Abstract
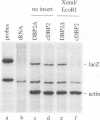
The human p68, Saccharomyces cerevisiae DBP2 and Schizosaccharomyces pombe dbp2 genes are closely related members of the 'DEAD-box' RNA helicase superfamily. All three genes contain an intron at a conserved site in RNA helicase motif V. The S.cerevisiae intron is unusual both for its position near the 3'-end of the open reading frame and for its size, 1001 nucleotides. We show here that precise deletion of the intron has no effect on cell viability but leads to an increase in Dbp2p protein expression. Inefficient splicing due to the size of the intron can not account for this difference because the intron is efficiently spliced in Dbp2p-deficient cells. Instead, there is a reciprocal relationship between the amount of Dbp2p in the cell and the efficiency with which DBP2 intron-containing genes are expressed. Inactive Dbp2p mutants are efficiently expressed from DBP2 intron-containing plasmids, and fragments of the DBP2 intron confer Dbp2p-responsiveness on heterologous reporter introns. This suggest that there is an intron-mediated negative feedback loop regulating DBP2 expression, and provides a possible explanation for the retention of such an unusual intron in S.cerevisiae.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balvay L., Libri D., Fiszman M. Y. Pre-mRNA secondary structure and the regulation of splicing. Bioessays. 1993 Mar;15(3):165–169. doi: 10.1002/bies.950150304. [DOI] [PubMed] [Google Scholar]
- Banroques J., Delahodde A., Jacq C. A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell. 1986 Sep 12;46(6):837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- Baserga S. J., Yang X. D., Steitz J. A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991 Sep;10(9):2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Burgess S. M., Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993 Oct;18(10):381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Burgess S., Couto J. R., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990 Mar 9;60(5):705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Fragapane P., Gehring C., Bozzoni I. The accumulation of mature RNA for the Xenopus laevis ribosomal protein L1 is controlled at the level of splicing and turnover of the precursor RNA. EMBO J. 1987 Nov;6(11):3493–3498. doi: 10.1002/j.1460-2075.1987.tb02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C., Legrain P. A novel gene, spp91-1, suppresses the splicing defect and the pre-mRNA nuclear export in the prp9-1 mutant. EMBO J. 1992 Sep;11(9):3279–3288. doi: 10.1002/j.1460-2075.1992.tb05406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Lin R. J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990 Nov 11;18(21):6447–6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991 Feb 7;349(6309):487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Dabeva M. D., Post-Beittenmiller M. A., Warner J. R. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5854–5857. doi: 10.1073/pnas.83.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Abelson J. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991 May 31;65(5):797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Fragapane P., Prislei S., Michienzi A., Caffarelli E., Bozzoni I. A novel small nucleolar RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J. 1993 Jul;12(7):2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguel V., Wang Y., Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol. 1993 Nov;13(11):6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995 Feb 15;9(4):437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- He F., Peltz S. W., Donahue J. L., Rosbash M., Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Iggo R. D., Jamieson D. J., MacNeill S. A., Southgate J., McPheat J., Lane D. P. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol. 1991 Mar;11(3):1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R. D., Lane D. P. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989 Jun;8(6):1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R., Picksley S., Southgate J., McPheat J., Lane D. P. Identification of a putative RNA helicase in E.coli. Nucleic Acids Res. 1990 Sep 25;18(18):5413–5417. doi: 10.1093/nar/18.18.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994 Nov 10;372(6502):193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- Ishioka C., Frebourg T., Yan Y. X., Vidal M., Friend S. H., Schmidt S., Iggo R. Screening patients for heterozygous p53 mutations using a functional assay in yeast. Nat Genet. 1993 Oct;5(2):124–129. doi: 10.1038/ng1093-124. [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Kiledjian M., Kadesch T. Post-transcriptional regulation of the human liver/bone/kidney alkaline phosphatase gene. J Biol Chem. 1991 Mar 5;266(7):4207–4213. [PubMed] [Google Scholar]
- Larson G. P., Itakura K., Ito H., Rossi J. J. Saccharomyces cerevisiae actin--Escherichia coli lacZ gene fusions: synthetic-oligonucleotide-mediated deletion of the 309 base pair intervening sequence in the actin gene. Gene. 1983 Apr;22(1):31–39. doi: 10.1016/0378-1119(83)90061-6. [DOI] [PubMed] [Google Scholar]
- Leeds P., Peltz S. W., Jacobson A., Culbertson M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- MacArthur H., Walter G. Monoclonal antibodies specific for the carboxy terminus of simian virus 40 large T antigen. J Virol. 1984 Nov;52(2):483–491. doi: 10.1128/jvi.52.2.483-491.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matviw H., Yu G., Young D. Identification and genetic analysis of Schizosaccharomyces pombe cDNAs that suppress deletion of IRA1 in Saccharomyces cerevisiae. Gene. 1993 Jul 15;129(1):147–152. doi: 10.1016/0378-1119(93)90711-b. [DOI] [PubMed] [Google Scholar]
- Prislei S., Michienzi A., Presutti C., Fragapane P., Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993 Dec 25;21(25):5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Schmid S. R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992 Feb;6(3):283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Schärer E., Iggo R. Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res. 1992 Apr 11;20(7):1539–1545. doi: 10.1093/nar/20.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993 Nov 5;75(3):403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- Strauss E. J., Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991 Apr;5(4):629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Warner J. R. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994 Jan;8(2):211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991 Feb 7;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]