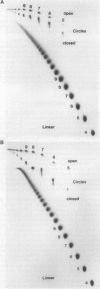
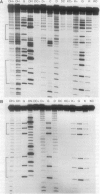
Abstract
Recent experiments have exposed significant discrepancies between experimental data and predictive models for DNA structure. These results strongly suggest that DNA structural parameters incorporated in the models are not always sufficient to account for the influence of sequence context and of specific ion effects. In an attempt to evaluate these two effects, we have investigated repetitive DNA sequences with the sequence motif GAGAG.CTCTC located in different helical phasing arrangements with respect to poly(A) tracts and GGGCCC.GGGCCC sequence motifs. Methods used are ligase-mediated cyclization and gel mobility experiments along with DNase I cutting and chemical probe studies. The results provide new evidence for curvature in poly(A) tracts. They also show that the sequence context in which bending and flexible sequence elements are found is an important aspect of sequence-dependent DNA conformation. Although dinucleotide models generally have good predictive power, this work demonstrates that in some instances sequence elements larger than the dinucleotide must be taken into account, and hence it provides a starting point for the appropriate modification and refinement of existing structural models for DNA.
Full text
PDF
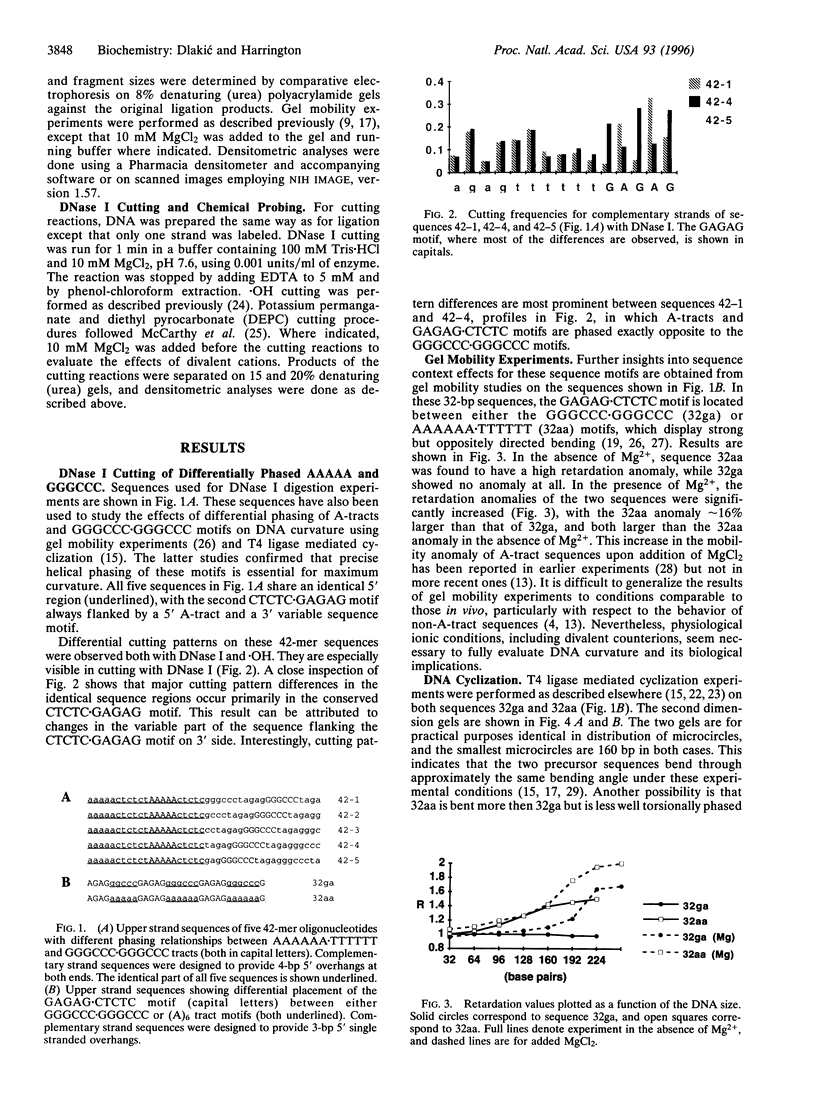
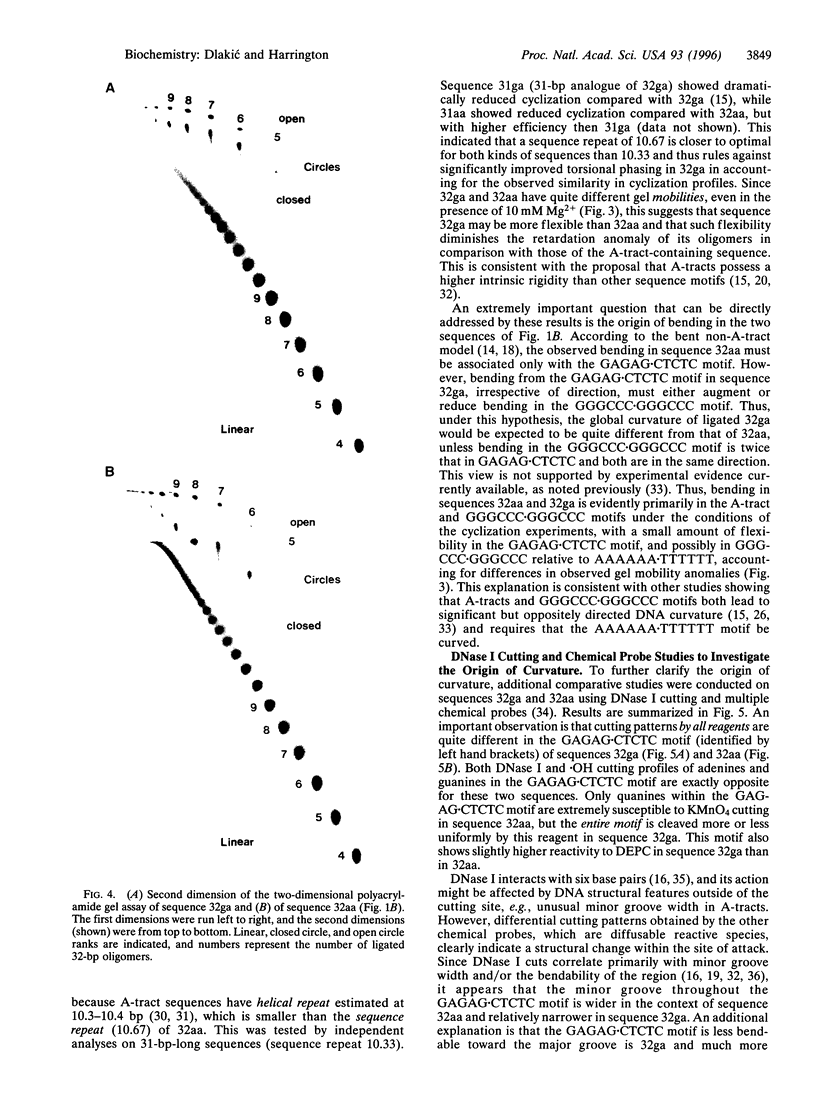



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I., Dlakic M., Savic A., Susic S., Pongor S., Suck D. Evidence for opposite groove-directed curvature of GGGCCC and AAAAA sequence elements. Nucleic Acids Res. 1993 Feb 25;21(4):1025–1029. doi: 10.1093/nar/21.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I., Susic S., Dlakic M., Savic A., Pongor S. Physiological concentration of magnesium ions induces a strong macroscopic curvature in GGGCCC-containing DNA. J Mol Biol. 1994 Feb 11;236(1):26–32. doi: 10.1006/jmbi.1994.1115. [DOI] [PubMed] [Google Scholar]
- Brukner I., Sánchez R., Suck D., Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995 Apr 18;14(8):1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R., McCall M. J. The intrinsic curvature of DNA in solution. J Mol Biol. 1988 May 5;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Drak J., Kahn J. D., Levene S. D. DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- De Santis P., Palleschi A., Savino M., Scipioni A. Validity of the nearest-neighbor approximation in the evaluation of the electrophoretic manifestations of DNA curvature. Biochemistry. 1990 Oct 2;29(39):9269–9273. doi: 10.1021/bi00491a023. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M., Harrington R. E. Bending and torsional flexibility of G/C-rich sequences as determined by cyclization assays. J Biol Chem. 1995 Dec 15;270(50):29945–29952. doi: 10.1074/jbc.270.50.29945. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Dickerson R. E. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994 Dec 11;22(24):5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Kaczor-Grzeskowiak M., Dickerson R. E. The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T-A steps. J Mol Biol. 1994 May 27;239(1):79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Kahn J. D., Crothers D. M. Sequence elements responsible for DNA curvature. J Mol Biol. 1994 Nov 25;244(2):135–143. doi: 10.1006/jmbi.1994.1713. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. DNA curving and bending in protein-DNA recognition. Mol Microbiol. 1992 Sep;6(18):2549–2555. doi: 10.1111/j.1365-2958.1992.tb01431.x. [DOI] [PubMed] [Google Scholar]
- Harrington R. E. Studies of DNA bending and flexibility using gel electrophoresis. Electrophoresis. 1993 Aug;14(8):732–746. doi: 10.1002/elps.11501401116. [DOI] [PubMed] [Google Scholar]
- Harrington R. E., Winicov I. New concepts in protein-DNA recognition: sequence-directed DNA bending and flexibility. Prog Nucleic Acid Res Mol Biol. 1994;47:195–270. doi: 10.1016/s0079-6603(08)60253-6. [DOI] [PubMed] [Google Scholar]
- Kahn J. D., Yun E., Crothers D. M. Detection of localized DNA flexibility. Nature. 1994 Mar 10;368(6467):163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Drak J., Rice J. A., Crothers D. M. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990 May 1;29(17):4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Probes of DNA structure. Methods Enzymol. 1992;212:133–139. doi: 10.1016/0076-6879(92)12009-f. [DOI] [PubMed] [Google Scholar]
- Lyubchenko Y., Shlyakhtenko L., Chernov B., Harrington R. E. DNA bending induced by Cro protein binding as demonstrated by gel electrophoresis. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5331–5334. doi: 10.1073/pnas.88.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroun R. C., Olson W. K. Base sequence effects in double-helical DNA. III. Average properties of curved DNA. Biopolymers. 1988 Apr;27(4):585–603. doi: 10.1002/bip.360270404. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Williams L. D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990 Jun 26;29(25):6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Shliakhtenko L. S., Liubchenko Iu L., Chernov B. K., Zhurkin V. B. Vliianie temperatury i ionnoi sily na élektroforeticheskuiu podvizhnost' sinteticheskikh fragmentov DNK. Mol Biol (Mosk) 1990 Jan-Feb;24(1):79–95. [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S. A., Lahm A., Suck D. X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol. 1992 Aug 20;226(4):1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]