Abstract
There is an extraordinary need to describe the structures of intrinsically disordered proteins (IDPs) due to their role in various biological processes involved in signaling and transcription. However, general study of IDPs by NMR spectroscopy is limited by the poor 1H-amide chemical shift dispersion typically observed in their spectra. Recently, 13C direct-detected NMR spectroscopy has been recognized as enabling broad structural study of IDPs. Most notably, multi-dimensional experiments based on the 15N,13C-CON spectrum make complete chemical shift assignment feasible. Here we document a collection of NMR based tools that efficiently lead to chemical shift assignment of IDPs, motivated by a case study of the C-terminal disordered region from the human pancreatic transcription factor Pdx1. Our strategy builds on the combination of two 3D experiments, (HN-flip)N(CA)CON and 3D (HN-flip)N(CA)NCO, that enable daisy-chain connections to be built along the IDP backbone, facilitated by acquisition of amino-acid specific 15N,13C-CON detected experiments. Assignments are completed through carbon-detected, TOCSY based side chain chemical shift measurement. Conducting our study required producing valuable modifications to many previously published pulse sequences, motivating us to announce the creation of a database of our pulse programs, which we make freely available through the web.
Keywords: intrinsically disordered proteins, carbon detect NMR, amino-acid filtered NMR, CAS-NMR, Pdx1
Introduction
Intrinsically Disordered Proteins (IDPs) partially or completely lack a cooperatively folded structure under native conditions. Even so, intrinsically disordered regions are abundant in proteins executing various biological processes requiring protein-protein interactions, such as molecular signaling and transcription.[1] This prevalent functional role for intrinsic disorder led Wright and Dyson to recognize that protein function derives from native structure, regardless of whether or not folding occurs.[2] Therefore, even in the absence of spatial and temporal order, native structure must always be considered significant. Recent NMR-derived structure models of disordered regions from the tumor suppressor protein p53[3] and the cyclin-dependent kinase inhibitor Sic1[4] have provided critical insight into the functional role of protein disorder for these systems. Success stories like these are not common, however, because most IDPs yield solution NMR spectra characterized by severe spectral overlap that precludes high-resolution study. Recent work done in our laboratory demonstrates that the problem with studying IDPs is not inherent to the proteins, but rather rests on the decision to employ NMR methods that have been optimized for cooperatively folding systems to evaluate disordered systems.[5–8] In the course of our efforts to systematically study IDP structure-function relationships by NMR, we have established a purely carbon-detected NMR protocol for generating IDP chemical shift assignments, which we demonstrate here through application to the C-terminal disordered region of the pancreatic duodenal homeobox protein 1 (Pdx1-C).
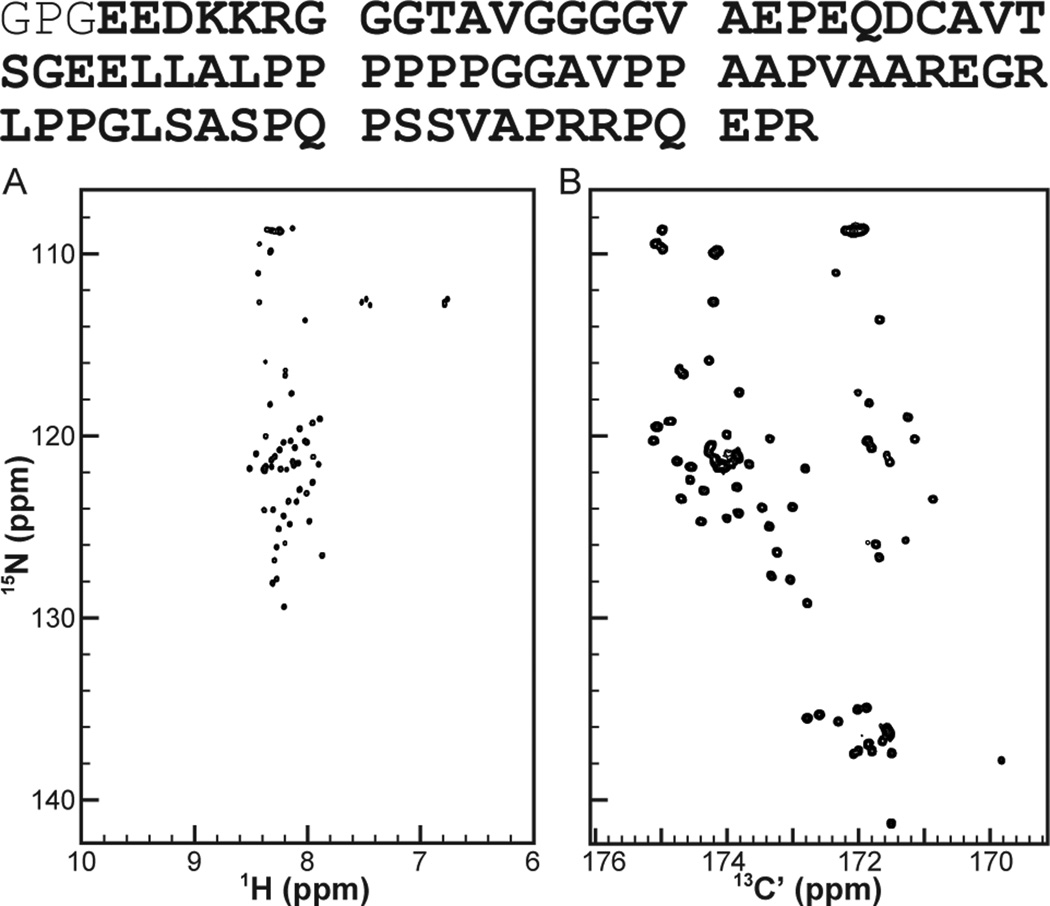
Solution NMR spectroscopy, which is well suited to studying biopolymers possessing heterogeneous conformational ensembles, stands as the most frequently successful method for IDP structure analysis. Two-dimensional heteronuclear correlation experiments have become a mainstay of biomolecular NMR spectroscopy, because they generally provide good peak dispersion, serve readily as a component for building multidimensional experiments that yield chemical shifts and structure constraints; and provide a valuable detection platform for the measurement of spin relaxation, scalar couplings, or dipolar couplings. In particular, the 1H,15NHSQC has become a workhorse experiment for protein NMR, because it provides nominally one-to-one mapping between resonances and residues (the absence of proline being the major exception) and can be rapidly acquired with inexpensive samples. For most small and cooperatively folding proteins, the 1H,15N-HSQC features an extremely well dispersed set of resonances. In contrast, IDPs almost always display problematically finite chemical shift dispersion in the proton dimension of this experiment. This problem is seen quite clearly in the 1H,15N-HSQC of Pdx1-C (Fig. 1A), for which the extreme spectral crowding in the random-coil region (near 8.5 ppm on the 1H-axis) makes it impossible to unambiguously identify most individual resonances in the spectrum.
Figure 1.
2D-heteronuclear correlation spectra provide the foundation for all biomolecular NMR studies, but traditional 1H-detected methods do not work as well for IDPs as newer 13C-detected methods. (A) The 1H,15N-HSQC of Pdx1-C (B) The 15N,13C-CON of Pdx1-C. Of special value is the presence of peaks corresponding to proline residues, seen downfield shifted in the nitrogen dimension of the 15N,13C-CON. Note that the amino acid sequence of Pdx1-C is provided at the top of the figure, with the N-terminal GPG cloning artifact shown in the thinner font. A larger version of panel (B) is provided in Fig. S1, including complete annotation of chemical shift assignments.
The principle advantage to choosing the 1H,15N-HSQC as a detection platform for protein NMR is that it provides a simple spectrum with one 2D-resonance per amino acid residue, excepting proline. Similar mapping is achievable using, e.g., the 13C´-15N or 13C´-13Cα as a reporting pair, but with the consequence 13C is a far less sensitive detection nucleus than 1H. Fortunately, recent improvements in instrumentation, most notably the development of stable cryogenically cooled probes, has largely mitigated this concern, rendering carbon-detected biomolecular NMR practical for routine applications. This, coupled with intense interest in the inner workings of IDPs, has propelled a sudden burst of development in the field of carbondetected biomolecular NMR, which can be used to report on the structural aspects of IDPs in atomic detail.[5, 8–13] These carbon-detect methods make the study of IDPs feasible because the resulting 2D heteronuclear correlation spectra generally feature excellent chemical shift dispersion in both dimensions. In particular, we and others have found the 15N,13C-CON spectrum, which records the correlation between the carbonyl carbon and amide nitrogen in the peptide plane of the protein backbone, to be a generally successful detection platform for IDPs. For Pdx1-C, the degree of improvement over conventional 1H,15N-HSCQ detection is clearly evident in Fig. 1B.
Successful investigation of biomolecular structure and dynamics by NMR always rests on the comprehensive and accurate generation of chemical shift assignments for the system of interest. The excellent quality of 15N,13C-CON spectra, like the one shown for Pdx1-C in Fig. 1B, suggests that achieving this goal should be straight forward using carbon-detected NMR (which is demonstrated by the full-page and fully annotated 15N,13C-CON of Pdx1-C in Fig. S1). Even so, few efficient and generally practical protocols for generating chemical shift assignments, based purely on the collection of carbon-detected spectra, have been reported to date. Here we report a protocol, based on a suite of four 3D experiments and additional amino-acid specific 2D spectra[5, 9, 11] that efficiently yields chemical shift assignments for IDPs that are soluble and stable under mildly acidic conditions. Our recommended strategy is based completely on carbon-detected spectra and yet required only two weeks of NMR time to complete for Pdx-C (BMRB accession number 19596). Furthermore, acquisition does not rely on high dimensional (i.e., 4D or 5D) experiments or on non-uniform sampling, making it accessible to users without access to these techniques. Finally, our analysis strategy builds upon traditional nearest-neighbor daisy-chaining through the protein backbone that will be familiar to anyone who has previously assigned a protein using conventional triple-resonance methods.[14, 15] In order to facilitate broader adoption of this highly efficient strategy, we have made our modified versions of each pulse program used available through the web,*along with representative parameter sets.
Materials and Methods
Purification of PDX1-C
A synthetic Pdx1 gene with codons optimized for Escherichia coli was purchased from Geneart and the Pdx1 C-Terminus (amino acids 204–283 of the human sequence; subsequently referred to as Pdx1-C), was subcloned by PCR into pET49b (Novagen) encoding a Glutathione-S-Transferase tag, a 6x His tag and a 3C protease recognition site upstream of the cloning site. The recombinant plasmid was transformed into BL21(DE3) competent cells for protein overexpression. Cell growth conditions and the protein purification protocol for Pdx1-C were identical to our previously reported procedures for the Pdx1-homeodomain,[16] except as noted below. As a final step to ensure full purification, Pdx1-C was subjected to size exclusion chromatography using a HiPrep 26/60 Sephacryl S-200 HR column (GE Life Sciences) in 50 mM Tris pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, and 1 mM EDTA. Following concentration using an Amicon Ultra centrifugal filter device (Millipore) that contained a PES 3000 MWCO membrane, Pdx1-C was buffer exchanged into 50 mM cacodylate pH 6.5, 50 mM KCl, and 1 mM DTT. Protein concentration was determined by Direct Detect FT-IR (Millipore) using the molecular weight of 8089 g/mol.
NMR spectroscopy
All the NMR experiments were recorded at 11.6 T and 14.0 T on Bruker AVANCE-3 spectrometers, operating at 1H frequencies of 500.13 MHz and 600.07 MHz, respectively, and equipped with TCI cryoprobes. All spectra were acquired on uniformly 15N and 13C isotope-enriched samples of Pdx1-C at concentrations ranging from 0.6–0.8 mM in 50 mM cacodylate buffer pH 6.5, 50 mM KCl, 1 mM DTT and 10% D2O. All spectra were collected at 298K.
Typical pulse times were 9.79 µs and 31 µs for hard 1H and 15N 90° pulses, respectively, with some sample-based variation in the 1H pulse time. All PFGs used in the experiments were applied for 1ms with a sine shape. In all pulse sequences, unless otherwise noted, the 90° band-selective 13C pulses have the Q5 shape (or time reversed, Q5tr) and the band-selective 13C 180° pulses use the Q3 shape[17] with durations of 384 µs and 307 µs, respectively, at 11.6 T; 320 µs and 256 µs, respectively, at 14.0 T. The timing and phase parameters specific to pulse sequences associated with the amino acid filtered NMR experiments are reported in the relevant figure captions in the supplementary information. The 1H and 15N carriers were placed at 4.7 and 124 ppm respectively. The pulses for excitation of carbonyl carbon and alpha carbon were centered at 172 ppm and 54 ppm, respectively, while the 13C carrier was changed at the positions indicated by vertical arrows to 13Cα/ali(β) = 39 ppm, 13Cα = 54 ppm, and 13C´ = 172 ppm. In all figures, the carbon pulses represented by solid shapes were applied on-resonance and those marked with slanted stripes were off-resonance pulses, centered on the aliphatic region. The adiabatic inversion pulse (grey pulse) that inverts both C´ and Cα magnetizations, used during the nitrogen chemical shift labeling period, was a 500 ms CHIRP pulse with 60 Hz sweep and 25% smoothing.[18] Composite pulse decoupling of 1H and 15N was achieved by the use of 3.57 kHz waltz-65 and 1.25 kHz garp- sequences, respectively. In all experiments, the recycle delay was set to 1.3 s, except for the HN-flip versions, where a 1.0 s delay was used.
Direct-detection on 13C´ requires that steps be taken to refocus the active 13C´-13Cα coupling during acquisition. In all pulse sequences used here, the carbonyl dimension is collected in the in-phase anti-phase manner (IPAP), enabling 13C´-13Cαcouplings to be “virtually decoupled” as a post-acquisition processing step.[9] The 3D (HN-flip)N(CA)CON experiment was performed with spectral widths of 20×40×40 ppm, with matrix size of 1024(C´)×64(N)×128(N) points, and 16 scans; amounting to total acquisition time of 2 days 2 hrs. Owing to the lower intensities from 3D (HN-flip)N(CA)NCO, the number of scans was increased to 32, yielding a total acquisition time of 4 days 4 hrs. The standard CCCON spectrum from the Topspin pulse program library was collected over two days, with a matrix size of 1024(C´)×64(N)×128(C) points, with spectral widths of 20×40×70 ppm, and 16 scans of signal averaging. The aliphatic protons in the side chains were assigned using H(CC)CON,[5] collected over two days with matrix size of 1024(C´)×64(N)×128(H) points, spectral widths of 20×40×40 ppm, and 16 scans of signal averaging. All amino acid filtered CON-based experiments were carried out with spectral widths of 20×40 ppm and matrix sizes of 1024(C´)×256(N). The details of the pulses and delays used in these experiments are described in the figure captions below each amino acid filtered experiment in the Supporting Information (Fig. S4–S18). All NMR data were processed in TOPSPIN 2.1 and converted to Sparky format[19] for data analysis.
Pdx1-C Secondary Structure Analysis
Secondary Structure Propensity (SSP) was calculated from secondary chemical shifts to determine the per-residue probability of finding α-helix and β-sheet conformations in Pdx1-C. [20] The data are represented here by calculating the difference in 13Cα (ΔδCα) and 13Cβ (ΔδCβ) chemical shifts measured from those of random coils and plotting ΔδCα - ΔδCβ. In addition, calculations were performed using the recently published δ2D methodology for predicting the secondary structures of disordered proteins using NMR chemical shifts.[21] This analysis was performed using the δ2D webserver (http://www-vendruscolo.ch.cam.ac.uk/d2D/) that takes assigned 1Hα, 13Cα, 13Cβ, 13C´, and 15N chemical shifts of each residue as inputs, predicting as output the relative populations of α-helix, extended β-sheets, polyproline II and random coil present in the ensemble.
Results and Discussion
Model System
Despite intense interest in evaluating structure-function relationships involving IDPs, there remains a significant composition difference[22] between databases of known protein sequences (e.g., the UniProt knowledgebase [23]) and the sequences found in known three-dimensional structures deposited in the Protein Data Bank,[24] thus reinforcing the assertion that disordered proteins are systematically under-represented in structural studies. In order to understand why IDPs pose such a challenge to structural biology, and to see the utility of the chemical shift assignment protocol we report here, it is helpful to understand how IDPs differ from cooperatively folding proteins on the molecular level. Bioinformatic studies have established a series of amino acid sequence hallmarks common to IDPs that render them distinct from known globular proteins, including the possession of long “medium complexity” sequence segments and a tendency towards repeats.[22] Typically, IDPs are also depleted in non-polar residues and enriched in polar and charged residues, while many display a pronounced enrichment in proline.[25]
Each of these sequence hallmarks of disorder is found in the Pdx1 C-terminal domain (Fig. 1, top), which makes this protein an excellent model system for NMR method development. Pdx1 is a transcription factor, known to play an important role in pancreatic development and β-cell maturation, as well as the regulation of insulin production in mature organisms.[26–31] The well folded homeodomain component of Pdx1 is well studied,[16, 29–31] but only represents a small fraction of the total polypeptide. Full length Pdx1 includes both Nand C-terminal intrinsically disordered regions that are responsible for mediating known protein-protein interactions[32, 33] and which include sites of polymorphism that correlate with type-2 diabetes in humans.[34–37] Consistent with the patterns reviewed above, the disordered segments of Pdx1 possess a low abundance of non-polar residues, have low-complexity amino acid sequences, and feature repetitive glycine and proline rich segments. Our laboratory has previously succeeded in using a combination of proton-detected and carbon-detected NMR spectra to fully assign and structurally characterize the intrinsically disordered C-terminus of the human protein FCP1.[6–8, 38] In contrast, the prevalence of proline residues and otherwise extreme amino acid bias in Pdx1-C proved too much for effective analysis using our original methods,[6] leading us to establish the protocol presented here for generating chemical shift assignments from purely carbon-detected NMR experiments.
Carbon-detected NMR approach for chemical shift assignment
Acquisition of essentially complete NMR chemical shift assignments is prerequisite for any NMR based approach characterizing the structures of biomolecules and the mechanisms driving their interactions with their cognate partners. For cooperatively folded proteins, the task of chemical shift assignment is usually accomplished by correlating the backbone carbon resonances (e.g., 13Cα, 13Cβ, 13C´) associated with a residue of interest to those of the nearest neighbor residue on the N-terminal side, in the context of a suite of triple-resonance NMR experiments detected through the 1H,15N-HSQC platform.[14, 15] When coupled with prior knowledge of the amino acid sequence, this procedure is generally sufficient on three grounds: 1) the nearest-neighbor connections bridge the peptide bond; 2) there is sufficient redundancy built into such a large chemical shift set; and 3) identifying the amino acid type of most residues based on statistical similarity to tabulated chemical shift values is straight forward and amenable to automation. In most cases, this strategy fails for IDPs because the heterogeneous nature of their structure causes a collapse in the 1HN chemical shift dimension of the spectra and, in many cases, the prevalence of proline residues blocks the walk-along-the-backbone procedure that depends on all residues possessing an amide proton.
As discussed above, we work around the limitations inherent to proton-detected NMR spectroscopy of IDPs by recording the correlation between the carbonyl carbon and amide nitrogen in the peptide plane of the protein backbone, which gives rise to the 15N,13C-CON spectrum. For Pdx1-C, the degree of improvement over conventional 1H,15N-HSQC detection (Fig. 1A) is clearly evident in Fig. 1B, which displays the 15N,13C-CON for this 80 residue IDP. It is worth noting that the 15N,13C-CON also yields sharper line widths (at a given magnetic field strength) than the 1H,15N-HSQC, further enhancing the practical resolution of the experiment. Finally, the 15N,13C-CON has the convenient property of containing a resonance for each peptide bond involving a proline 15N (easily identified by the downfield shift in the 15N-dimension).
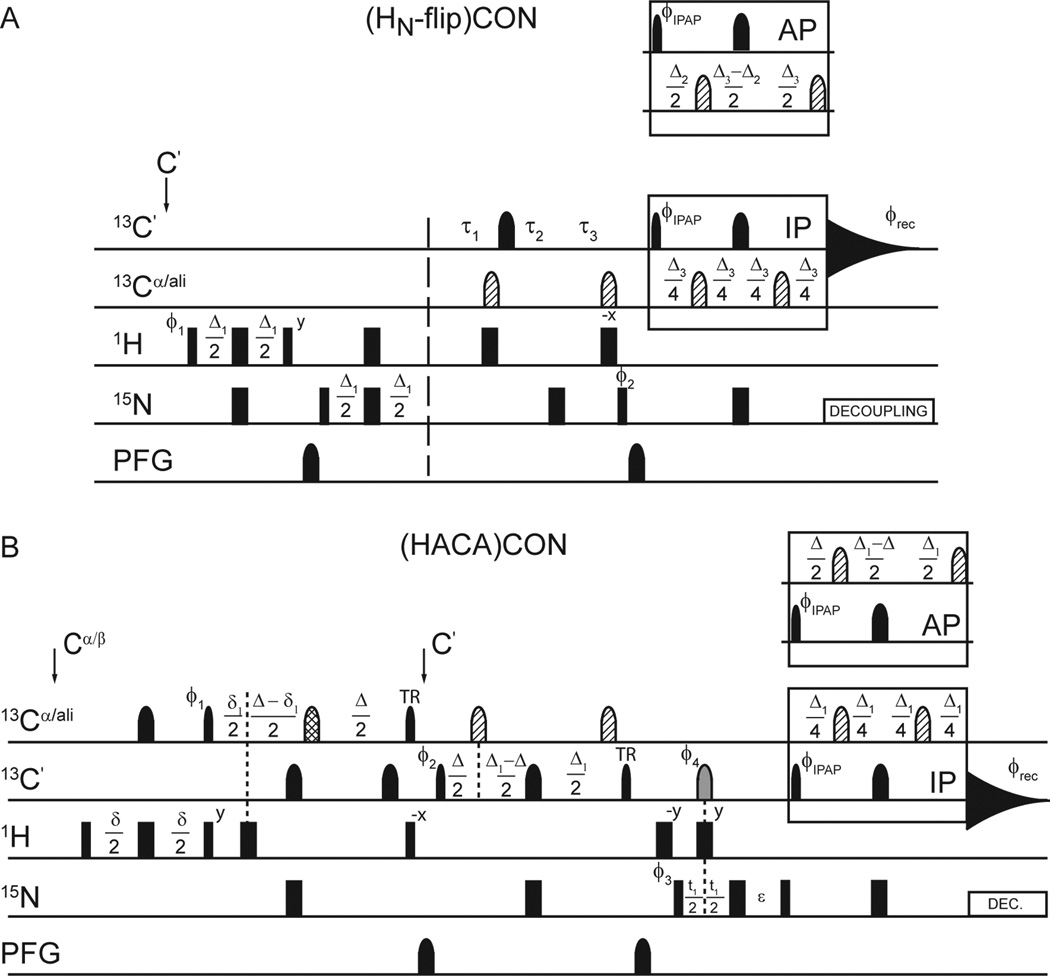
One limitation of carbon-detected NMR spectroscopy is its inherently lower sensitivity, on a per scan basis, compared with proton-detected NMR, which originates in the reduced gyromagnetic ratio of the 13C-nucleus. While much of the early carbon-detected NMR literature focused on so called “protonless” experiments, a wide range of “H-start” experiments have been developed more recently, yielding enhanced sensitivity. For proteins like Pdx1-C, which can be studied under mildly acidic conditions, we have found “HN-flip” variants of the 15N,13C-CON experiment to be especially effective, because the increased per-scan sensitivity is coupled to a decrease in the necessary inter-scan delay created by a more rapid recovery of the amide protons to equilibrium.[39, 40] A representative HN-flip 15N,13C-CON pulse sequence is shown in Fig. 2A and forms the basis for constructing the 3D pulse programs we will discuss below. It is important to note that exclusive reliance on HN-start experiments is not feasible for most IDPs, due to their tendency to be enriched in proline, which is lost from the CON when the HN-start format is used. Additionally, fast solvent exchange caused by enhanced solvent exposure in the extended state renders IDPs highly susceptible to resonance intensity loss when exchangeable protons are included in the magnetization transfer pathway. For this reason, application of HNstart CON techniques works best for IDPs that are soluble under mildly basic solution conditions. For proteins where these factors become limiting, and for the carbon-detected amino acid selective (CAS) NMR experiments we also use here, initial excitation on aliphatic proton resonances is the preferred means to enhancing sensitivity. Nearly all of the CAS-NMR experiments we will discuss are formally similar to the Hα-start 15N,13C-(HACA)CON, shown in Fig. 2B, which can also be used as a starting point for modification of other carbon-start or HN-start CON-based experiments.
Figure 2.
H-start versions of the 15N,13C-CON form the basis of nearly all experiments used in this study. In both panels, narrow and wide rectangular pulses correspond to 90° and 180° hard pulses, respectively, while band-selective pulses on the 13C-channel are similarly represented with narrow (90°) and wide (180°) shapes. Pulsed field gradients (PFG) are also indicated by shapes. (A) Pulse sequence for the (HN-flip) CON-IPAP experiment. The delays are Δ = 5 ms, Δ1 = 4.6 ms, Δ2 = 9.0 ms, and Δ3 = 25 ms. 15N chemical shift evolution is measured during t1 using a semi-constant time period with delays τ1 = (Δ3 + t1)/2, τ2 = (1 - Δ3/t1max)t1/2, and τ3 = (1 - t1/t1max)Δ3/2; where t1max is the maximum duration of the t1 labeling period.[47, 48] The phase cycle is φ1 = x, −x; φ2 = y, y, −y, −y; φIPAP(IP) = x, x, x, x, −x, −x, −x, −x; φIPAP(AP) = −y, −y, −y, −y, y, y, y, y; and φrec = x, −x, −x, x, x, −x, −x, x. Quadrature detection in the indirect dimension was obtained by States-TPPI incrementation of φ2. (B) Pulse sequence for the (HACA)CON-IPAP experiment. The delays are φ = 3.6 ms, δ1 = 2.2 ms, Δ = 9.0 ms, Δ1 = 25 ms, and ε = t1(0) + pC180. The phase cycle is φ1 = 4(x), 4(−x); φ2 = 2(x), 2(−x); φ3 = x, −x; φ4 = 8(x), 8(−x); φIPAP(IP) = x, φIPAP(AP) = −y; φREC = x, −x, −x, x, −x, x, x, −x. Quadrature detection in the indirect dimension is obtained by States-TPPI incrementation of φ3. In both experiments, pulses are applied at the frequency indicated on the left of each line, with narrow and wide rectangles or shapes representing 90° and 180° pulses, respectively. All pulses are applied with x-phase unless otherwise indicated. The gray pulse on 13C indicates a band-selective 13C´ and 13Cαinversion pulse. Pulses filled with diagonal lines are off-resonance 180° Q3 shaped pulses, centered at 54 ppm. The pulse filled with hashed lines is a higher-selectivity 180° Q3 shaped pulse applied on resonance with a duration of 1200 µs on a system opperating at 11.7 T static field strength. Pulses labeld ‘TR’ are time-reversed versions of the standard 90° Q5 shaped pulse.
Summarizing the procedure we will outline here, our strategy is to combine 3D NMR experiments that help establish nearest-neighbor connections to adjacent residues with 3D NMR experiments fully characterizing the side chain chemical shifts. Acquisition of 2D amino-acid filtered experiments, which identify the amino-acid types of as many resonances as possible through their spin topologies, further reduces the spectral degeneracy commonly associated with low-complexity protein sequences and yields numerous unambiguous entry points from which the process of assigning the backbone can be initiated.
3D-NMR spectroscopy: connecting the backbone
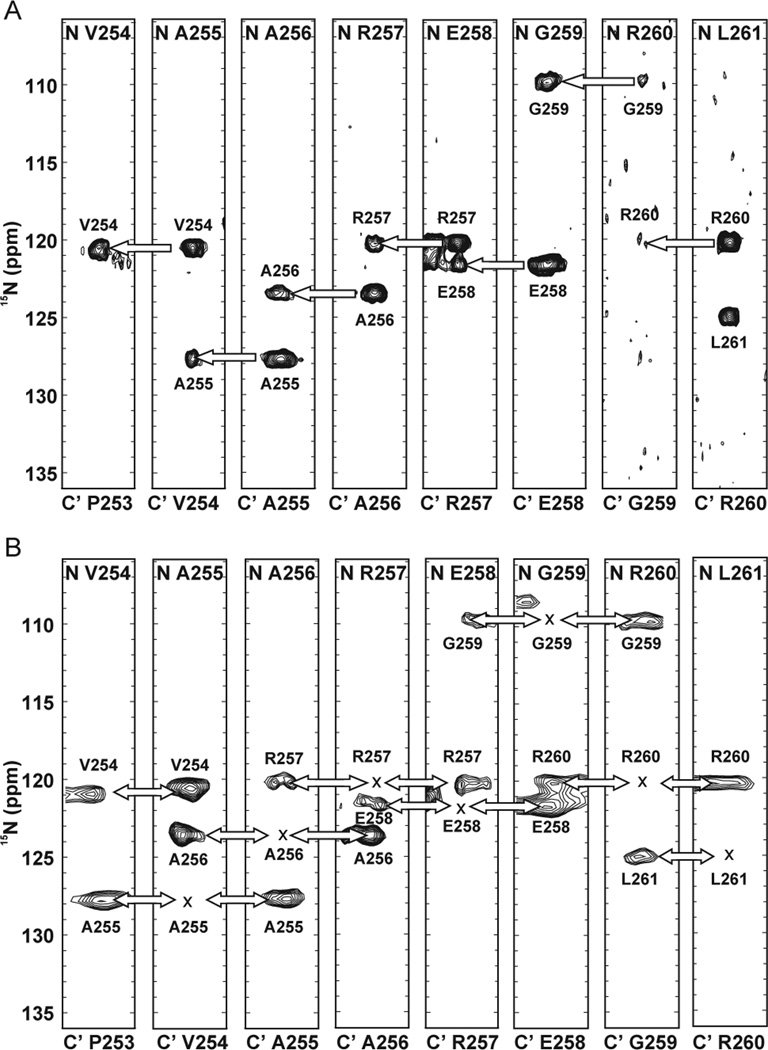
The HN-flip principle has been implemented in a pair of 3D carbon-detected NMR experiments described as (HN-flip)N(CA)CON and (HN-flip)N(CA)NCO,[11] which collectively are sufficient to provide nearest-neighbor linkages throughout the main chain. In our experience, optimal signal intensity in these two experiments was realized using the updated pulse sequences shown in Fig. S2 and S3. The 3D (HN-flip)N(CA)CON experiment is used to build nitrogen-nitrogen correlations between each signal observed in the CON and the amide nitrogen of the preceding residue in the chain. As displayed by the strips in Fig. 3A, the F1 dimension of the spectrum provides two correlations: the auto-correlation peak records the “self” amide nitrogen chemical shift, while the cross-peak records the nitrogen chemical shift of the preceding residue in the chain. For many systems, this one experiment should be sufficient to assign the observed magnetic resonances by daisy-chain connecting each residue with its nearest neighbor to the N-terminal side. Unfortunately, owing to the HN-start format, this experiment suffers from the same complication of proline breaking the chain as conventional proton-detected triple resonance experiments. Note, however, that the strips corresponding to non-proline residues that precede a proline do display their amide nitrogen correlations to the CON correlation featuring the proline, thus providing some guidance for their placement. If the obstacle presented by proline-induced breaks in the walk along the backbone proves to be too great, the 3D (HN-flip)N(CA)CON is amenable to conversion into an CANCO format.
Figure 3.
Strips corresponding to the labeled Pdx1-C resonances obtained from (A) 3D (HNflip) N(CA)CON and (B) 3D (HN-flip)N(CA)NCO. The bottom labels of the strips correspond to the carbonyl chemical shifts while the top labels to that of amide nitrogen in the F2 dimension. Nitrogen shifts in the F1 dimension are provided adjacent to each peak. The autocorrelation peak is rarely present in the 3D (HN-flip)N(CA)NCO, so its position is marked with an×in the figure. The pulse sequences used to generate this data are presented in Fig. S2 and S3.
For systems that are challenging to assign based on the 3D (HN-flip)N(CA)CON alone, the 3D (HN-flip)N(CA)NCO offers powerful complementary information, by correlating each of the resonances in a CON with the amide chemical shift of the preceding, self, and following residue in the F1 dimension (Fig. 3B). As the self and preceding amide nitrogen resonances are typically both known from (HN-flip)N(CA)CON, it is usually quite easy to deduce which cross-peak corresponds to the amide resonance of the residue on the C-terminal side of the residue providing the CON peak. Note that the intensity of signals obtained from the (HN-flip)N(CA)NCO is weaker as compared to the (HN-flip)N(CA)CON; but, because this experiment allows the spectroscopist the liberty of moving either forward or back in the polypeptide daisy-chain, the (HN-flip)N(CA)NCO offers an especially powerful tool for difficult to assign proteins. When spectrometer time and sample stability are not limiting factors, the lower signal to noise ratio can generally be overcome with an increase in the number of scans acquired. As a starting point, we generally collect the (HN-flip)N(CA)NCO with double the number of scans used for the (HNflip) N(CA)CON, which does not provide equal intensity, but is generally sufficient to provide a useable signal-to-noise.
The necessary task of correlating each peak in the 15N,13C-CON with its position in the amino acid sequence of the protein chain is not sufficient to complete the assignment process for most purposes. In addition, it is desirable to record the 13Cαand 13Cαchemical shifts for the purpose of evaluating whether the IDP being studied possesses any secondary structure biases. Furthermore, full side chain chemical shift assignment can facilitate amino acid type assignment, among other objectives. The most efficient means we have found towards completing the goal of comprehensive side chain chemical shift assignment is to collect an additional pair of 3D experiments: the 3D CCCON, used to obtain the aliphatic carbon side chain assignments;[9] and the 3D H(CC)CON experiment, used to obtain the aliphatic proton side chain assignments.[5] In general, IDPs are sufficiently depleted in aromatic amino acids to render additional effort aimed at assigning the aromatic chemical shifts unnecessary, although the addition of extra experiments aiming to fill this gap remains an option.
2D-NMR spectroscopy: carbon amino acid selective (CAS) experiments
In many cases, collection of a high-resolution 15N,13C-CON and the four 3D experiments discussed above is sufficient to assign the backbone of a target IDP. For each CON signal, 3D CCCON yields the complete aliphatic carbon resonances of the preceding residue, narrowing down the amino acid type, and the 3D (HN-flip)N(CA)CON provides the nitrogen chemical shift of the preceding residue as well. Sometimes, however, this process leads to regions in the CON spectrum where, for the same nitrogen chemical shift, there is more than one resonance present, causing ambiguity. In these instances, the use of amino acid filtered 2D experiments can provide the additional information needed to break the impasse at very little additional cost in terms of spectrometer time.[12, 41–44] These experiments capitalize on amino acid-type specific spin physics, band-selective pulses exciting very unique spectral regions, and specific coupling transfer steps to yield even greater amino acid filtering than is achievable by inference from the set of aliphatic carbon chemical shifts recorded in the CCCON alone. Originally designed by the Oschkinat group in an amide proton-detected format,[41–44] these experiments have recently been modified for 15N,13C-CON readout as carbon-detected amino acid specific spectra (CAS-NMR).[12] As was the case with the 3D pulse sequences discussed above, we have found that in many cases minor modifications to the originally reported pulse sequences were required for optimal performance. Pulse programs for the entire suite of carbon-detected amino acid specific 2D experiments that we employ in our laboratory are provided in the Supplementary Information as Fig. S4–18.
In general, the CAS-NMR data sets are composed of two experiments, collected as a 15N,13C-CACON and the closely related 15N,13C-CANCO. The CACON type experiment makes use of amino acid filters to select the desired amino acid type, followed by intra-residue transfer from the 13Cαto the 13C´, via the 1JCαC’ scalar coupling. The experiment concludes with standard readout of the 13C´-15N correlation in the 2D CON format. The CANCO type experiment also makes use of the same amino-acid filters, but employs a different set of echo times such that both the intra-residue 1JCαC’ and the inter-residue 2JCαC’ are active during transfer to the 13C´ prior to detection of 13C´-15N pairs in a format visually indistinguishable from the 2D CON. Note, however, that the longer delay times required for refocusing both one- and two- bond coupling at the end of the CANCO experiment, along with splitting the initial magnetization between two correlations, leads to lower signal to noise, which is usually circumvented by an increase in the number of scans. The key to the CAS-NMR strategy is the following: for a given selected amino acid type, the CAS-filtered 15N,13C-CACON only contains peaks corresponding to the CON correlations of residues adjacent to the selected amino acid type, on the C-terminal side; the CAS-filtered 15N,13C-CANCO recaptures this same set of peaks and also the set of CON correlations directly corresponding to the amino acid type selected for. While it is generally not possible to know a priori which self-peak corresponds to which neighbor-peak in the 15N,13C-CANCO, the spectrum is greatly simplified and any ambiguity present in the 3D spectra discussed above is generally resolved.
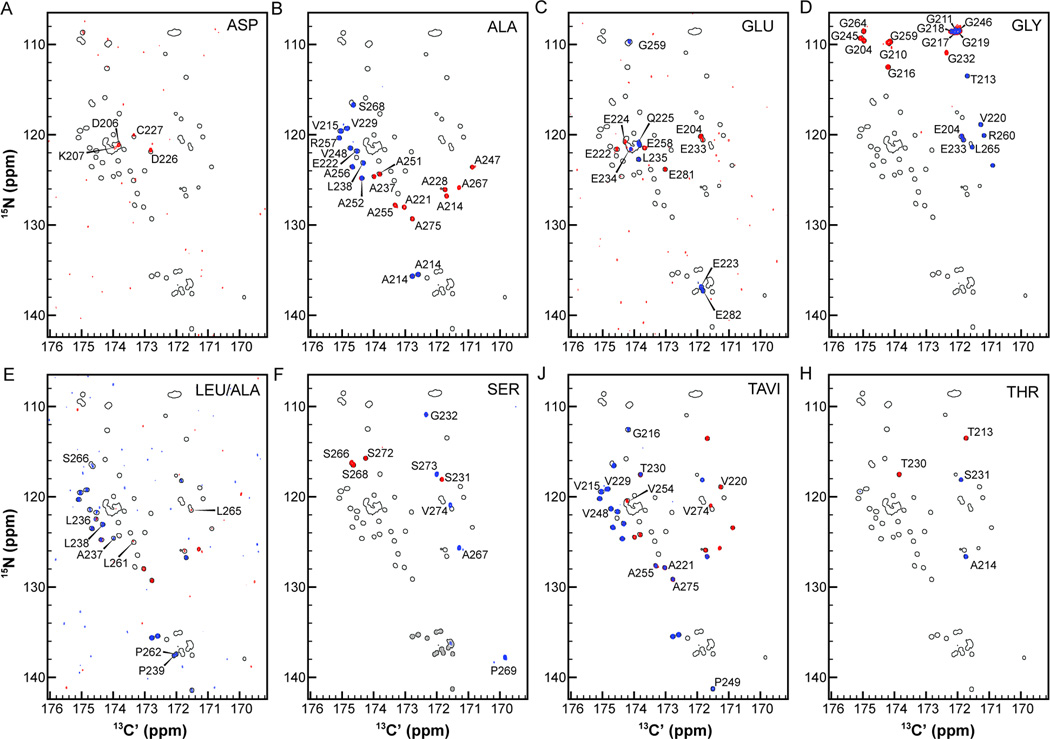
To provide an example of how this procedure works, we present Pdx1-C CAS-NMR experiments acquired with eight different amino acid filters in Fig. 4 (the pulse sequences used are presented in Fig. S4–S18). In each panel, the shadow of the full 15N,13C-CON is displayed, with the CAS-CACON overlaid in blue and the CAS-CANCO in red. All CAS-NMR peaks have been annotated with an assignment corresponding to the residue contributing the 15N-resonance to the 2D correlation. Note that in all panels, half of the CAS-CANCO resonances are obscured by the CAS-CACON resonances. These spectra are satisfyingly free of bleed-through artifacts in all cases but one; all of the peaks arising from a proline amide nitrogen are present as artifacts in the Ser-filtered CAS-CANCO (and are colored grey in Fig. 4 for clarity). Note that this artifact poses little practical impediment to assignment.
Figure 4.
Suite of carbon-detected amino acid selective (CAS-NMR) experiments collected as CACON/CANCO for (A) Asp (B) Ala (C) Glu (D) Gly (E) Leu/Ala (F) Ser (G) Thr-Ala-Val-Ile and (H) Thr. In each panel, the 15N,13C-CON is shown as outlined peaks, with the CACON resonances overlaid in blue and the CANCO resonances in red. Note that the Ser-filtered CAS-CANCO contains artifact peaks arising from poor suppression of proline resonances (colored grey in panel F). The pulse sequences used to generate this data are presented in Fig. S4–18.
The simplest illustration of how these experiments are used is provided by considering the Thr-filtered spectra in Fig. 4H (bottom right). Pdx1-C contains two threonine residues (T213 and T230), resulting in the two annotated correlations from the Thr-filtered CAS-CANCO (red peaks in Fig. 4H); and the two inter-residue correlations annotated as A214 and S231, respectively (blue peaks in Fig. 4H). The identity of the blue peak appearing downfield in the 15N-dimension, annotated as A214 in Fig. 4H, was established by noting that it is also recorded in the Ala-filtered CAS-CANCO (see the red peak in the same position of the Ala panel, Fig. 4B). The dipeptide Thr-Ala only appears once in the sequence of Pdx1-C, providing unambiguous assignment of this resonance. Similarly, the blue resonance in the Thr-filtered CAS-CACON labeled as S231 is also present in the Ser-filtered CAS-CANCO, where it appears in red at the same location in Fig. 4F. Because the Thr-Ser dipeptide sequence is also unique, this resonance is unambiguously assigned. Next, the two red autocorrelations identified as threonine residues must be unambiguously assigned. This is similarly straight forward because T213 also is found in the Gly-filtered CAS-CACON (blue resonance in Fig. 4D), yielding positive identification; T230 is identified because all other possibilities are eliminated (although its assignment can also be verified from the TAVI-filtered CAS-CACON in Fig. 4J upon additional inspection).
It is similarly straight forward to unambiguously identify the residue specific assignments for all five resonances in the Ser-filtered CAS-CON, because each serine in Pdx1-C is followed by an amino acid of a different type (G231, A267, P269, S273, and V274), all of which are used here to filter a CAS-CANCO spectrum. Following this procedure to its completion yields unambiguous assignment for a large fraction of peaks without any reference to 3D spectra and at least provides amino acid typing for many more. In other words, this procedure yields multiple robust starting points for analysis of the (HN-flip)N(CA)CON experiment, generally resulting in in efficient resonance assignments of IDPs.
To summarize, a total of four 3D carbon-detected spectra and 15 2D carbon-detected spectra were used to generate thorough chemical shift assignment for the C-terminal disordered region of Pdx1 (the assignments are deposited in the BMRB, under the accession number 19596), as a result of investing two weeks of NMR time. While all of the pulse programs we used for this project have been previously published,[5, 9, 11, 12] we had to modify a large number of them for use on our spectrometers; few others were included in the standard, manufacturer provided, pulse program library. In order to facilitate adoption of the protocol recommended in this paper, we have made Bruker formatted versions of the pulse sequences used for this project (and others written by our laboratory) available through our website.
Outcomes for Pdx1-C
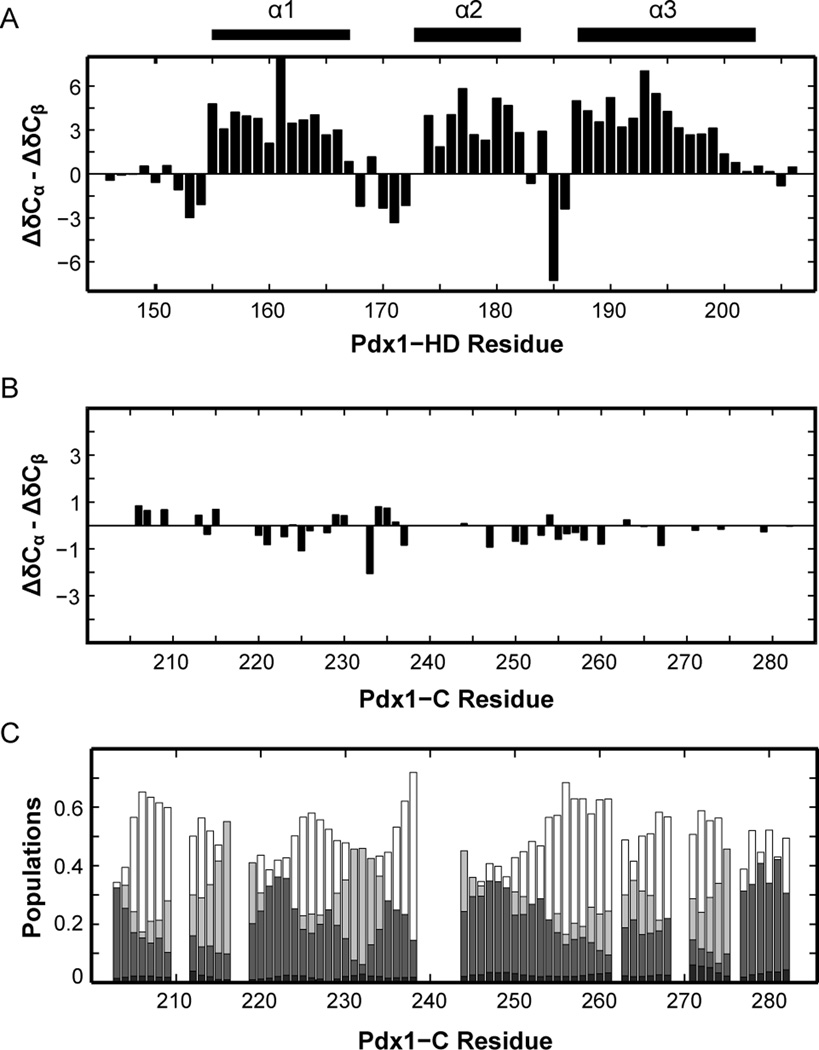
The experiments described herein led efficiently to complete assignment of Pdx1-C, despite its biased amino acid composition and the presence of long glycine and proline repeats. This project was motivated by a desire to study structure-function relationships for Pdx1-C, which is known to mediate interactions with other transcription factors and to harbor amino acid polymorphisms that correlate with the early onset of type-2 diabetes in humans.[33, 36] In contrast to natively folding proteins, IDPs like Pdx1-C do not adopt a single conformation under native conditions, but instead remain in a potentially heterogeneous ensemble of disordered conformations.[45] As a first step towards evaluating the structure of Pdx1-C, we have calculated its secondary structure propensity (SSP), [20] based on the chemical shift assignments we report here, and compared the results to those we have previously reported for the cooperatively folding homeodomain of Pdx1.[16] Figure 5 shows the difference between the offset from random coil for the 13Cαand 13Cβ nuclei (ΔδCα - ΔδCβ) for each residue in Pdx1-homeodomain (Fig. 5A) and Pdx1-C (Fig. 5B). When chemical shift data is represented in this way, stretches of values greater than or less than ~2 ppm correspond to α-helix and β-strand character, respectively, making it easy to see the three α-helical segments composing the homeodomain (Fig. 5A). In contrast, the graph for Pdx1-C is almost completely devoid of features, suggesting that its conformation in solution is highly random. This assignment is confirmed through calculation of secondary structure tendencies in Pdx1-C using the recently published δ2D method,[21] which predicts the ensemble to be a mixture of coil, polyproline II, and extended-β conformation (Fig. 5C). The nearly complete absence of secondary structure bias predicted for Pdx1-C is somewhat unexpected, because disordered fragments that mediate protein-protein interactions are often identified by secondary-structural bias in the unbound state.[46] It is therefore intriguing to speculate that Pdx1-C may bind its partners in a highly extended conformation.
Figure 5.
Secondary chemical shifts of Pdx1-C indicate that this protein adopts an extended coil-like structure. (A) For comparison, the secondary structure of Pdx1-HD (BMRB accession number 19227) is shown as the difference between 13Cα and 13Cβ secondary chemical shifts, indicating the presence of an α-helix when long stretches of positive values are encountered. The secondary structure from the co-crystal of Pdx1 with a consensus DNA duplex is represented by bars above the figure for comparison. (B) Pdx1-C displays virtually no tendency towards secondary structure, based on the difference between 13Cα and 13Cβ secondary chemical shifts. (C) δ2D calculations for Pdx1-C (see Materials and Methods) provide additional insight into the nature of the extended structure found. For each residue, α-helix is shown in black, extended-β in dark grey, polyproline II in light grey, and coil structure as white (non-filled) bars.
Conclusions
Using a suite of exclusively carbon-detected NMR experiments, we were able to successfully complete chemical shift assignment for the intrinsically disordered polypeptide chain of Pdx1-C. Our strategy combines nearest-neighbor assignment, facilitated by 3D experiments that correlate amide nitrogen pairs and triples, with carbon-detected amino-acid selective NMR experiments composed of pairs of 2D CACON and CANCO spectra. The amalgamation of these two experiment types yielded a chemical shift assignment strategy that only required 2 weeks of instrument time and that was sufficient for the complete assignment of an 80 residue IDP, shown by secondary chemical shift information to be almost completely devoid of secondary structure. This chemical shift assignment of Pdx1-C now serves as the stepping stone for future structural and mechanistic studies. Of broad importance for the community, all of the pulse programs used to generate these results, as well as many others our laboratory has used and/or published, are now freely available through our website, making adoption of the protocol used here straight forward.
Supplementary Material
Acknowledgments
This work was supported by a NSF-CAREER award (MCB-0953918) and start-up funds from the Pennsylvania State University to SAS. MB is the recipient of a NIH predoctoral fellowship (F31GM101936). We would like to thank the Penn State ScholarSphere team (http://scholarsphere.psu.edu) for their help establishing permanent and stable web-hosting of our pulse programs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
Contributor Information
Debashish Sahu, Email: dzs12@psu.edu.
Monique Bastidas, Email: mob5256@psu.edu.
References
- 1.Tantos A, Han KH, Tompa P. Intrinsic disorder in cell signaling and gene transcription. Mol. Cell. Endocrinol. 2012;348:457–465. doi: 10.1016/j.mce.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 3.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Nat. Acad. Sci. USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Hare B, Benesi AJ, Showalter SA. Incorporating 1H-chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J. Magn. Reson. 2009;200:354–358. doi: 10.1016/j.jmr.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Showalter SA. NMR Assignment of the Intrinsically Disordered C-terminal Region of Homo sapiens FCP1 in the Unbound State. Biomol. NMR Assign. 2009;3:179–181. doi: 10.1007/s12104-009-9169-1. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence CW, Bonny A, Showalter SA. The disordered C-terminus of the RNA Polymerase II phosphatase FCP1 is partially helical in the unbound state. Biochem. Biophys. Res. Commun. 2011;410:461–465. doi: 10.1016/j.bbrc.2011.05.160. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence CW, Showalter SA. Carbon-Detected N-15 NMR Spin Relaxation of an Intrinsically Disordered Protein: FCP1 Dynamics Unbound and in Complex with RAP74. J. Phys. Chem. Lett. 2012;3:1409–1413. doi: 10.1021/jz300432e. [DOI] [PubMed] [Google Scholar]
- 9.Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R. 13C-detected protonless NMR spectroscopy of proteins in solution. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:25–45. [Google Scholar]
- 10.Novacek J, Haba NY, Chill JH, Zidek L, Sklenar V. 4D non-uniformly sampled HCBCACON and (1)J(NCalpha)-selective HCBCANCO experiments for the sequential assignment and chemical shift analysis of intrinsically disordered proteins. J. Biomol. NMR. 2012;53:139–148. doi: 10.1007/s10858-012-9631-8. [DOI] [PubMed] [Google Scholar]
- 11.Bermel W, Bertini I, Felli IC, Gonnelli L, Kozminski W, Piai A, Pierattelli R, Stanek J. Speeding up sequence specific assignment of IDPs. J. Biomol. NMR. 2012;53:293–301. doi: 10.1007/s10858-012-9639-0. [DOI] [PubMed] [Google Scholar]
- 12.Bermel W, Bertini I, Chill J, Felli IC, Haba N, Kumar MVV, Pierattelli R. Exclusively heteronuclear (13) C-detected amino-acid-selective NMR experiments for the study of intrinsically disordered proteins (IDPs) Chembiochem. 2012;13:2425–2432. doi: 10.1002/cbic.201200447. [DOI] [PubMed] [Google Scholar]
- 13.Pantoja-Uceda D, Santoro J. Direct correlation of consecutive C'-N groups in proteins: a method for the assignment of intrinsically disordered proteins. J. Biomol. NMR. 2013;57:57–63. doi: 10.1007/s10858-013-9765-3. [DOI] [PubMed] [Google Scholar]
- 14.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ, Rance M. Protein NMR Spectroscopy: Principles and Practice. 2nd ed. Burlington: Academic Press; 2005. [Google Scholar]
- 15.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- 16.Bastidas M, Showalter SA. Thermodynamic and structural determinants of differential Pdx1 binding to elements from the insulin and IAPP promoters. J. Mol. Biol. 2013;425:3360–3377. doi: 10.1016/j.jmb.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emsley L, Bodenhausen G. Optimization of Shaped Selective Pulses for NMR Using a Quaternion Description of Their Overall Propagators. J. Magn. Reson. 1992;97:135–148. [Google Scholar]
- 18.Bohlen JM, Bodenhausen G. Experimental Aspects of CHIRP NMR-Spectroscopy. J. Magn. Reson. A. 1993;102:293–301. [Google Scholar]
- 19.Goddard T, Kneller D. SPARKY 3.113. 2006 [Google Scholar]
- 20.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 22.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 23.The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero P, Obradovic Z, Li XH, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins Struct. Funct. Dyn. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol. Cell. Endocrinol. 2008;294:1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Glick E, Leshkowitz D, Walker MD. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J. Biol. Chem. 2000;275:2199–2204. doi: 10.1074/jbc.275.3.2199. [DOI] [PubMed] [Google Scholar]
- 28.Cissell MA, Zhao L, Sussel L, Henderson E, Stein R. Transcription factor occupancy of the insulin gene in vivo. Evidence for direct regulation by Nkx2.2. J. Biol. Chem. 2003;278:751–756. doi: 10.1074/jbc.M205905200. [DOI] [PubMed] [Google Scholar]
- 29.Ohneda K, Mirmira RG, Wang J, Johnson JD, German MS. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serup P, Jensen J, Andersen FG, Jorgensen MC, Blume N, Holst JJ, Madsen OD. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc. Natl. Acad. Sci. USA. 1996;93:9015–9020. doi: 10.1073/pnas.93.17.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longo A, Guanga GP, Rose RB. Structural basis for induced fit mechanisms in DNA recognition by the Pdx1 homeodomain. Biochemistry. 2007;46:2948–2957. doi: 10.1021/bi060969l. [DOI] [PubMed] [Google Scholar]
- 32.Liu A, Oliver-Krasinski J, Stoffers DA. Two conserved domains in PCIF1 mediate interaction with pancreatic transcription factor PDX-1. FEBS Lett. 2006;580:6701–6706. doi: 10.1016/j.febslet.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 2002;22:412–420. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hani EH, Stoffers DA, Chevre JC, Durand E, Stanojevic V, Dina C, Habener JF, Froguel P. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in lateonset type 2 diabetes mellitus. J. Clin. Invest. 1999;104:R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng J, Macfarlane WM, Lehto M, Gu HF, Shepherd LM, Ivarsson SA, Wibell L, Smith T, Groop LC. Functional consequences of mutations in the MODY4 gene (IPF1) and coexistence with MODY3 mutations. Diabetologia. 2001;44:249–258. doi: 10.1007/s001250051608. [DOI] [PubMed] [Google Scholar]
- 36.Cockburn BN, Bermano G, Boodram LL, Teelucksingh S, Tsuchiya T, Mahabir D, Allan AB, Stein R, Docherty K, Bell GI. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J. Clin. Endocrinol. Metab. 2004;89:971–978. doi: 10.1210/jc.2003-031282. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane WM, Frayling TM, Ellard S, Evans JC, Allen LI, Bulman MP, Ayres S, Shepherd M, Clark P, Millward A, Demaine A, Wilkin T, Docherty K, Hattersley AT. Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J. Clin. Invest. 1999;104:R33–R39. doi: 10.1172/JCI7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Showalter SA, Noid WG. Native-based simulations of the binding interaction between RAP74 and the disordered FCP1 peptide. J. Phys. Chem. B. 2013;117:3074–3085. doi: 10.1021/jp310293b. [DOI] [PubMed] [Google Scholar]
- 39.Bermel W, Bertini I, Felli IC, Pierattelli R. Speeding Up 13C Direct Detection Biomolecular NMR Spectroscopy. J. Am. Chem. Soc. 2009;131:15339–15345. doi: 10.1021/ja9058525. [DOI] [PubMed] [Google Scholar]
- 40.Bertini I, Felli IC, Gonnelli L, Kumar MVV, Pierattelli R. High-Resolution Characterization of Intrinsic Disorder in Proteins: Expanding the Suite of (13)C-Detected NMR Spectroscopy Experiments to Determine Key Observables. Chembiochem. 2011;12:2347–2352. doi: 10.1002/cbic.201100406. [DOI] [PubMed] [Google Scholar]
- 41.Schmieder P, Leidert M, Kelly M, Oschkinat H. Multiplicity-Selective Coherence Transfer Steps for the Design of Amino Acid-Selective Experiments-A Triple-Resonance Experiment Selective for Asn and Gln. J. Magn. Reson. 1998;131:199–202. doi: 10.1006/jmre.1997.1348. [DOI] [PubMed] [Google Scholar]
- 42.Schubert M, Smalla M, Schmieder P, Oschkinat H. MUSIC in triple-resonance experiments: amino acid type-selective (1)H-(15)N correlations. J. Magn. Reson. 1999;141:34–43. doi: 10.1006/jmre.1999.1881. [DOI] [PubMed] [Google Scholar]
- 43.Schubert M, Ball LJ, Oschkinat H, Schmieder P. Bridging the gap: A set of selective 1H-15N-correlations to link sequential neighbors of prolines. J. Biomol. NMR. 2000;17:331–335. doi: 10.1023/a:1008362904205. [DOI] [PubMed] [Google Scholar]
- 44.Schubert M, Oschkinat H, Schmieder P. MUSIC and aromatic residues: amino acid type-selective (1)H-(15)N correlations, III. J. Magn. Reson. 2001;153:186–192. doi: 10.1006/jmre.2001.2447. [DOI] [PubMed] [Google Scholar]
- 45.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 47.Grzesiek S, Bax A. Amino-Acid Type Determination in the Sequential Assignment Procedure of Uniformly C-13/N-15-Enriched Proteins. J. Biomol. NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 48.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A General-Method for Assigning NMR-Spectra of Denatured Proteins Using 3D HC(CO)NH-TOCSY Triple Resonance Experiments. J. Biomol. NMR. 1993;3:225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.