Abstract
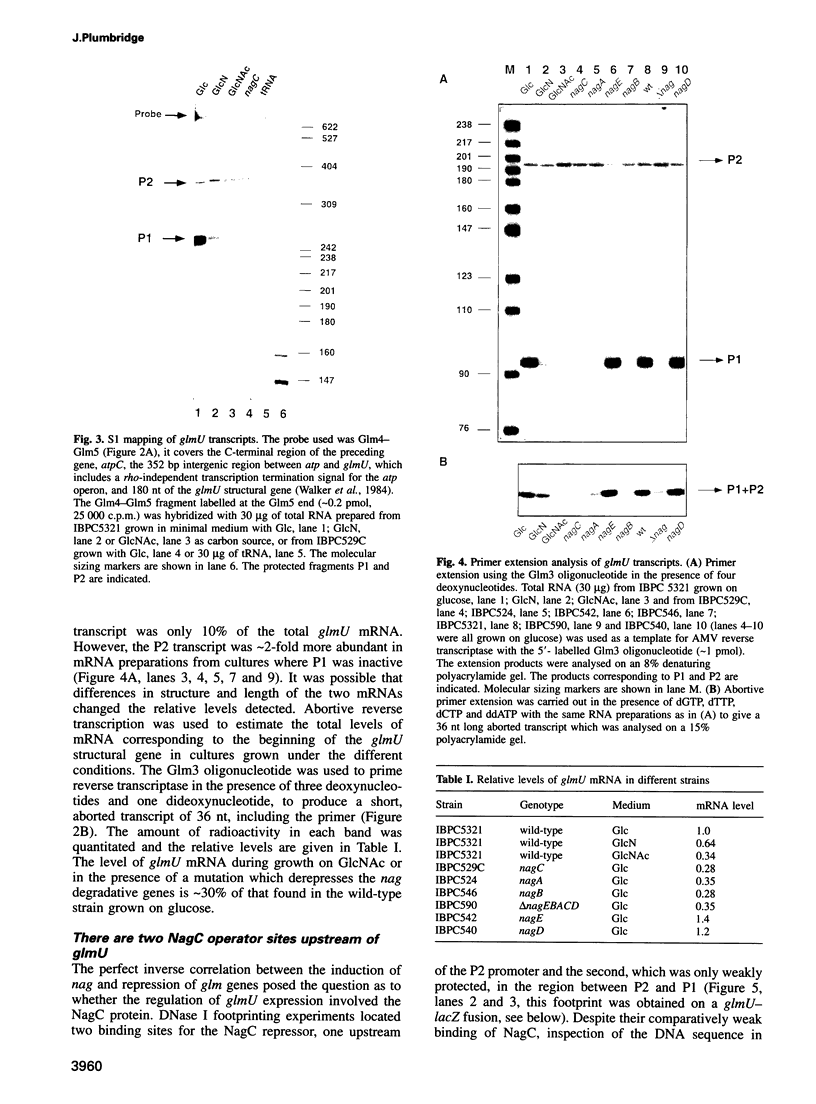
The NagC repressor controls the expression of the divergently transcribed nagE-nagBACD operons involved in the uptake and degradation of the amino sugars, N-acetyl-D-glucosamine (GlcNAc) and glucosamine (GlcN). The glmUS operon, encoding proteins necessary for the synthesis of GlcN (glmS) and the formation of UDP-GlcNAc (glmU), is transcribed from two promoters located upstream of glmU. In the absence of amino sugars both promoters are active. However, in the presence of GlcNAc, the glmU proximal promoter, P1, is inactive while the upstream promoter, P2, is subject to weak induction. Two binding sites for the NagC repressor are located at -200 and -47 bp upstream of P1. Mutations which prevent NagC binding to either of these sites eliminate expression from the P1 promoter. This shows that binding of NagC is necessary for expression of the glmU P1 promoter and implies that NagC is playing the role of activator for this promoter. Moreover, the location of the distal NagC site suggests that this site is behaving like an upstream activating sequence (UAS).
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Busby S., Ebright R. H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994 Dec 2;79(5):743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Busby S., West D., Lawes M., Webster C., Ishihama A., Kolb A. Transcription activation by the Escherichia coli cyclic AMP receptor protein. Receptors bound in tandem at promoters can interact synergistically. J Mol Biol. 1994 Aug 19;241(3):341–352. doi: 10.1006/jmbi.1994.1511. [DOI] [PubMed] [Google Scholar]
- Chin A. M., Feucht B. U., Saier M. H., Jr Evidence for regulation of gluconeogenesis by the fructose phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1987 Feb;169(2):897–899. doi: 10.1128/jb.169.2.897-899.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992 Oct 20;227(4):996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerse R. H., van der Pluijm J., Postma P. W. The repressor of the PEP:fructose phosphotransferase system is required for the transcription of the pps gene of Escherichia coli. Mol Gen Genet. 1989 Aug;218(2):348–352. doi: 10.1007/BF00331288. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Transcriptional control--lessons from an E. coli promoter data base. Cell. 1991 Aug 9;66(3):415–418. doi: 10.1016/0092-8674(81)90001-5. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zalkin H. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J Bacteriol. 1992 Nov;174(22):7121–7127. doi: 10.1128/jb.174.22.7121-7127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M. F., Cronan J. E., Jr Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J Mol Biol. 1991 Dec 20;222(4):843–849. doi: 10.1016/0022-2836(91)90574-p. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J. K., Le L. U., Hochschild A. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):3083–3087. doi: 10.1073/pnas.90.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnke G., Theres C., Fritz H. J., Ehring R. RNA polymerase and gal repressor bind simultaneously and with DNA bending to the control region of the Escherichia coli galactose operon. EMBO J. 1989 Apr;8(4):1247–1255. doi: 10.1002/j.1460-2075.1989.tb03498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S., North A. K., Weiss D. S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991 Nov;16(11):397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- Law S. M., Bellomy G. R., Schlax P. J., Record M. T., Jr In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J Mol Biol. 1993 Mar 5;230(1):161–173. doi: 10.1006/jmbi.1993.1133. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Schleif R. F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989 Jan;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal N., Su W., Haber R., Adhya S., Echols H. DNA looping in cellular repression of transcription of the galactose operon. Genes Dev. 1990 Mar;4(3):410–418. doi: 10.1101/gad.4.3.410. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994 Sep;176(18):5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993 Oct;175(19):6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard A. J., Davidson B. E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991 Jul;5(7):1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A., Cochet O., Souza J. M., Altamirano M. M., Calcagno M. L., Badet B. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J Bacteriol. 1993 Aug;175(16):4951–4956. doi: 10.1128/jb.175.16.4951-4956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J Bacteriol. 1990 May;172(5):2728–2735. doi: 10.1128/jb.172.5.2728-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991 Aug;5(8):2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J., Kolb A. CAP and Nag repressor binding to the regulatory regions of the nagE-B and manX genes of Escherichia coli. J Mol Biol. 1991 Feb 20;217(4):661–679. doi: 10.1016/0022-2836(91)90524-a. [DOI] [PubMed] [Google Scholar]
- Plumbridge J., Kolb A. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol Microbiol. 1993 Dec;10(5):973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J., Kolb A. Nag repressor-operator interactions: protein-DNA contacts cover more than two turns of the DNA helix. J Mol Biol. 1995 Jun 23;249(5):890–902. doi: 10.1006/jmbi.1995.0346. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W., Jacobson G. R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993 Sep;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin R. S., Collins L. A., Stewart V. In vivo requirement of integration host factor for nar (nitrate reductase) operon expression in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8701–8705. doi: 10.1073/pnas.89.18.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramseier T. M., Nègre D., Cortay J. C., Scarabel M., Cozzone A. J., Saier M. H., Jr In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1993 Nov 5;234(1):28–44. doi: 10.1006/jmbi.1993.1561. [DOI] [PubMed] [Google Scholar]
- Reeder T., Schleif R. AraC protein can activate transcription from only one position and when pointed in only one direction. J Mol Biol. 1993 May 20;231(2):205–218. doi: 10.1006/jmbi.1993.1276. [DOI] [PubMed] [Google Scholar]
- Schröder I., Darie S., Gunsalus R. P. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J Biol Chem. 1993 Jan 15;268(2):771–774. [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Stewart V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol. 1993 Aug;9(3):425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- Titgemeyer F., Reizer J., Reizer A., Saier M. H., Jr Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994 Sep;140(Pt 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- Vogler A. P., Lengeler J. W. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol Gen Genet. 1989 Oct;219(1-2):97–105. doi: 10.1007/BF00261163. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Gay N. J., Saraste M., Eberle A. N. DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J. 1984 Dec 15;224(3):799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968 Feb;106(4):847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]