Abstract
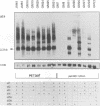
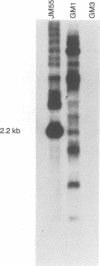
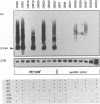
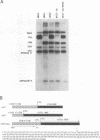
Expression of the Saccharomyces cerevisiae mitochondrial COX1 locus, which contains several introns and is co-transcribed with the downstream genes AAP1, OLI2 and ENS2, is controlled by at least 18 nuclear-encoded proteins. The PET309 gene, encoding one of these proteins, was cloned, sequenced and shown to contain an open reading frame of 965 codons. Isonuclear PET309+ and delta pet309::URA3 strains carrying mitochondrial genomes that differ in the number of COX1 introns, were generated. Analysis of RNA species from these strains demonstrated an inverse relationship between the number of introns present in the precursor RNA and the amount of COX1 and AAP1/OLI2/ENS2 RNAs accumulated in a pet309 mutant. Hence, PET309 plays a role either in transcription of intron-containing primary transcripts from the COX1-AAP1-OLI2-ENS2 transcription unit or in stabilization of primary transcripts. PET309 is also required in translation of COX1 mRNA. A mitochondrial bypass suppressor of the pet309 deletion mutation was isolated, and shown to consist of a DNA rearrangement at the COX1 locus, such that the 5' untranslated leader region (UTR) of the COB gene was fused to COX1 at nucleotide -174 of its 5' UTR. This result suggests that Pet309p acts through the COX1 5' UTR to activate initiation of translation of the COX1 coding region.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asher E. B., Groudinsky O., Dujardin G., Altamura N., Kermorgant M., Slonimski P. P. Novel class of nuclear genes involved in both mRNA splicing and protein synthesis in Saccharomyces cerevisiae mitochondria. Mol Gen Genet. 1989 Feb;215(3):517–528. doi: 10.1007/BF00427051. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bousquet I., Dujardin G., Poyton R. O., Slonimski P. P. Two group I mitochondrial introns in the cob-box and coxI genes require the same MRS1/PET157 nuclear gene product for splicing. Curr Genet. 1990 Aug;18(2):117–124. doi: 10.1007/BF00312599. [DOI] [PubMed] [Google Scholar]
- Brown N. G., Costanzo M. C., Fox T. D. Interactions among three proteins that specifically activate translation of the mitochondrial COX3 mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Feb;14(2):1045–1053. doi: 10.1128/mcb.14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M. Molecular genetics of group I introns: RNA structures and protein factors required for splicing--a review. Gene. 1988 Dec 20;73(2):273–294. doi: 10.1016/0378-1119(88)90493-3. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A. Structure and function of cytochrome c oxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- Carignani G., Groudinsky O., Frezza D., Schiavon E., Bergantino E., Slonimski P. P. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S. cerevisiae. Cell. 1983 Dec;35(3 Pt 2):733–742. doi: 10.1016/0092-8674(83)90106-x. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cherniack A. D., Garriga G., Kittle J. D., Jr, Akins R. A., Lambowitz A. M. Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell. 1990 Aug 24;62(4):745–755. doi: 10.1016/0092-8674(90)90119-y. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986 Nov;6(11):3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Mueller P. P., Strick C. A., Fox T. D. Primary structure of wild-type and mutant alleles of the PET494 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 Feb;202(2):294–301. doi: 10.1007/BF00331654. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 1986 Dec 20;5(13):3637–3641. doi: 10.1002/j.1460-2075.1986.tb04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. The PET54 gene of Saccharomyces cerevisiae: characterization of a nuclear gene encoding a mitochondrial translational activator and subcellular localization of its product. Genetics. 1989 Jun;122(2):297–305. doi: 10.1093/genetics/122.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster E., Simon M., Hatat D., Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet. 1990 Oct;224(1):111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Staples R. R. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int Rev Cytol. 1994;152:145–181. doi: 10.1016/s0074-7696(08)62556-5. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. The transcription termination site at the end of the early region of bacteriophage T7 DNA. Nucleic Acids Res. 1980 May 24;8(10):2119–2132. doi: 10.1093/nar/8.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Simon M. Analysis of a yeast nuclear gene involved in the maturation of mitochondrial pre-messenger RNA of the cytochrome oxidase subunit I. Cell. 1983 Jan;32(1):77–87. doi: 10.1016/0092-8674(83)90498-1. [DOI] [PubMed] [Google Scholar]
- Fernández A. On how hydrolysis at the 3' end is prevented in the splicing of a sequentially folded group I intron. FEBS Lett. 1992 Feb 3;297(1-2):201–204. doi: 10.1016/0014-5793(92)80360-s. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Kelly J. L., Lehman I. R. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudinsky O., Bousquet I., Wallis M. G., Slonimski P. P., Dujardin G. The NAM1/MTF2 nuclear gene product is selectively required for the stability and/or processing of mitochondrial transcripts of the atp6 and of the mosaic, cox1 and cytb genes in Saccharomyces cerevisiae. Mol Gen Genet. 1993 Sep;240(3):419–427. doi: 10.1007/BF00280396. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Arnberg A. C., Roosendaal E., van der Horst G., van der Veen R., van Ommen G. J., Grivell L. A. Variation, transcription and circular RNAs of the mitochondrial gene for subunit I of cytochrome c oxidase. J Mol Biol. 1983 Feb 15;164(1):35–58. doi: 10.1016/0022-2836(83)90086-4. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Base-pairing interactions involving the 5' and 3'-terminal nucleotides of group II self-splicing introns. J Mol Biol. 1990 Jun 5;213(3):437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- Jang S. H., Jaehning J. A. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J Biol Chem. 1991 Nov 25;266(33):22671–22677. [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene is Saccharomyces cerevisiae. J Bacteriol. 1988 Mar;170(3):1399–1402. doi: 10.1128/jb.170.3.1399-1402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae: multiple trans-acting nuclear genes exert specific effects on expression of each of the cytochrome c oxidase subunits encoded on mitochondrial DNA. Curr Genet. 1987;12(5):311–322. doi: 10.1007/BF00405753. [DOI] [PubMed] [Google Scholar]
- Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol Gen Genet. 1990 Nov;224(2):209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Perlman P. S. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem Sci. 1990 Nov;15(11):440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989 Oct 20;59(2):283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Lisowsky T. Molecular analysis of the mitochondrial transcription factor mtf2 of Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jan;220(2):186–190. doi: 10.1007/BF00260480. [DOI] [PubMed] [Google Scholar]
- Luft R. The development of mitochondrial medicine. Proc Natl Acad Sci U S A. 1994 Sep 13;91(19):8731–8738. doi: 10.1073/pnas.91.19.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., McCarron R. J. Hybridization of the in vitro products of bacteriop&hage T7 RNA polymerase to restriction fragments of T7 DNA. Virology. 1977 Oct 15;82(2):288–298. doi: 10.1016/0042-6822(77)90004-6. [DOI] [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem. 1986 Sep 5;261(25):11872–11879. [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989 Oct 15;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Mishra N. C. Genetics and molecular biology of Neurospora crassa. Adv Genet. 1991;29:1–62. [PubMed] [Google Scholar]
- Mittelmeier T. M., Dieckmann C. L. In vivo analysis of sequences required for translation of cytochrome b transcripts in yeast mitochondria. Mol Cell Biol. 1995 Feb;15(2):780–789. doi: 10.1128/mcb.15.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monéger F., Smart C. J., Leaver C. J. Nuclear restoration of cytoplasmic male sterility in sunflower is associated with the tissue-specific regulation of a novel mitochondrial gene. EMBO J. 1994 Jan 1;13(1):8–17. doi: 10.1002/j.1460-2075.1994.tb06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J. V., Mecklenburg K. L., Sass P., Belcher S. M., Mahnke D., Lewin A., Perlman P. Splicing defective mutants of the COXI gene of yeast mitochondrial DNA: initial definition of the maturase domain of the group II intron aI2. Nucleic Acids Res. 1994 Jun 11;22(11):2057–2064. doi: 10.1093/nar/22.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero J. J., Fox T. D. Alteration of the Saccharomyces cerevisiae COX2 mRNA 5'-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993 Dec;4(12):1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero J. J., Fox T. D. PET111 acts in the 5'-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics. 1993 Mar;133(3):509–516. doi: 10.1093/genetics/133.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P. P., Reif M. K., Zonghou S., Sengstag C., Mason T. L., Fox T. D. A nuclear mutation that post-transcriptionally blocks accumulation of a yeast mitochondrial gene product can be suppressed by a mitochondrial gene rearrangement. J Mol Biol. 1984 Jun 5;175(4):431–452. doi: 10.1016/0022-2836(84)90178-5. [DOI] [PubMed] [Google Scholar]
- Pel H. J., Tzagoloff A., Grivell L. A. The identification of 18 nuclear genes required for the expression of the yeast mitochondrial gene encoding cytochrome c oxidase subunit 1. Curr Genet. 1992 Feb;21(2):139–146. doi: 10.1007/BF00318473. [DOI] [PubMed] [Google Scholar]
- Pelissier P. P., Camougrand N. M., Manon S. T., Velours G. M., Guerin M. G. Regulation by nuclear genes of the mitochondrial synthesis of subunits 6 and 8 of the ATP synthase of Saccharomyces cerevisiae. J Biol Chem. 1992 Feb 5;267(4):2467–2473. [PubMed] [Google Scholar]
- Poutre C. G., Fox T. D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987 Apr;115(4):637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putrament A., Baranowska H., Ejchart A., Jachymczyk W. Manganese mutagenesis in yeast. VI. Mn2+ uptake, mitDNA replication and ER induction: comparison with other divalent cations. Mol Gen Genet. 1977 Feb 28;151(1):69–76. doi: 10.1007/BF00446914. [DOI] [PubMed] [Google Scholar]
- Rödel G., Fox T. D. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5' untranslated leader. Mol Gen Genet. 1987 Jan;206(1):45–50. doi: 10.1007/BF00326534. [DOI] [PubMed] [Google Scholar]
- Rödel G. Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5'-untranslated COB leader. Curr Genet. 1986;11(1):41–45. doi: 10.1007/BF00389424. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schinkel A. H., Koerkamp M. J., Touw E. P., Tabak H. F. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J Biol Chem. 1987 Sep 15;262(26):12785–12791. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sengupta D., Chakravarti D., Maitra U. Relative efficiency of utilization of promoter and termination sites by bacteriophage T3 RNA polymerase. J Biol Chem. 1989 Aug 25;264(24):14246–14255. [PubMed] [Google Scholar]
- Seraphin B., Simon M., Jacq C., Faye G. Sequence of the yeast mitochondrial OX13/OL12 promoter region. Nucleic Acids Res. 1989 Jun 26;17(12):4886–4886. doi: 10.1093/nar/17.12.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Faye G. Steps in processing of the mitochondrial cytochrome oxidase subunit I pre-mRNA affected by a nuclear mutation in yeast. Proc Natl Acad Sci U S A. 1984 Jan;81(1):8–12. doi: 10.1073/pnas.81.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Faye G. MSS18, a yeast nuclear gene involved in the splicing of intron aI5 beta of the mitochondrial cox1 transcript. EMBO J. 1988 May;7(5):1455–1464. doi: 10.1002/j.1460-2075.1988.tb02963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman J. W., Capaldi R. A. Purification of yeast cytochrome c oxidase with a subunit composition resembling the mammalian enzyme. J Biol Chem. 1992 Nov 5;267(31):22481–22485. [PubMed] [Google Scholar]
- Tzagoloff A., Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990 Sep;54(3):211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencik M. L., Kloeckener-Gruissem B., Poyton R. O., McEwen J. E. Disruption of the yeast nuclear PET54 gene blocks excision of mitochondrial intron aI5 beta from pre-mRNA for cytochrome c oxidase subunit I. EMBO J. 1989 Dec 1;8(12):3899–3904. doi: 10.1002/j.1460-2075.1989.tb08569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencik M. L., McEwen J. E. Genetic evidence that different functional domains of the PET54 gene product facilitate expression of the mitochondrial genes COX1 and COX3 in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2399–2405. doi: 10.1128/mcb.11.5.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992 May 1;256(5057):628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- Wallis M. G., Groudinsky O., Slonimski P. P., Dujardin G. The NAM1 protein (NAM1p), which is selectively required for cox1, cytb and atp6 transcript processing/stabilisation, is located in the yeast mitochondrial matrix. Eur J Biochem. 1994 May 15;222(1):27–32. doi: 10.1111/j.1432-1033.1994.tb18837.x. [DOI] [PubMed] [Google Scholar]
- Zeviani M. Nucleus-driven mutations of human mitochondrial DNA. J Inherit Metab Dis. 1992;15(4):456–471. doi: 10.1007/BF01799604. [DOI] [PubMed] [Google Scholar]
- de Winde J. H., Grivell L. A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]