least squares (NIPALS) algorithm61. In order to avoid overfitting the model, the number of significant principal components was determined based on both cross-validation (Q2Y) and permutation testing of each Y variable, with the background Q2Y less than 0.05 considered significant. Predictions were determined based on cross-validation, with standard error of predicted values determined via jack-knifing, and the goodness of fit for the predicted data was calculated from coefficient of determination (R2)62. Statistical significance testing was conducted by calculating the p-values from Pearson's r correlation coefficients of the predicted and observed data. Predictability was calculated from the root mean squared error of estimation (RMSEE)63.
Statistical analysis
All experiments conducted with replicate data are represented as the mean of independent replicates +/− standard error, unless otherwise noted, with the experimental size (n) individually detailed in the corresponding figure legends. Prior to statistical analysis, all data were pre-processed using a box-cox power transform, in order to normalize data according to a Gaussian distribution. Statistical tests were conducted between groups and time points using one-way or two-way ANOVA, combined with either a post-hoc Tukey or Mann-Whitney U test for comparison of individual samples, depending on the results from Levene's equality of variances test, with p-values less than 0.05 considered significant.
Author Contributions
Conceived and designed the experiments: M.A.K., R.S., T.C.M. Performed the experiments: M.A.K., R.S. Analyzed the data: M.A.K., R.S. Contributed reagents, materials, and analysis tools: M.A.K., T.C.M. Wrote the paper: M.A.K., T.C.M.
Supplementary Material
Supplemental Information
Acknowledgments
The authors thank Dr. Andrés García, Dr. Melissa Kemp, and Dr. Todd Streelman for critical review of this manuscript. In addition, we would like to thank Dr. Manu Platt and Keon-Young Park at Georgia Institute of Technology for helpful discussions regarding PLS and Dr. Tracy Hookway for assistance with immunohistochemical techniques. This work was supported by the National Institutes of Health (NIH R01 EB010061) and the National Science Foundation Emergent Behavior of Integrated Cellular Systems (EBICS) Science and Technology Center (CBET 093511). M.A.K. is currently supported by an American Heart Association (AHA) Pre-Doctoral Fellowship and previously by an NSF Graduate Research Fellowship.
References
- Scadden D. T. The stem-cell niche as an entity of action. Nature 441, 1075–1079 (2006). [DOI] [PubMed] [Google Scholar]
- Morrison S. J. & Spradling A. C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Tumbar T. & Guasch G. Socializing with the neighbors: stem cells and their niche. Cell 116, 769–778 (2004). [DOI] [PubMed] [Google Scholar]
- Chen S., Lewallen M. & Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development 140, 255–265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. & Neaves W. B. Normal stem cells and cancer stem cells: the niche matters. Cancer Res 66, 4553–4557 (2006). [DOI] [PubMed] [Google Scholar]
- Tam P. P. L. & Loebel D. A. F. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet 8, 368–381 (2007). [DOI] [PubMed] [Google Scholar]
- Burdsal C. A., Damsky C. H. & Pedersen R. A. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development 118, 829–844 (1993). [DOI] [PubMed] [Google Scholar]
- Ciruna B. & Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell 1, 37–49 (2001). [DOI] [PubMed] [Google Scholar]
- Thiery J. P., Acloque H., Huang R. Y. J. & Nieto M. A. Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- Keller R., Davidson L. A. & Shook D. R. How we are shaped: the biomechanics of gastrulation. Differentiation 71, 171–205 (2003). [DOI] [PubMed] [Google Scholar]
- Behrndt M. et al. Forces driving epithelial spreading in zebrafish gastrulation. Science 338, 257–260 (2012). [DOI] [PubMed] [Google Scholar]
- Krieg M. et al. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol 10, 429–436 (2008). [DOI] [PubMed] [Google Scholar]
- Kim H. Y. & Davidson L. A. Punctuated actin contractions during convergent extension and their permissive regulation by the non-canonical Wnt-signaling pathway. J Cell Sci 124, 635–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Kim H. Y. & Davidson L. A. Actomyosin stiffens the vertebrate embryo during crucial stages of elongation and neural tube closure. Development 136, 677–688 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J. D., Montague R. A. & Kiehart D. P. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol 15, 2208–2221 (2005). [DOI] [PubMed] [Google Scholar]
- Aegerter-Wilmsen T., Aegerter C. M., Hafen E. & Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev 124, 318–326 (2007). [DOI] [PubMed] [Google Scholar]
- Diez-Roux G. et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9, e1000582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M. A. & Chen C. S. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol 10, 34–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- Dassow M., Strother J. A. & Davidson L. A. Surprisingly simple mechanical behavior of a complex embryonic tissue. PLoS ONE 5, e15359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S.-I., Jakt L. M. & Era T. Embryonic stem-cell culture as a tool for developmental cell biology. Nat Rev Mol Cell Biol 8, 502–507 (2007). [DOI] [PubMed] [Google Scholar]
- Murry C. E. & Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680 (2008). [DOI] [PubMed] [Google Scholar]
- Discher D. E., Mooney D. J. & Zandstra P. W. Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673–1677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. & Discher D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- Shukla S. et al. Synthesis and organization of hyaluronan and versican by embryonic stem cells undergoing embryoid body differentiation. J Histochem Cytochem 58, 345–358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney M. A., Saeed R. & McDevitt T. C. Systematic analysis of embryonic stem cell differentiation in hydrodynamic environments with controlled embryoid body size. Integr Biol 4, 641–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B. M. & Wiles M. V. Evidence for involvement of activin A and bone morphogenetic protein 4 in mammalian mesoderm and hematopoietic development. Mol Cell Biol 15, 141–151 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Lü S., Vida L., Thomson J. A. & Honig G. R. Bone morphogenetic protein 4 induces efficient hematopoietic differentiation of rhesus monkey embryonic stem cells in vitro. Blood 98, 335–342 (2001). [DOI] [PubMed] [Google Scholar]
- Masszi A. et al. Central role for Rho in TGF-beta1-induced alpha-smooth muscle actin expression during epithelial-mesenchymal transition. Am J Physiol Renal Physiol 284, F911–24 (2003). [DOI] [PubMed] [Google Scholar]
- Sachlos E. & Auguste D. T. Embryoid body morphology influences diffusive transport of inductive biochemicals: a strategy for stem cell differentiation. Biomaterials 29, 4471–4480 (2008). [DOI] [PubMed] [Google Scholar]
- Kardash E. et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol 12, 47–53– sup pp 1–11 (2010). [DOI] [PubMed] [Google Scholar]
- Ishizaki T. et al. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol 57, 976–983 (2000). [PubMed] [Google Scholar]
- Gomez E. W., Chen Q. K., Gjorevski N. & Nelson C. M. Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J Cell Biochem 110, 44–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A. & Beier F. RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J Biol Chem 281, 13134–13140 (2006). [DOI] [PubMed] [Google Scholar]
- Wakatsuki T., Schwab B., Thompson N. C. & Elson E. L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci 114, 1025–1036 (2001). [DOI] [PubMed] [Google Scholar]
- Saha K. et al. Substrate modulus directs neural stem cell behavior. Biophys J 95, 4426–4438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K. & Chen C. S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6, 483–495 (2004). [DOI] [PubMed] [Google Scholar]
- Wobus A. M. & Boheler K. R. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol Rev 85, 635–678 (2005). [DOI] [PubMed] [Google Scholar]
- Jensen J., Hyllner J. & Björquist P. Human embryonic stem cell technologies and drug discovery. J Cell Physiol 219, 513–519 (2009). [DOI] [PubMed] [Google Scholar]
- Baraniak P. R. & McDevitt T. C. Stem cell paracrine actions and tissue regeneration. Regen Med 5, 121–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt S. et al. The Bmp gradient of the zebrafish gastrula guides migrating lateral cells by regulating cell-cell adhesion. Curr Biol 17, 475–487 (2007). [DOI] [PubMed] [Google Scholar]
- Yeung T. et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton 60, 24–34 (2005). [DOI] [PubMed] [Google Scholar]
- Ducibella T. & Anderson E. Cell shape and membrane changes in the eight-cell mouse embryo: prerequisites for morphogenesis of the blastocyst. Dev Biol 47, 45–58 (1975). [DOI] [PubMed] [Google Scholar]
- Nair R., Ngangan A. V., Kemp M. L. & McDevitt T. C. Gene expression signatures of extracellular matrix and growth factors during embryonic stem cell differentiation. PLoS ONE 7, e42580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O. et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat Methods (2013). 10.1038/nmeth.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek G. et al. Mechanical characterization of differentiated human embryonic stem cells. J Biomech Eng 131, 061011 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E., Janmey P. & Wang Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005). [DOI] [PubMed] [Google Scholar]
- Swift J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski J. D., Dahl K. N., Zhong F. L., Sammak P. J. & Discher D. E. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA 104, 15619–15624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M. et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci USA 102, 11594–11599 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiser M. D. et al. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci USA 107, 610–615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Cruz R. D., Fonseca V. C. & Darling E. M. Cellular mechanical properties reflect the differentiation potential of adipose-derived mesenchymal stem cells. Proc Natl Acad Sci USA 109, E1523–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak P. R. et al. Stiffening of human mesenchymal stem cell spheroid microenvironments induced by incorporation of gelatin microparticles. J Mech Behav Biomed Mater 11, 63–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F.-J. et al. A bioinformatic assay for pluripotency in human cells. Nat Methods 8, 315–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T. & Ingber D. E. Mechanical control of tissue and organ development. Development 137, 1407–1420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L. & Smith A. G. Defined conditions for neural commitment and differentiation. Meth Enzymol 365, 327–341 (2003). [DOI] [PubMed] [Google Scholar]
- Purpura K. A., Morin J. & Zandstra P. W. Analysis of the temporal and concentration-dependent effects of BMP-4, VEGF, and TPO on development of embryonic stem cell-derived mesoderm and blood progenitors in a defined, serum-free media. Exp Hematol 36, 1186–1198 (2008). [DOI] [PubMed] [Google Scholar]
- Watanabe K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnol 25, 681–686 (2007). [DOI] [PubMed] [Google Scholar]
- Leipzig N. D. & Athanasiou K. A. K. A. Unconfined creep compression of chondrocytes. J Biomech 38, 77–85 (2005). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geladi P. & Kowalski B. R. Partial least-squares regression: a tutorial. Analytica Chimica Acta 185, 1–17 (1986). [Google Scholar]
- Kumar N., Wolf-Yadlin A., White F. M. & Lauffenburger D. A. Modeling HER2 effects on cell behavior from mass spectrometry phosphotyrosine data. PLoS Comput Biol 3, e4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.-Y., Li W. A. & Platt M. O. Patient specific proteolytic activity of monocyte-derived macrophages and osteoclasts predicted with temporal kinase activation states during differentiation. Integr Biol 4, 1459–1469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information