Abstract
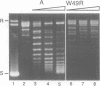
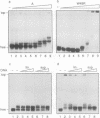
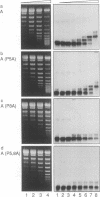
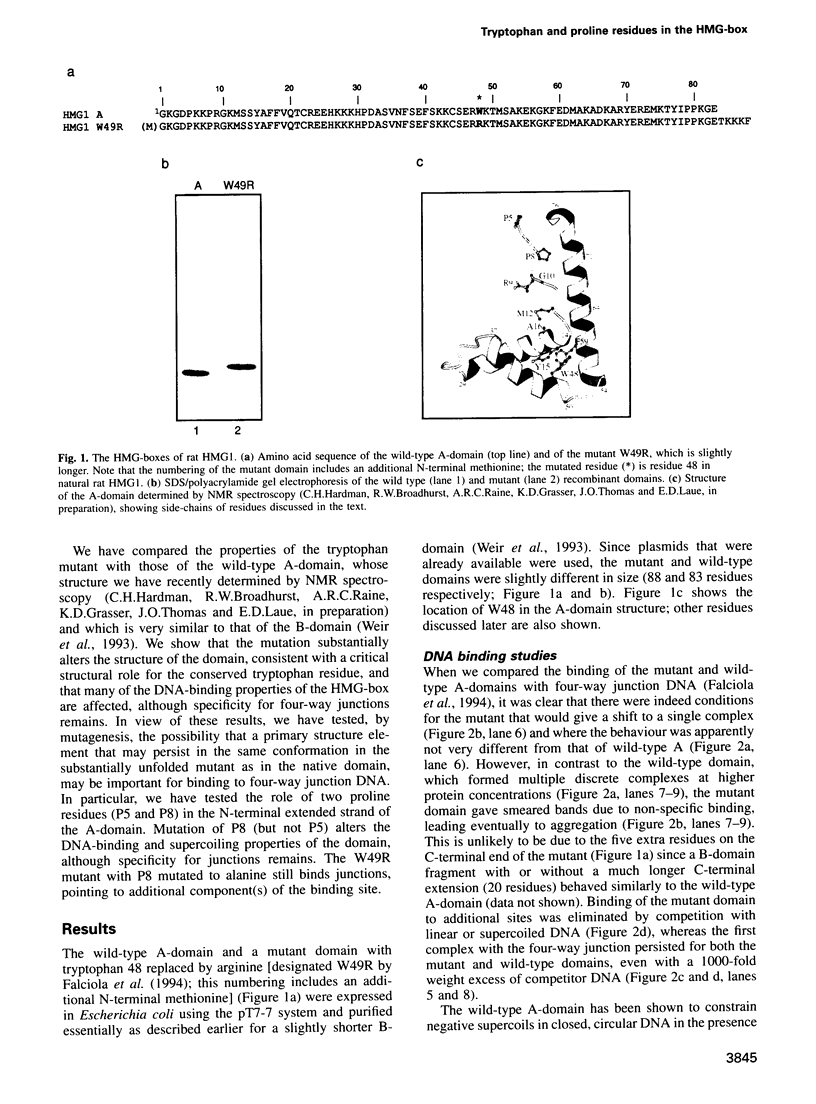
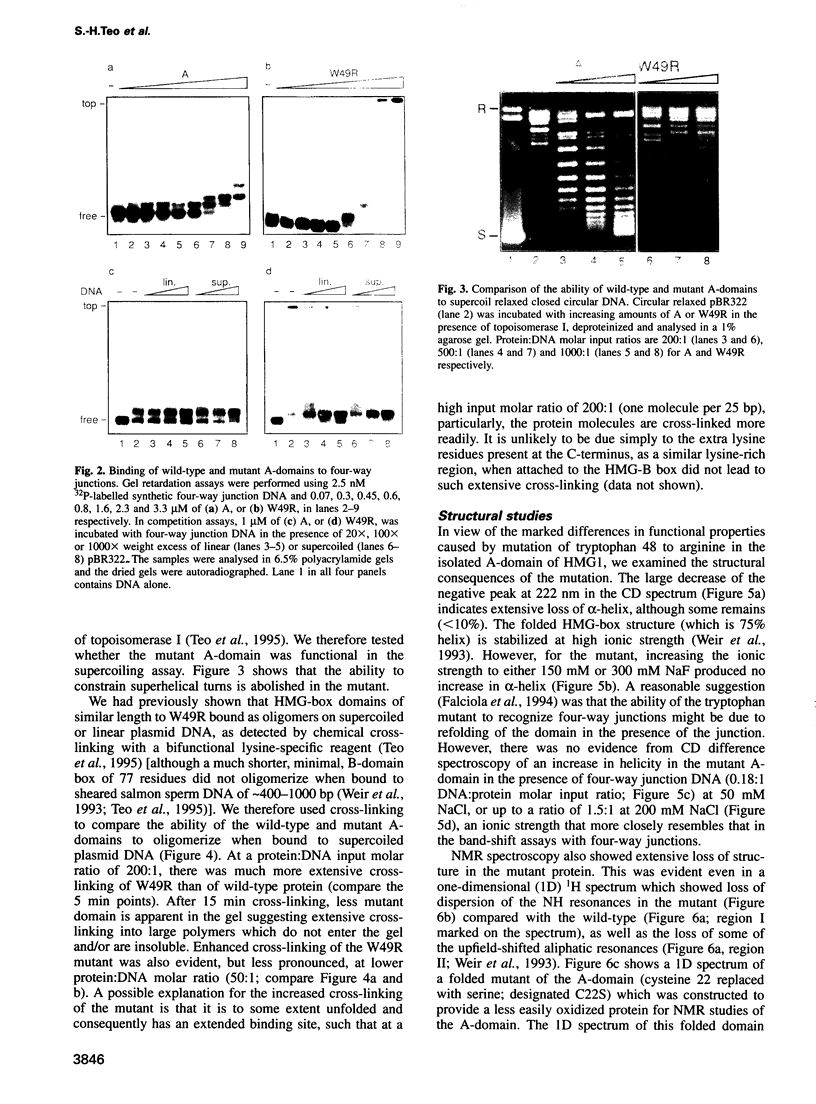
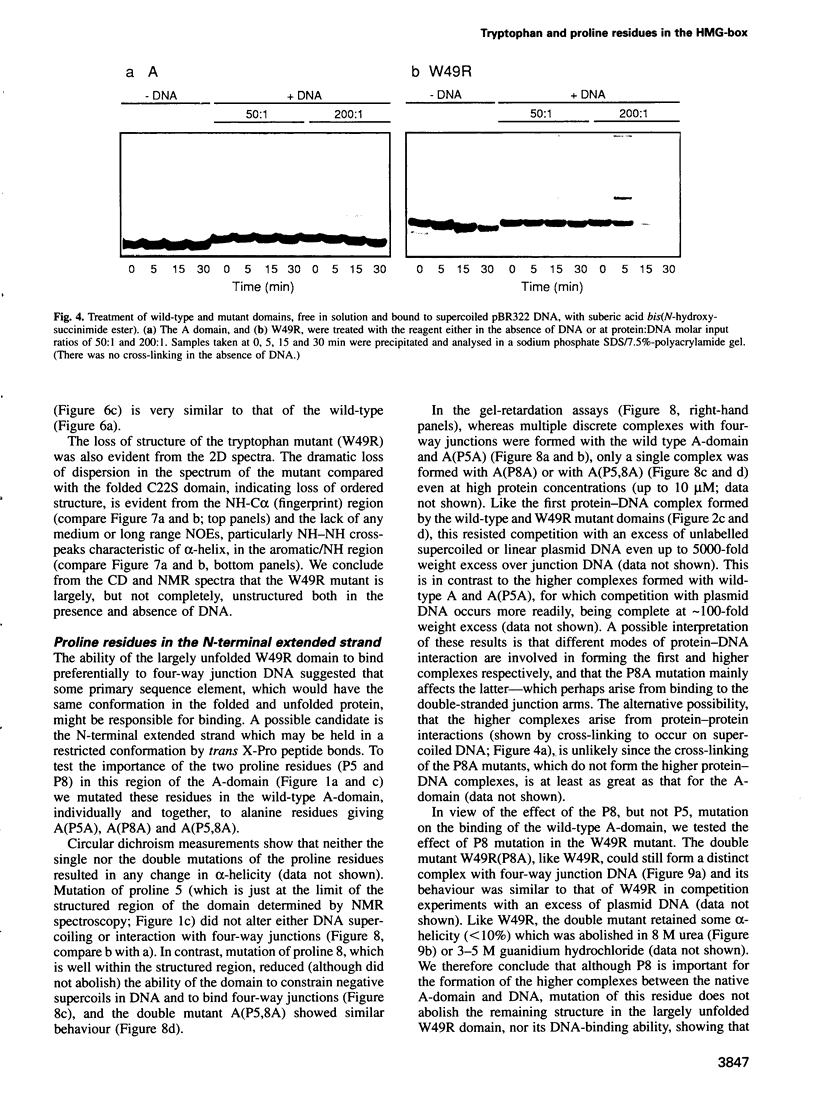
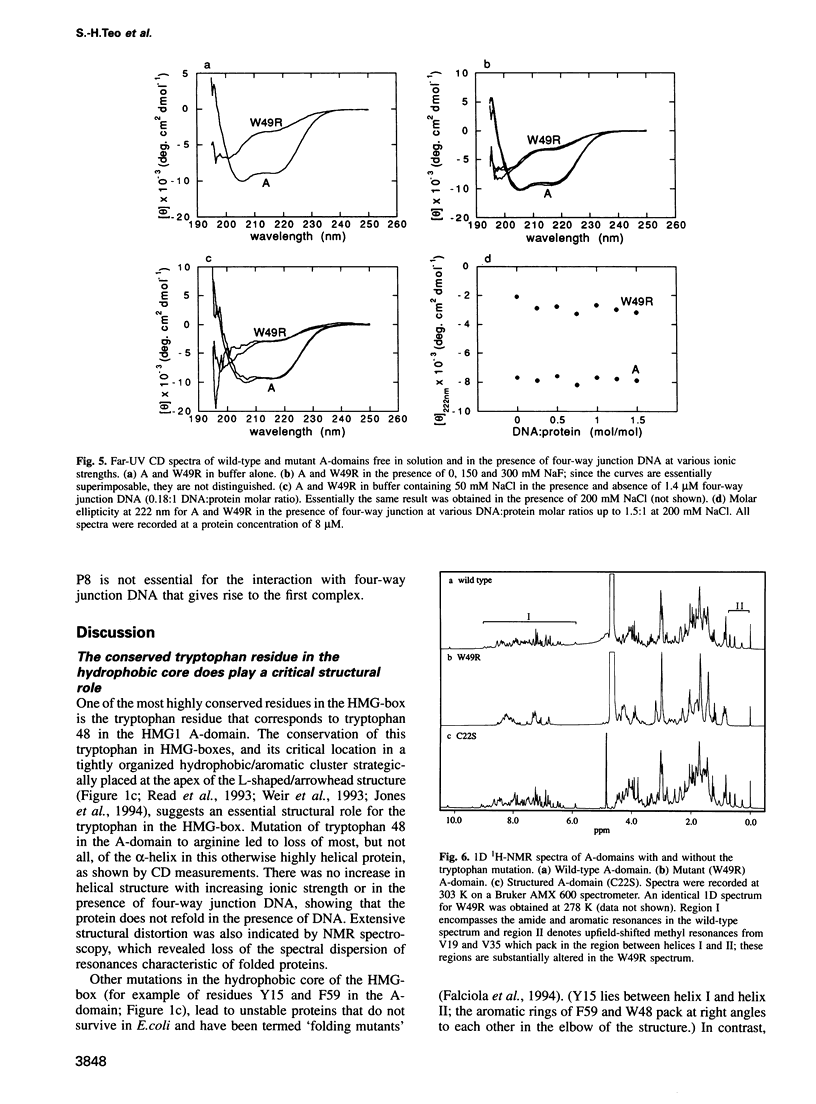
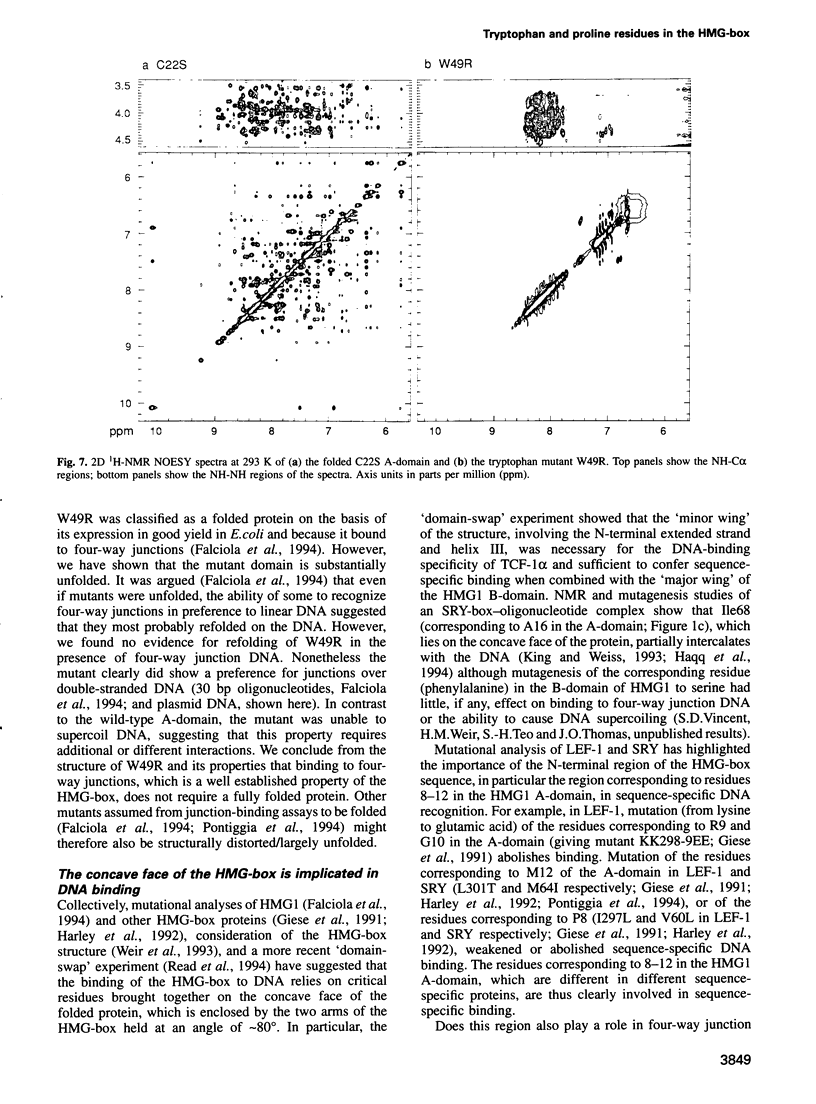
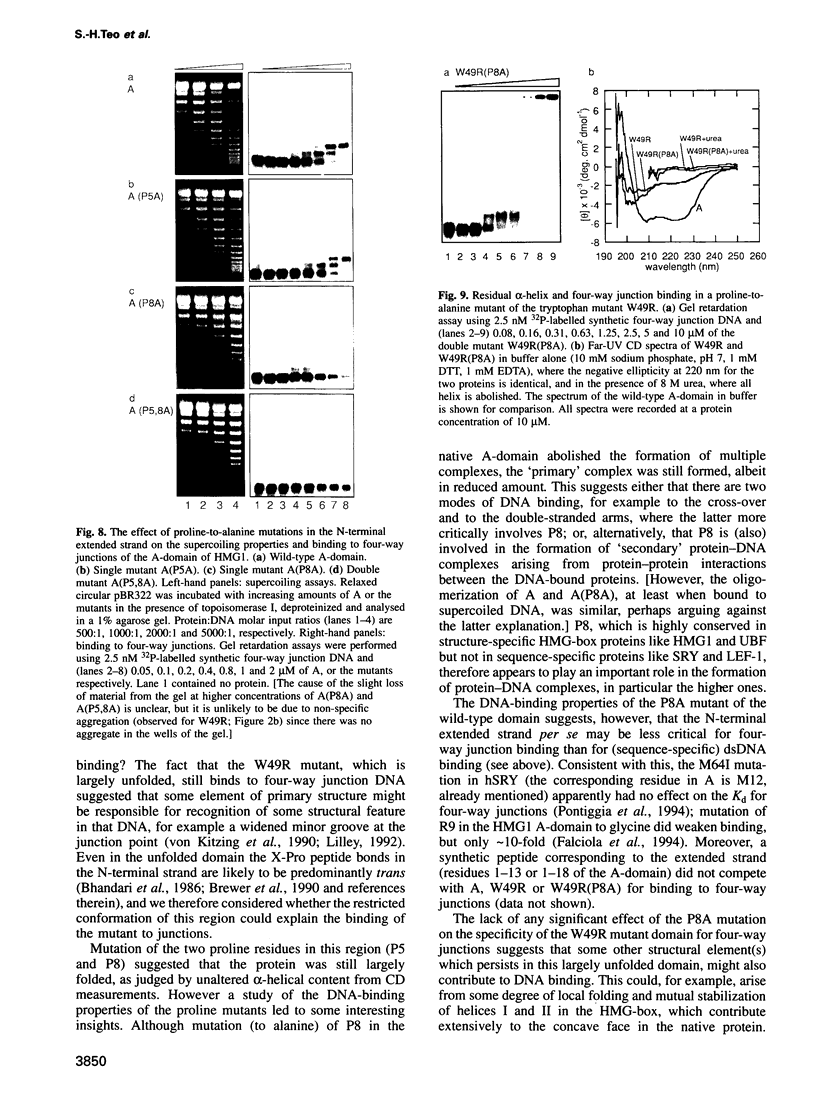
Mutation of the highly conserved tryptophan residue in the A-domain HMG-box of HMG1 largely, but not completely, destroys the protein tertiary structure and abolishes its supercoiling ability, but does not abolish structure-specific DNA binding to four-way junctions. Circular dichroism shows that the protein has some residual alpha-helix (< 10%) and does not re-fold in the presence of DNA. Structure-specific DNA binding might therefore be a property of some primary structure element, for example the N-terminal extended strand, which even in the unfolded protein would be held in a restricted conformation by two, largely trans, X-Pro peptide bonds. However, mutation of P5 or P8 of the A-domain to alanine does not abolish the formation of the (first) complex in a gel retardation assay, which probably arises from binding to the junction cross-over, although the P8 mutation does affect the formation of higher complexes which may arise from binding to the junction arms. Since mutation of P8 in the W49R mutant has no effect on structure-specific junction binding, we propose that some residual alpha-helix in the protein might be involved, implicating this element in the interactions of HMG-boxes generally with DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdul-Razzak K. K., Denton M. L., Cox D. J., Reeck G. R. Isolation and characterization of folded fragments released by Staphylococcal aureus proteinase from the non-histone chromosomal protein HMG-1. Biochim Biophys Acta. 1989 Jun 13;996(1-2):125–131. doi: 10.1016/0167-4838(89)90104-0. [DOI] [PubMed] [Google Scholar]
- Bachvarov D., Moss T. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 1991 May 11;19(9):2331–2335. doi: 10.1093/nar/19.9.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D. G., Levine B. A., Trayer I. P., Yeadon M. E. 1H-NMR study of mobility and conformational constraints within the proline-rich N-terminal of the LC1 alkali light chain of skeletal myosin. Correlation with similar segments in other protein systems. Eur J Biochem. 1986 Oct 15;160(2):349–356. doi: 10.1111/j.1432-1033.1986.tb09978.x. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Falciola L., Ferrari S., Lilley D. M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992 Mar;11(3):1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E., Takahashi M., Yaniv J. R. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J Mol Biol. 1994 Sep 16;242(2):116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- Brewer S., Tolley M., Trayer I. P., Barr G. C., Dorman C. J., Hannavy K., Higgins C. F., Evans J. S., Levine B. A., Wormald M. R. Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J Mol Biol. 1990 Dec 20;216(4):883–895. doi: 10.1016/S0022-2836(99)80008-4. [DOI] [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Carballo M., Puigdomènech P., Palau J. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 1983;2(10):1759–1764. doi: 10.1002/j.1460-2075.1983.tb01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary P. D., Turner C. H., Mayes E., Crane-Robinson C. Conformation and domain structure of the non-histone chromosomal proteins, HMG 1 and 2. Isolation of two folded fragments from HMG 1 and 2. Eur J Biochem. 1983 Mar 15;131(2):367–374. doi: 10.1111/j.1432-1033.1983.tb07272.x. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Architectural elements in nucleoprotein complexes. Curr Biol. 1993 Oct 1;3(10):675–676. doi: 10.1016/0960-9822(93)90065-v. [DOI] [PubMed] [Google Scholar]
- Falciola L., Murchie A. I., Lilley D. M., Bianchi M. Mutational analysis of the DNA binding domain A of chromosomal protein HMG1. Nucleic Acids Res. 1994 Feb 11;22(3):285–292. doi: 10.1093/nar/22.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Amsterdam A., Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991 Dec;5(12B):2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Grasser K. D., Krech A. B., Feix G. The maize chromosomal HMGa protein recognizes structural features of DNA and increases DNA flexibility. Plant J. 1994 Sep;6(3):351–358. doi: 10.1046/j.1365-313x.1994.06030351.x. [DOI] [PubMed] [Google Scholar]
- Griess E. A., Rensing S. A., Grasser K. D., Maier U. G., Feix G. Phylogenetic relationships of HMG box DNA-binding domains. J Mol Evol. 1993 Aug;37(2):204–210. doi: 10.1007/BF02407357. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Haqq C. M., King C. Y., Ukiyama E., Falsafi S., Haqq T. N., Donahoe P. K., Weiss M. A. Molecular basis of mammalian sexual determination: activation of Müllerian inhibiting substance gene expression by SRY. Science. 1994 Dec 2;266(5190):1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- Harley V. R., Jackson D. I., Hextall P. J., Hawkins J. R., Berkovitz G. D., Sockanathan S., Lovell-Badge R., Goodfellow P. N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992 Jan 24;255(5043):453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- Hughes E. N., Engelsberg B. N., Billings P. C. Purification of nuclear proteins that bind to cisplatin-damaged DNA. Identity with high mobility group proteins 1 and 2. J Biol Chem. 1992 Jul 5;267(19):13520–13527. [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P., Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990 Apr 26;344(6269):830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- Jones D. N., Searles M. A., Shaw G. L., Churchill M. E., Ner S. S., Keeler J., Travers A. A., Neuhaus D. The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure. 1994 Jul 15;2(7):609–627. doi: 10.1016/s0969-2126(00)00063-0. [DOI] [PubMed] [Google Scholar]
- King C. Y., Weiss M. A. The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: a topological mechanism of sequence specificity. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11990–11994. doi: 10.1073/pnas.90.24.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman D., Bustin M. A signature for the HMG-1 box DNA-binding proteins. Bioessays. 1993 Aug;15(8):539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- Laudet V., Stehelin D., Clevers H. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 1993 May 25;21(10):2493–2501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. DNA--protein interactions. HMG has DNA wrapped up. Nature. 1992 May 28;357(6376):282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Paonessa G., Frank R., Cortese R. Nucleotide sequence of rat liver HMG1 cDNA. Nucleic Acids Res. 1987 Nov 11;15(21):9077–9077. doi: 10.1093/nar/15.21.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Pil P. M., Chow C. S., Lippard S. J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pil P. M., Lippard S. J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992 Apr 10;256(5054):234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- Pontiggia A., Rimini R., Harley V. R., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994 Dec 15;13(24):6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read C. M., Cary P. D., Crane-Robinson C., Driscoll P. C., Norman D. G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993 Jul 25;21(15):3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read C. M., Cary P. D., Preston N. S., Lnenicek-Allen M., Crane-Robinson C. The DNA sequence specificity of HMG boxes lies in the minor wing of the structure. EMBO J. 1994 Dec 1;13(23):5639–5646. doi: 10.1002/j.1460-2075.1994.tb06902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeck G. R., Isackson P. J., Teller D. C. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982 Nov 4;300(5887):76–78. doi: 10.1038/300076a0. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Wang T., Wu M., Bowen J., Cook R. G., Gorovsky M. A., Allis C. D. Macronuclei and micronuclei in Tetrahymena thermophila contain high-mobility-group-like chromosomal proteins containing a highly conserved eleven-amino-acid putative DNA-binding sequence. Mol Cell Biol. 1991 Jan;11(1):166–174. doi: 10.1128/mcb.11.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros M., Stokrová J., Thomas J. O. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994 Mar 25;22(6):1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thomas J. O. Chemical cross-linking of histones. Methods Enzymol. 1989;170:549–571. doi: 10.1016/0076-6879(89)70064-1. [DOI] [PubMed] [Google Scholar]
- Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Allis C. D., Sweet M. T., Cook R. G., Thatcher T. H., Gorovsky M. A. Four distinct and unusual linker proteins in a mitotically dividing nucleus are derived from a 71-kilodalton polyprotein, lack p34cdc2 sites, and contain protein kinase A sites. Mol Cell Biol. 1994 Jan;14(1):10–20. doi: 10.1128/mcb.14.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]