Abstract
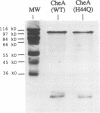
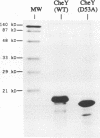
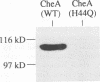
Regulated phosphorylation of proteins has been shown to be a hallmark of signal transduction mechanisms in both Eubacteria and Eukarya. Here we demonstrate that phosphorylation and dephosphorylation are also the underlying mechanism of chemo- and phototactic signal transduction in Archaea, the third branch of the living world. Cloning and sequencing of the region upstream of the cheA gene, known to be required for chemo- and phototaxis in Halobacterium salinarium, has identified cheY and cheB analogs which appear to form part of an operon which also includes cheA and the following open reading frame of 585 nucleotides. The CheY and CheB proteins have 31.3 and 37.5% sequence identity compared with the known signal transduction proteins CheY and CheB from Escherichia coli, respectively. The biochemical activities of both CheA and CheY were investigated following their expression in E.coli, isolation and renaturation. Wild-type CheA could be phosphorylated in a time-dependent manner in the presence of [gamma-32P]ATP and Mg2+, whereas the mutant CheA(H44Q) remained unlabeled. Phosphorylated CheA was dephosphorylated rapidly by the addition of wild-type CheY. The mutant CheY(D53A) had no effect on phosphorylated CheA. The mechanism of chemo- and phototactic signal transduction in the Archaeon H.salinarium, therefore, is similar to the two-component signaling system known from chemotaxis in the eubacterium E.coli.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Hazelbauer G. L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991 Sep;173(18):5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Lebert M., Oesterhelt D., Hazelbauer G. L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989 Feb;8(2):631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R., Eisenbach M. Fumarate or a fumarate metabolite restores switching ability to rotating flagella of bacterial envelopes. J Bacteriol. 1992 Jan;174(2):643–645. doi: 10.1128/jb.174.2.643-645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff D. S., Ordal G. W. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992 Jan;6(1):23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- Bischoff D. S., Ordal G. W. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem. 1991 Jul 5;266(19):12301–12305. [PubMed] [Google Scholar]
- Bourret R. B., Davagnino J., Simon M. I. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol. 1993 Apr;175(7):2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., Bourret R. B., Simon M. I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993 May;4(5):469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Buss J. E., Stull J. T. Measurement of chemical phosphate in proteins. Methods Enzymol. 1983;99:7–14. doi: 10.1016/0076-6879(83)99035-3. [DOI] [PubMed] [Google Scholar]
- Dons L., Olsen J. E., Rasmussen O. F. Characterization of two putative Listeria monocytogenes genes encoding polypeptides homologous to the sensor protein CheA and the response regulator CheY of chemotaxis. DNA Seq. 1994;4(5):301–311. doi: 10.3109/10425179409020856. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Krah M., Marwan W., Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993 Aug;12(8):2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greck M., Platzer J., Sourjik V., Schmitt R. Analysis of a chemotaxis operon in Rhizobium meliloti. Mol Microbiol. 1995 Mar;15(6):989–1000. doi: 10.1111/j.1365-2958.1995.tb02274.x. [DOI] [PubMed] [Google Scholar]
- Hain J., Reiter W. D., Hüdepohl U., Zillig W. Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 1992 Oct 25;20(20):5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Matsumura P., Simon M. I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Kirsch M. L., Peters P. D., Hanlon D. W., Kirby J. R., Ordal G. W. Chemotactic methylesterase promotes adaptation to high concentrations of attractant in Bacillus subtilis. J Biol Chem. 1993 Sep 5;268(25):18610–18616. [PubMed] [Google Scholar]
- Krah M., Marwan W., Oesterhelt D. A cytoplasmic domain is required for the functional interaction of SRI and HtrI in archaeal signal transduction. FEBS Lett. 1994 Oct 24;353(3):301–304. doi: 10.1016/0014-5793(94)01068-4. [DOI] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994 May 1;13(9):2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987 Mar;169(3):1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lukat G. S., Lee B. H., Mottonen J. M., Stock A. M., Stock J. B. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991 May 5;266(13):8348–8354. [PubMed] [Google Scholar]
- Marwan W., Alam M., Oesterhelt D. Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J Bacteriol. 1991 Mar;173(6):1971–1977. doi: 10.1128/jb.173.6.1971-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwan W., Bibikov S. I., Montrone M., Oesterhelt D. Mechanism of photosensory adaptation in Halobacterium salinarium. J Mol Biol. 1995 Mar 3;246(4):493–499. doi: 10.1006/jmbi.1994.0101. [DOI] [PubMed] [Google Scholar]
- Matsumura P., Rydel J. J., Linzmeier R., Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984 Oct;160(1):36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Zusman D. R. Purification and characterization of the Myxococcus xanthus FrzE protein shows that it has autophosphorylation activity. J Bacteriol. 1990 Dec;172(12):6661–6668. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrone M., Marwan W., Grünberg H., Musseleck S., Starostzik C., Oesterhelt D. Sensory rhodopsin-controlled release of the switch factor fumarate in Halobacterium salinarium. Mol Microbiol. 1993 Dec;10(5):1077–1085. doi: 10.1111/j.1365-2958.1993.tb00978.x. [DOI] [PubMed] [Google Scholar]
- Morrison T. B., Parkinson J. S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh N., Simon M. I. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol. 1986 Jan;165(1):161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa E. G., Atkinson M. R., Kamberov E. S., Ninfa A. J. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993 Nov;175(21):7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann B., Lebert M. R., Alam M., Nitz S., Kollmannsberger H., Oesterhelt D., Hazelbauer G. L. Identification of volatile forms of methyl groups released by Halobacterium salinarium. J Biol Chem. 1994 Jun 10;269(23):16449–16454. [PubMed] [Google Scholar]
- Olson K. D., Spudich J. L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993 Dec;65(6):2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. R., Daniels C. J. In vivo definition of an archaeal promoter. J Bacteriol. 1995 Apr;177(7):1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rothärmel T., Wagner G. Isolation and characterization of a calmodulin-like protein from Halobacterium salinarium. J Bacteriol. 1995 Feb;177(3):864–866. doi: 10.1128/jb.177.3.864-866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J., Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995 Feb 15;14(4):667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. C., Swanson R. V., Alex L. A., Bourret R. B., Simon M. I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993 Sep 23;365(6444):343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- Segall J. E., Manson M. D., Berg H. C. Signal processing times in bacterial chemotaxis. Nature. 1982 Apr 29;296(5860):855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Hasselbacher C. A., Spudich J. L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988 Sep;170(9):4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Photosensitive phosphoproteins in Halobacteria: regulatory coupling of transmembrane proton flux and protein dephosphorylation. J Cell Biol. 1981 Dec;91(3 Pt 1):895–900. doi: 10.1083/jcb.91.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993 Aug 5;268(22):16095–16097. [PubMed] [Google Scholar]
- Spudich E. N., Takahashi T., Spudich J. L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Color sensing in the Archaea: a eukaryotic-like receptor coupled to a prokaryotic transducer. J Bacteriol. 1993 Dec;175(24):7755–7761. doi: 10.1128/jb.175.24.7755-7761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Stoeckenius W. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J Biol Chem. 1980 Jun 25;255(12):5501–5503. [PubMed] [Google Scholar]
- Starnbach M. N., Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992 Feb;6(4):459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Mottonen J. M., Stock J. B., Schutt C. E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989 Feb 23;337(6209):745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Wylie D. C., Mottonen J. M., Lupas A. N., Ninfa E. G., Ninfa A. J., Schutt C. E., Stock J. B. Phosphoproteins involved in bacterial signal transduction. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 1):49–57. doi: 10.1101/sqb.1988.053.01.009. [DOI] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Lebert M., Spudich J. L., Oesterhelt D., Hazelbauer G. L. Characterization of Halobacterium halobium mutants defective in taxis. J Bacteriol. 1990 May;172(5):2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R. V., Bourret R. B., Simon M. I. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993 May;8(3):435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- Tawa P., Stewart R. C. Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry. 1994 Jun 28;33(25):7917–7924. doi: 10.1021/bi00191a019. [DOI] [PubMed] [Google Scholar]
- Thoelke M. S., Kirby J. R., Ordal G. W. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry. 1989 Jun 27;28(13):5585–5589. doi: 10.1021/bi00439a037. [DOI] [PubMed] [Google Scholar]
- Volz K., Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991 Aug 15;266(23):15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- West A. H., Martinez-Hackert E., Stock A. M. Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB. J Mol Biol. 1995 Jul 7;250(2):276–290. doi: 10.1006/jmbi.1995.0376. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Stewart R. C. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]