Background: Dpo4 and Dbh translesion polymerases show distinct mutagenic signatures and lesion bypass abilities.
Results: Three residues in the interdomain linker are major determinants of polymerase conformation and nucleotide incorporation rate.
Conclusion: Residues remote from the active site strongly influence single-base deletion and mispair extension efficiency by controlling polymerase conformation.
Significance: This work suggests new possibilities for regulating translesion synthesis activity through altered conformation.
Keywords: DNA Polymerase, DNA Replication, Enzyme Mutation, Kinetics, Protein Conformation, Structural Biology, Fidelity, Translesion Synthesis, Y-family Polymerase
Abstract
Dpo4 and Dbh are from two closely related Sulfolobus species and are well studied archaeal homologues of pol IV, an error prone Y-family polymerase from Escherichia coli. Despite sharing 54% amino acid identity, these polymerases display distinct mutagenic and translesion specificities. Structurally, Dpo4 and Dbh adopt different conformations because of the difference in relative orientation of their N-terminal catalytic and C-terminal DNA binding domains. Using chimeric constructs of these two polymerases, we have previously demonstrated that the interdomain linker is a major determinant of polymerase conformation, base-substitution fidelity, and abasic-site translesion synthesis. Here we find that the interdomain linker also affects the single-base deletion frequency and the mispair extension efficiency of these polymerases. Exchanging just three amino acids in the linkers of Dbh and Dpo4 is sufficient to change the fidelity by up to 30-fold, predominantly by altering the rate of correct (but not incorrect) nucleotide incorporation. Additionally, from a 2.4 Å resolution crystal structure, we have found that the three linker amino acids from Dpo4 are sufficient to allow Dbh to adopt the standard conformation of Dpo4. Thus, a small region of the interdomain linker, located more than 11 Å away from the catalytic residues, determines the fidelity of these Y-family polymerases, by controlling the alignment of substrates at the active site.
Introduction
DNA polymerases are the enzymatic workhorses that ensure effective genome duplication through multiple rounds of nucleotide addition. Translesion synthesis (TLS)3 polymerases have the ability to effectively bypass sites of DNA damage, an activity that is crucial to cell survival (1). Functionally, TLS polymerases lack a 3′ to 5′ proofreading activity, and structurally, they lack the extensive protein-DNA contacts found in their replicative counterparts that ensure a tightly constrained active site. Thus, these polymerases are unusually mutagenic on undamaged DNA.
Most TLS polymerases belong to the Y-family of DNA polymerases (2). These can be further categorized based on sequence similarity into six types that include the ubiquitous DinB family, the two UmuC families found only in bacteria, and the Rad30A (pol η), Rad30B (pol ι), and Rev1 families found only in eukaryotes. These enzymes display individual substrate specificities in the preferential bypass of certain types of DNA damage (1). TLS can be error-free or error-prone, depending on the polymerase and the lesion. Even when replicating undamaged DNA, the Y-family polymerases each display unique mutagenic signatures (see, for example, Refs. 3–8).
In the past decade, a large number of studies have focused on understanding the correlation between structure and function in the Y-family TLS polymerases (9). These enzymes share a core structure consisting of an N-terminal catalytic domain, with palm, fingers, and thumb subdomains (as are found in other families of DNA polymerases), and a C-terminal domain (unique to the Y-family) that is known as the “little finger” or polymerase-associated domain (LF/PAD) (10, 11). Although much is known about which specific DNA lesions are bypassed by individual polymerases, we still do not have a comprehensive understanding of which features of polymerase architecture are responsible for providing lesion bypass selectivity and mutagenic specificity. This knowledge is highly significant, because mutations made by Y-family polymerases predispose cells to developing cancer or antibiotic resistance.
In 2004, Boudsocq et al. reported that many of the lesion bypass and mutational activities of two closely related archaeal DinB homologues, Dbh and Dpo4, were largely dependent on sequences outside of the catalytic domain (12). Dbh and Dpo4 are from two different strains of Sulfolobus and share 54% sequence identity, yet Dpo4 is able to bypass abasic sites and thymidine dimers, whereas Dbh cannot. Dpo4 makes more base-substitution mutations, whereas Dbh has a higher propensity for making single-base deletion mutations (12–14).
We have shown recently that the 15-residue linker connecting the polymerase and LF/PAD domains of Dbh and Dpo4 (Fig. 1) can alone influence the enzymatic activity and selectivity of these polymerases (15). A ternary complex structure of a chimera with the Dbh polymerase core and LF/PAD but the Dpo4 linker (Dbh-Dpo4-Dbh) adopts a Dpo4-like conformation. Like Dpo4, the LF/PAD is in contact with the fingers domain, docking into the major groove of the DNA duplex and positioning the primer-template junction at the active site for efficient catalysis. This chimera was also found to bypass an abasic site and display single nucleotide incorporation fidelity similar to Dpo4.
FIGURE 1.
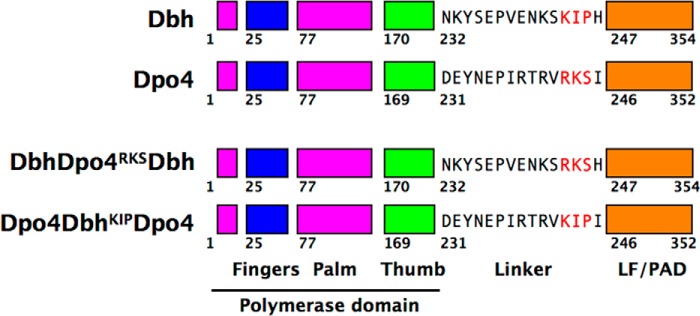
Parental Y-family polymerases and chimeric constructs. Both Dbh and Dpo4 have a polymerase core consisting of palm (magenta), fingers (blue), and thumb (green) domains in addition to a C-terminal LF/PAD (orange). The amino acid sequence of the linker connecting the polymerase core to the LF/PAD is shown. Residues highlighted in red have been interchanged to create chimeras. Chimeric polymerases are named by the parental source of each domain, in the order polymerase-linker-LF/PAD. Superscript indicates the swapped residues present in the chimeric polymerase.
Here we extend our previous studies of chimeric constructs of Dbh and Dpo4 and find that the single-base deletion activity of these polymerases is also dependent on the linker identity. Furthermore, just three residues in the interdomain linker control the enzyme conformation and influence fidelity by affecting the rate of nucleotide incorporation, without being in the vicinity of the active site.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Dbh expression and purification were performed as described before (16). Dpo4 and all the chimeric constructs used had C-terminal His6 tags, and growth and purification steps were performed as described before (15, 17).
Primer-Template DNA
All DNA substrates used in this study are listed in Table 1 and were synthesized from Integrated DNA Technologies. The primer used for extension assays was synthesized with a 5′-6-carboxyfluorescein (FAM) label; the DNA used for crystallization was unlabeled. Primer was annealed to template in annealing buffer containing 10 mm HEPES (pH 7.5) and 50 mm NaCl.
TABLE 1.
DNA substrate sequences
FAM refers to the 6-carboxyfluorescein label at the 5′ ends of primers for substrates 1–6, used for primer-extension assays. Substrate 7, used for crystallization, was unlabeled. The deletion hotspot sequence and variations thereof are shown in bold type.
| Substrate | Sequence | |
|---|---|---|
| 1 | 4C-G | 5′-(FAM)-AGG CAC TGA TCG GG-3′ |
| 3′-CC GTG ACT AGC CCC GCA TT-5′ | ||
| 2 | 1T-G | 5′-(FAM)-AGG CAC TGA TCG GG-3′ |
| 3′-CC GTG ACT AGC CCT GCA TT-5′ | ||
| 3 | 2T-G | 5′-(FAM)-AGG CAC TGA TCG G G-3′ |
| 3′-CC GTG ACT AGC CTC GCA TT-5′ | ||
| 4 | 3T-G | 5′-(FAM)-AGG CAC TGA TCG GG-3′ |
| 3′-CC GTG ACT AGC TCC GCA TT-5′ | ||
| 5 | 4T-G | 5′-(FAM)-AGG CAC TGA TC GGG-3′ |
| 3′-CC GTG ACT AGT CCC GCA TT-5′ | ||
| 6 | C-C mispair | 5′-(FAM)-AGG CAC TGA TCG GGC-3′ |
| 3′-CC GTG ACT AGC CCC GCA TT-5′ | ||
| 7 | X-4T-G | 5′- GG CAC TGA TC GGG-3′ |
| 3′- CC GTG ACT AGT CCC GCA TT-5′ |
Deletion Assays
Reaction mixtures contained a final concentration of 40 nm annealed primer-template DNA, 4 μm polymerase, 20 mm HEPES (pH 7.0), 85 mm NaCl, 5 mm MgCl2, 1 mm DTT, and 1 mm dCTP or dGTP. Reactions were incubated at room temperature and quenched after 1, 2, 4, 8, 12, or 20 min by mixing them with an equal volume of stopping solution (80% formamide, 100 mm EDTA, with bromphenol blue and xylene cyanol dyes). Samples were separated by electrophoresis on a 17.5% polyacrylamide (19:1), 7.5 m urea, 1× Tris-borate-EDTA (TBE) sequencing gel. Gels were imaged using a Typhoon 9400 scanner, and the fluorescence intensity of the unextended and extended primer bands was quantified with ImageQuant software (GE Healthcare). Percentage of primer extension was determined by measuring the relative intensity of the band corresponding to the extended primer with respect to the total labeled DNA. Data were fit to an exponential equation (Equation 1),
where A is amplitude, k1 is the observed rate of product formation, t is the time after which reaction was quenched, and c is a constant. All of the graphs and nonlinear regressions were done using GraphPad Prism, version 6.0a (GraphPad Software). Experiments were performed in triplicate with error bars representing the S.E. of the data collected.
KDdNTP and kpol Determination
Primer extension assays were performed using final concentrations of 4 μm polymerase and 40 nm primer-template DNA in reaction buffer (25 mm HEPES (pH 7.5), 85 mm NaCl, 10 mm MgCl2, 2 mm DTT). Reactions were started by adding various deoxynucleotide triphosphate (dNTP) concentrations ranging from 5–2000 μm (depending on polymerase) and allowed to proceed for appropriate time intervals. Experiments were performed in duplicate. Time courses of primer extension reactions were fit to a single- exponential equation (Equation 1). The observed rates (k1) thus obtained were further plotted as a function of dNTP concentration and then fit to a hyperbolic equation,
where kpol is the maximum rate of product formation, KDdNTP is the dissociation constant for dNTP binding to the enzyme-DNA binary complex and [dNTP] is the concentration of dNTP added.
Crystallization and Structure Determination
Complexes were prepared at room temperature by combining 200 μm DbhDpo4RKSDbh (Fig. 1) and 240 μm DNA (X-4T-G substrate, Table 1) in 25 mm HEPES (pH 7.0), 5 mm Ca(OAc)2, 85 mm NaCl, 1 mm DTT, and 1 mm dCTP (final concentrations). Catalysis was prevented by including Ca2+ as the divalent metal ion instead of Mg2+. Crystals were grown at room temperature by hanging-drop vapor diffusion by mixing equal volumes of complex and well solution containing 14% PEG-3350, 100 mm MES-Tris (pH 6.0), 100 mm Ca(OAc)2, 2.5% glycerol, and 250 mm sucrose. Crystals were stabilized and cryoprotected by the addition of a solution containing 20% PEG-3350, 100 mm MES-Tris (pH 6.5), 100 mm Ca(OAc)2, 20% w/v sucrose, and 1 mm dCTP. Crystals were flash-cooled in liquid nitrogen. X-ray diffraction data were collected at Brookhaven National Laboratory (BNL), National Synchrotron Light Source (NSLS) beamline X25, and were processed and scaled using HKL2000 (18). The structure was solved by molecular replacement using the Dbh-Dpo4-Dbh ternary complex (PDB 4F4Y (15)) as a search model and was refined using PHENIX (19), alternating with cycles of manual rebuilding in COOT (20). The geometry of the DNA was analyzed using Curves+ (21), and protein conformations were analyzed using DynDom (22). Structure figures were made using PyMOL (version 1.5.0.4; Schrödinger, LLC). Refined coordinates and structure factors have been deposited as PDB ID code 4NLG.
RESULTS
Linker Identity Determines −1 Deletion Frequency on a Repetitive Sequence
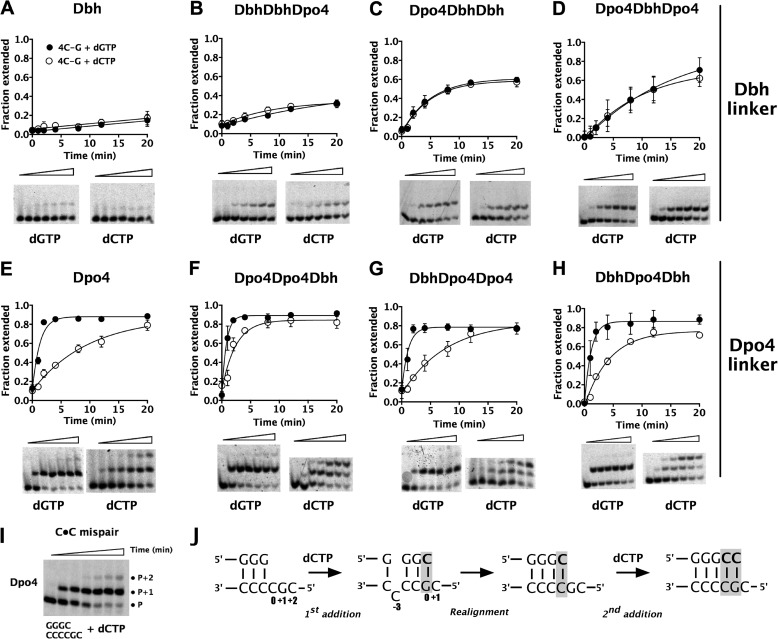
Both Dbh and Dpo4 generate single-base deletions on repetitive sequences using a template-slippage mechanism, but they do so at different frequencies (12–14), with Dbh being substantially more error-prone in this respect. To determine whether the linker is the major determinant of this characteristic, we performed single-nucleotide incorporation assays on a DNA substrate with a sequence that has previously been shown to be a deletion hotspot (3′-CCCCG-5′ in the template strand) for not only Dbh and Dpo4, but also other DinB homologues (13, 14, 23, 24). Primer extension assays using the 4C-G substrate (Table 1) included either dGTP or dCTP as the incoming nucleotide, to give either correct extension (dGTP) or to initiate a single-base deletion (dCTP). Nucleotide incorporation rates were determined for both parental enzymes and for six chimeras constructed from all possible combinations of the polymerase domain, 15-amino acid linker and LF/PAD (Fig. 2, A–H, and Table 2). Chimeric polymerases are named by the source enzyme in the order: polymerase core-linker-LF/PAD.
FIGURE 2.
Single-base deletion efficiency is dependent on the identity of the linker. A–H, primer extension by Dbh (A), Dpo4 (E), and the chimeric polymerases (Dbh linker, B--D; Dpo4 linker, F–H) on the 4C-G substrate, which contains the repetitive hotspot sequence (Table 1). Incorporation of dGTP (filled circles) shows the correct product formation whereas dCTP addition (open circles) initiates a single-base deletion. 40 nm annealed primer-template DNA was preincubated with 4 μm protein before 1 mm appropriate nucleotide was added to start the reaction. All concentrations given are final. Reactions were quenched after varying time intervals. Lower panels show primer-extension products over time (0–20 min) for each of the nucleotides. I, gel showing efficient mispair extension by Dpo4 on the hotspot sequence with a C-C mispair at the primer template junction (sequence shown below gel), through dCTP incorporation. J, schematic representation of possible mechanism of mispair formation and extension.
TABLE 2.
Summary of observed nucleotide incorporation rates for data shown in Fig. 2, using the 4C-G substrate
| Polymerase | dGTP (correct) k1 (min−1) | dCTP (incorrect) k1 (min−1) | k1dG/k1dC |
|---|---|---|---|
| Dbh | ∼0.02a | ∼0.01a | |
| DbhDbhDpo4 | 0.04 ± 0.02 | 0.11 ± 0.02 | 0.36 |
| Dpo4DbhDbh | 0.20 ± 0.04 | 0.21 ± 0.03 | 0.95 |
| Dpo4DbhDpo4 | 0.06 ± 0.01 | 0.10 ± 0.01 | 0.60 |
| Dpo4 | 0.77 ± 0.20 | 0.10 ± 0.02 | 7.70 |
| Dpo4Dpo4Dbh | 1.29 ± 0.09 | 0.40 ± 0.11 | 3.23 |
| DbhDpo4Dpo4 | 0.92 ± 0.21 | 0.13 ± 0.03 | 7.08 |
| DbhDpo4Dbh | 0.88 ± 0.09 | 0.23 ± 0.05 | 3.83 |
a These values are only approximate because of the large errors associated with these measurements.
We find that the rates of correct and incorrect nucleotide incorporation are dependent on the identity of the linker. As we have observed previously (16, 17), Dbh incorporates both nucleotides (correct, dG and incorrect, dC) at near equal rates (Fig. 2A) whereas Dpo4 adds the correct nucleotide 8-fold faster than the incorrect nucleotide (Fig. 2E and Table 2). The ratios of incorrect to correct nucleotide incorporation rates are consistent with Dbh having a higher single-base deletion frequency than Dpo4. Similarly, all chimeras containing the Dpo4 linker add the correct dG nucleotide 3–8-fold faster than the incorrect dC (Fig. 2, F–H, and Table 2) whereas the chimeras containing the Dbh linker incorporate dG up to 3-fold more slowly than dC (Fig. 2, B–D, and Table 2). Thus, the linker is a major determinant of single-base deletion activity.
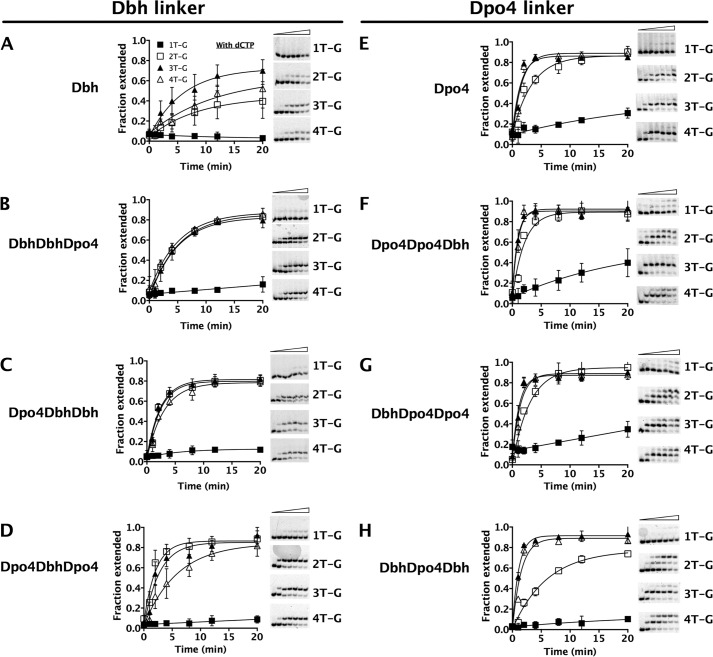
Using variations on the deletion hotspot sequence, with a T individually substituting for each C (Fig. 3 and Table 1, substrates 1T-G, 2T-G, 3T-G and 4T-G), we confirmed that all of the chimeras predominantly use a template-slippage deletion mechanism, as do the parental enzymes (16, 17, 25). Because the deletion hotspot sequence can adopt multiple conformations, with each of the Cs potentially being unpaired, these variant substrates are designed to favor an unpaired base at a single position, as shown in Table 1. The 1T-G substrate is used least efficiently in all cases (Fig. 3), ruling out both dNTP-stabilized misalignment and misincorporation-misalignment as the major mechanism by which single-base deletions are made. The 3T-G and 4T-G substrates are used most efficiently by all of the enzymes (Fig. 3), indicating use of a template-slippage mechanism with a preference for the unpaired base being 3 or 4 nucleotides upstream of the templating base. In fact, incorporation of dCTP on the 3T-G and 4T-G substrates by each polymerase is at least as rapid as the incorporation of the correct dGTP on the 4C-G substrate (Figs. 2 and 3).
FIGURE 3.
Chimeric polymerases use a template-slippage deletion mechanism. A–H, incorporation of dCTP by Dbh, Dpo4, and chimeras on primer-template DNA containing modified hotspot sequences (1T-G, 2T-G, 3T-G and 4T-G; Table 1). Fraction of primer DNA extended, as a function of time, is displayed graphically for the different DNA substrates shown: 1T-G (filled squares), 2T-G (open squares), 3T-G (filled triangles), and 4T-G (open triangles). The 2T-G, 3T-G, and 4T-G substrates are designed to favor an unpaired base at positions 2, 3, and 4 nucleotides 3′ of the templating base, mimicking the multiple conformations that can occur during template slippage. The 1T-G substrate is designed to inhibit slippage and mimic the substrate present if either a dNTP-stabilized misalignment or a misincorporation-misalignment mechanism was used to create deletions in repetitive sequences. The extremely slow nucleotide incorporation on the 1T-G substrate compared with the 2T-G, 3T-G, and 4T-G substrates indicates template slippage as the major deletion mechanism.
Linker Identity Determines Efficiency of Mispair Extension
Another difference among the polymerases is evident on inspection of the pattern of product formation: the enzymes containing the Dpo4 linker tend to efficiently incorporate a second dCTP (Fig. 2, E–H, and Fig. 3, E–H), but those containing the Dbh linker do not (Fig. 2, A–D, and Fig. 3, A–D). Because the polymerases use a template-slippage deletion mechanism, misincorporation of the first dC nucleotide occurs when the incoming dCTP pairs with the +1G as templating base in the hotspot sequence, skipping over one of the Cs in the template strand (Fig. 2J). We suspected that the second nucleotide added is likely to be templated by the same G, after isomerization of the primer-template DNA to form a C-C (or C-T) mispair at the primer terminus (Fig. 2J). To test this idea, we examined the ability of Dpo4 to extend from a C-C mispair substrate (Table 1), which would be the substrate for addition of the second nucleotide, and found that it can efficiently add dC to this primer-template junction (Fig. 2I). Altogether, these results suggest that Dpo4 and the chimeras containing the Dpo4 linker have a greater ability to realign slipped DNA and extend from a mispair than do those that contain the Dbh linker.
Amino Acid Trio in the Linker Determines Overall Polymerase Conformation
We have shown previously that the key determinant of the polymerase conformation for Dbh and Dpo4 is the interdomain linker (15). Furthermore, only 3 of the 15 amino acids in the linker, residues 242–244 (Arg-Lys-Ser) in Dpo4 and 243–245 (Lys-Ile-Pro) in Dbh (Fig. 1), are responsible for the base-substitution and abasic-site bypass properties of the two polymerases, suggesting that those 3 amino acids might be sufficient to control the polymerase conformation.
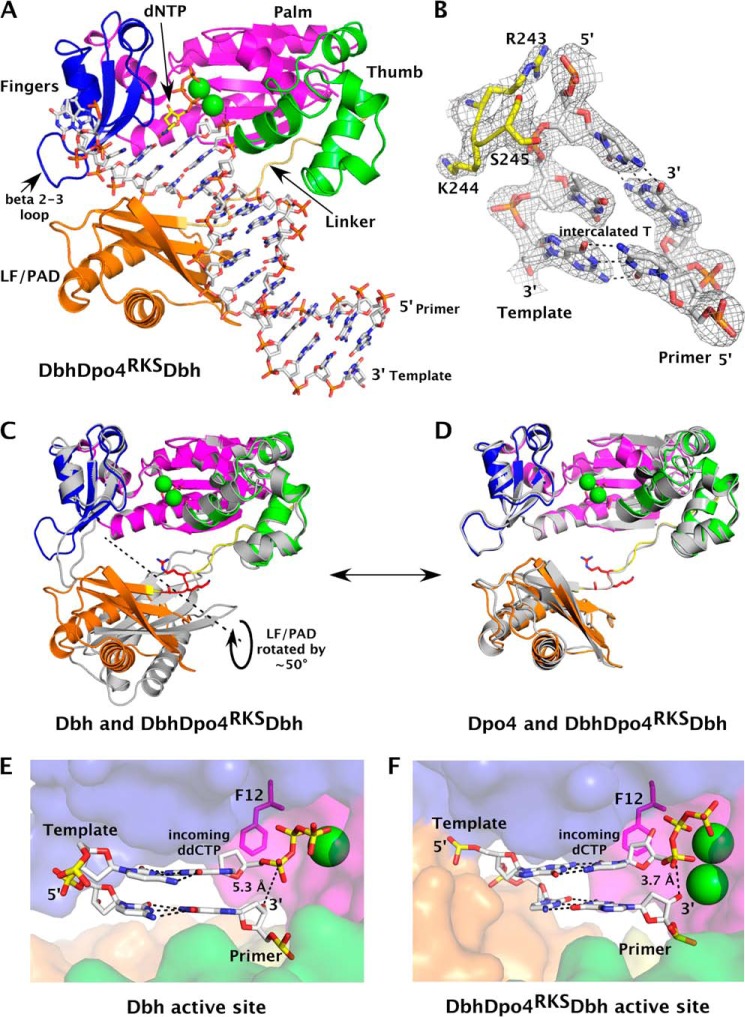
To test this hypothesis, we determined a 2.4 Å ternary complex crystal structure of Dbh containing just the three linker residues from Dpo4 (Fig. 4 and Table 3). Here we refer to this chimeric polymerase as DbhDpo4RKSDbh. We were unable to crystallize the complementary chimera, Dpo4DbhKIPDpo4. In the crystal structure, an incoming dCTP is correctly paired to the G in the deletion hotspot sequence (Fig. 4A), and the template contains an unpaired T four nucleotides 3′ to the templating G (Fig. 4B). The template slippage, enforced by the T replacing a C in the deletion hotspot sequence, allows the incorrect incoming nucleotide to form a standard Watson-Crick pair. DbhDpo4RKSDbh superimposes on Dbh (PDB code 3BQ1 (16)) with a r.m.s.d. of 5.08 Å (Fig. 4C), even though the polymerase and LF/PAD domains individually align with r.m.s.d. values of 1.62 and 0.5 Å, respectively. In contrast, DbhDpo4RKSDbh superimposes on Dpo4 (PDB code 3QZ7 (17)) with an overall r.m.s.d. of 1.34 Å (Fig. 4D).
FIGURE 4.
DbhDpo4RKSDbh adopts a Dpo4-like conformation. A, ternary complex of DbhDpo4RKSDbh chimeric polymerase is shown with domains color-coded as in Fig. 1 (linker is shown in yellow-orange). Both DNA substrate (white) and incoming dCTP (yellow) are shown as sticks, and the divalent Ca2+ ions are shown as green spheres. B, primer-template DNA around the intercalated −4T nucleotide is shown (white). Linker residues that are of Dpo4 origin (Arg243-Lys244-Ser245) in the chimera are shown in yellow, and a 2Fo − Fc map calculated from the final refined structure is shown contoured at 1.5 σ (gray mesh). C and D, superposition of DbhDpo4RKSDbh (color-coded as in A) on the polymerase core of Dbh (PDB ID code 3BQ1, chain A) (C) and Dpo4 (PDB ID code 3QZ7, chain A) (D) is shown. Parental enzymes are shown in gray. The LF/PAD of the chimeric polymerase would need to undergo a ∼50° rotation (22) compared with Dbh to adopt the Dpo4-like domain orientation. Linker residues that are of Dpo4 origin in the chimeric Dbh polymerase are shown in red. Carboxylates in the chimera palm are shown (magenta sticks). E and F, close up views show Dbh (E) and DbhDpo4RKSDbh (F) active site with domains colored as before. The steric gate residue (Phe12) is shown as sticks to reflect the stacking interaction established when substrates are positioned correctly in DbhDpo4RKSDbh, allowing selection against ribonucleotides.
TABLE 3.
Data collection and refinement statistics
| Ternary complex of | |
| Polymerase | DbhDpo4RKSDbh |
| DNA substrate | X-4T-G |
| Incoming nucleotide | dCTP |
| PDB ID | 4NLG |
| Data collection | |
| Beamline | X25 |
| Wavelength (Å) | 1.1 |
| Resolution range (Å) | 30.0–2.4 (2.43–2.40) |
| Space group | C 2 |
| Unit cell dimensions | |
| a,b,c (Å) | 120.19, 103.07, 53.05 |
| β (°) | 105.4 |
| Unique reflections | 23,003 (1,042) |
| Multiplicity | 2.7 (1.9) |
| Completeness (%) | 91.7 (80.7) |
| R-merge (%) | 4.4 (19.1) |
| Mean I/σ(I) | 23.2 (2.9) |
| Refinement statistics | |
| Resolution (Å) | 28.7–2.4 |
| Reflections | 22,579 |
| R-work (%) | 17.4 |
| R-free (%) | 23.1 |
| Number of nonhydrogen atoms | 3,822 |
| Number of | |
| Protein residues | 344 |
| Nucleotides (including dCTP) | 32 |
| Ca2+ ions | 2 |
| Water | 41 |
| r.m.s. (bonds) | 0.011 |
| r.m.s. (angles) | 1.19 |
| Ramachandran region | |
| Most favored (%) | 90.6 |
| Additional allowed (%) | 8.7 |
| Generously allowed (%) | 0.6 |
| Disallowed (%) | 0.0 |
| Average B-factor | 69.4 |
| Macromolecules | 69.8 |
| Ligands | 45 |
| Solvent | 53.2 |
Thus, the three amino acids are indeed sufficient to allow the Dbh LF/PAD to adopt the same conformation as in structures of Dpo4. With the polymerase domains superimposed, a rotation of ∼50° around an axis roughly parallel to the helical axis of the DNA would be required to bring the LF/PAD domain of Dbh into alignment with that of DbhDpo4RKSDbh (Fig. 4C). In the structure of DbhDpo4RKSDbh, the LF/PAD is positioned so that it is in contact with the β2–3 loop in the fingers domain, causing this loop to become ordered. This contrasts with the β2–3 loop being poorly ordered or completely disordered in all published Dbh structures (16, 26, 27).
Contact between the LF/PAD and β2–3 loop brings the substrates into alignment at the active site for catalysis, by constraining the width of the nascent base pair binding pocket. This is consistent with the DbhDpo4Dbh chimera adding a C very efficiently to the 4T-G variant of the deletion hotspot sequence (Fig. 3H). Compared with the Dbh ternary complex structure (Fig. 4E), the chimera (Fig. 4F) has two metal ions bound at the active site and the ribose of the dNTP sits directly on top of the steric gate residue (Phe12). Additionally the α-phosphate of the nucleotide is 3.7 Å from the 3′-OH of the primer terminus, compared with 5.3 Å in the parental Dbh structure. Thus, the active site in this structure has the characteristics associated with correct dNTP incorporation.
Interestingly, the unpaired T in the template DNA is not in an extrahelical conformation. Instead, it intercalates into the DNA duplex (Fig. 4B), causing a tilt of 18° between the flanking base pairs. In our previous structures of chimeric polymerases containing this same primer-template DNA, the unpaired T was also intercalated into the duplex when bound to the DbhDpo4Dbh chimera, whereas it was in an extrahelical conformation when bound to the DbhDpo4Dpo4 chimera (15).
dNTP Selection: Linker-dependent Alteration of Nucleotide Incorporation Rate
Next, we wanted to understand how the enzyme conformation conferred by the Dbh and Dpo4 linkers influence the steps involved in nucleotide incorporation. To investigate this we performed single-turnover primer extension assays using the 4C-G substrate (Table 1) to determine the nucleotide binding affinity (KDdNTP) and rate of polymerization (kpol) for the parental proteins, Dbh and Dpo4, as well as for the two chimeras, DbhDpo4RKSDbh and Dpo4DbhKIPDpo4. The results are summarized in Table 4.
TABLE 4.
Kinetic parameters for correct and incorrect nucleotide incorporation on a deletion hotspot sequence (substrate 4C-G)
| Polymerase | dGTP (correct) |
dCTP (incorrect) |
KDdGTP/KDdCTP | kpoldGTP/kpoldCTP | Fidelityb | ||||
|---|---|---|---|---|---|---|---|---|---|
| KDdNTP (μm) | kpol (min−1) | Efficiencya (×10−3) (μm−1min−1) | KDdNTP (μm) | kpol (min−1) | Efficiencya (×10−3) (μm−1min−1) | ||||
| Dbh | 822 ± 397 | 0.17 ± 0.04 | 0.21 | 1135 ± 367 | 0.17 ± 0.03 | 0.15 | 0.7 | 1 | 1.4 |
| Dpo4DbhKIP Dpo4 | 92 ± 29 | 1.9 ± 0.13 | 20.8 | 818 ± 198 | 2.85 ± 0.31 | 3.5 | 0.1 | 0.67 | 6 |
| Dpo4 | 199 ± 49 | 20.4 ± 2.2 | 102 | 893 ± 121 | 2.39 ± 0.1 | 2.6 | 0.2 | 8.5 | 39 |
| DbhDpo4RKS Dbh | 74 ± 29 | 16.5 ± 1.9 | 222 | 222 ± 84 | 1.17 ± 0.13 | 5.3 | 0.3 | 14 | 42 |
a Efficiency = kpol/KDdNTP.
b Fidelity = efficiency dGTP/efficiency dCTP.
Comparison of the four polymerases shows that the linker residues strongly influence the overall fidelity of the enzyme. Dpo4 fidelity is reduced by 6.5-fold (from 39 to 6) when Dbh residues are present in the linker of Dpo4 (Dpo4DbhKIPDpo4). Conversely, Dpo4 residues in the linker of Dbh (DbhDpo4RKSDbh) increase fidelity of Dbh by ∼30-fold (from 1.4 to 42).
The major contribution to fidelity comes from differences in the maximum rate of nucleotide incorporation, kpol (Table 4). Comparing the rates of correct versus incorrect nucleotide incorporation (kpoldGTP/kpoldCTP), the Dpo4 linker causes a 14-fold increase in the selectivity of nucleotide incorporation (1 versus 14), whereas the Dbh linker causes a 13-fold decrease (8.5 versus 0.67). Thus, the linker-dependent change in nucleotide incorporation rates contributes to the differences in single-base deletion fidelity, and this occurs by substantially influencing the rate of correct, but not incorrect, nucleotide incorporation (Table 4).
Nucleotide binding affinity also contributes to fidelity, but in a more complex way (Table 4). The ratio of the dissociation constants for correct and incorrect nucleotides (KDdGTP/KDdCTP) is decreased by ∼2-fold for both the Dbh and Dpo4 linkers (0.7 versus 0.3 for Dbh versus DbhDpo4RKSDbh; 0.2 versus 0.1 for Dpo4 versus Dpo4DbhKIPDpo4). Because increased affinity results in a lower KD, the linker sequences increase the preference of the enzyme for binding the correct nucleotide, which contributes to a modest increase in fidelity for both polymerases.
Overall, the linker-dependent changes to nucleotide binding and incorporation rate act in the same direction with the Dpo4 linker, increasing fidelity, but act in opposite directions with the Dbh linker, increasing the fidelity based on nucleotide binding but decreasing the fidelity based on nucleotide incorporation rate. It also appears that residues from the polymerase and/or LF/PAD domains contribute to nucleotide binding affinity, but this is difficult to dissect because of the large errors in measuring weak nucleotide binding.
DISCUSSION
Using two closely related Y-family DNA polymerases from Sulfolobus and their chimeric counterparts, we show here that three residues in the interdomain linker have a major impact on the replication of a repetitive sequence that is a deletion hotspot, changing the single-base deletion fidelity by up to 30-fold. The linker identity correlates with the maximal rate of correct versus incorrect nucleotide incorporation but not with nucleotide binding affinity (Table 4). On the deletion hotspot sequence, enzymes with the Dbh linker catalyze addition of both dGTP (the correct nucleotide) and dCTP (the nucleotide that initiates a single-base deletion) at approximately the same rates, whereas enzymes with the Dpo4 linker have a distinct preference for incorporating the correct nucleotide. Significantly, the rates of incorrect nucleotide incorporation do not vary much between the enzymes (either parent or chimera). Thus, the linker specifically alters the rate of correct nucleotide addition, which accounts for the changes in single-base deletion fidelity.
A correlation between polymerase fidelity and the efficiency of correct, but not incorrect, nucleotide incorporation has been noted previously for a wide range of high and low fidelity polymerases (28). Interestingly, the linker sequences affect single-base deletion and base-substitution fidelity in opposite directions: the Dpo4 linker increases the base- substitution frequency (15) but decreases the single-base deletion fidelity (Fig. 2) whereas the Dbh linker does the opposite.
Information about the identity of the linker sequences is propagated to the active site via the overall conformation of the polymerase. The structure of DbhDpo4RKSDbh demonstrates that replacing three amino acids in the Dbh linker with the equivalent residues from Dpo4 allows the Dbh LF/PAD to move into a position where it contacts the β2–3 loop of the catalytic domain (Fig. 4, C and D). This is in contrast to structures of Dbh that show a gap between the two domains at this location. Contacts between the two domains tightly constrain the width of the nascent base pair binding pocket. This brings the primer terminus and incoming nucleotide into alignment at the active site (Fig. 4, E and F), providing an explanation for the increased rate of correct nucleotide incorporation in enzymes containing the Dpo4 linker. Even though the crystallized substrate contains a misaligned primer-template, the unpaired base is located at a position where nucleotide incorporation occurs at a rate as fast as on correctly aligned primer-template with the correct incoming nucleotide.
The ability to extend from a C-C mismatched primer terminus, which is enhanced by the Dpo4 linker sequences, is comparable with what has been reported for human polymerase κ (3, 29). For pol κ, extension from the mismatch involves realigning the slipped DNA strands (29), and we presume that this is the case here. It remains to be determined whether the realignment is actively performed by the polymerase or occurs due to transient unpairing of the template DNA from the primer terminus. In either case, realignment and mispair extension abilities act to reduce the frequency of deletion mutations at the expense of increasing base-substitution mutations.
The results reported here, combined with our previous data (15), show that base-substitution frequency, single-base deletion frequency, mispair extension, and abasic-site bypass are all strongly affected by the linker-dependent conformation of the polymerase. These observations have two significant implications. First, they highlight how different substrate specificities can evolve as a result of just a few amino acid changes remote from the site of catalysis. The Y-family polymerases display an extraordinary variety of lesion bypass and mutagenic activities. It is easy to envision how these enzymes could readily diverge if, after a gene duplication event, just a few mutations could produce a polymerase with a new specificity that provides a selective advantage. Second, they suggest that Y-family polymerase fidelity and specificity could be regulated by altering polymerase conformation. This could, for example, be the mechanism by which the single-base deletion activity of Escherichia coli DNA polymerase IV (DinB) is suppressed when forming a complex with UmuD and RecA (30). Thus, it will be interesting to study the influence of protein partners on both the activity and structural conformation of TLS polymerases, which may shed light on the mechanism of regulating these polymerases in the cell.
Acknowledgments
We thank Jessica Olson for assistance in the early stages of this project and Joachim Jaeger for helpful advice throughout.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM080573 (to J. D. P.). This work was also supported by fellowship support from the Wadsworth Center (to P. M. and I. L.). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
The atomic coordinates and structure factors (code 4NLG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TLS
- translesion synthesis
- FAM
- 5′-6-carboxyfluorescein
- LF/PAD
- little finger/polymerase-associated domain
- PDB
- Protein Data Bank
- pol
- polymerase
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Sale J. E., Lehmann A. R., Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohmori H., Friedberg E. C., Fuchs R. P., Goodman M. F., Hanaoka F., Hinkle D., Kunkel T. A., Lawrence C. W., Livneh Z., Nohmi T., Prakash L., Prakash S., Todo T., Walker G. C., Wang Z., Woodgate R. (2001) The Y-family of DNA polymerases. Mol. Cell 8, 7–8 [DOI] [PubMed] [Google Scholar]
- 3. Carlson K. D., Johnson R. E., Prakash L., Prakash S., Washington M. T. (2006) Human DNA polymerase κ forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc. Natl. Acad. Sci. U.S.A. 103, 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson R. E., Prakash S., Prakash L. (2000) The human DINB1 gene encodes the DNA polymerase pol θ. Proc. Natl. Acad. Sci. U.S.A. 97, 3838–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson R. E., Washington M. T., Haracska L., Prakash S., Prakash L. (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 6. Nair D. T., Johnson R. E., Prakash S., Prakash L., Aggarwal A. K. (2004) Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 [DOI] [PubMed] [Google Scholar]
- 7. Washington M. T., Johnson R. E., Prakash S., Prakash L. (2000) Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 97, 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoon J. H., Prakash L., Prakash S. (2009) Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pata J. D. (2010) Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta 1804, 1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ling H., Boudsocq F., Woodgate R., Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 11. Trincao J., Johnson R. E., Escalante C. R., Prakash S., Prakash L., Aggarwal A. K. (2001) Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8, 417–426 [DOI] [PubMed] [Google Scholar]
- 12. Boudsocq F., Kokoska R. J., Plosky B. S., Vaisman A., Ling H., Kunkel T. A., Yang W., Woodgate R. (2004) Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 279, 32932–32940 [DOI] [PubMed] [Google Scholar]
- 13. Kokoska R. J., Bebenek K., Boudsocq F., Woodgate R., Kunkel T. A. (2002) Low fidelity DNA synthesis by a Y-family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 277, 19633–19638 [DOI] [PubMed] [Google Scholar]
- 14. Potapova O., Grindley N. D., Joyce C. M. (2002) The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem. 277, 28157–28166 [DOI] [PubMed] [Google Scholar]
- 15. Wilson R. C., Jackson M. A., Pata J. D. (2013) Y-family polymerase conformation is a major determinant of fidelity and translesion specificity. Structure 21, 20–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson R. C., Pata J. D. (2008) Structural insights into the generation of single-base deletions by the Y family DNA polymerase Dbh. Mol. Cell 29, 767–779 [DOI] [PubMed] [Google Scholar]
- 17. Wu Y., Wilson R. C., Pata J. D. (2011) The Y-family DNA polymerase Dpo4 uses a template slippage mechanism to create single-base deletions. J. Bacteriol. 193, 2630–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lavery R., Moakher M., Maddocks J. H., Petkeviciute D., Zakrzewska K. (2009) Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayward S., Kitao A., Berendsen H. J. (1997) Model-free methods of analyzing domain motions in proteins from simulation: a comparison of normal mode analysis and molecular dynamics simulation of lysozyme. Proteins 27, 425–437 [DOI] [PubMed] [Google Scholar]
- 23. Ohashi E., Bebenek K., Matsuda T., Feaver W. J., Gerlach V. L., Friedberg E. C., Ohmori H., Kunkel T. A. (2000) Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem. 275, 39678–39684 [DOI] [PubMed] [Google Scholar]
- 24. Foti J. J., Walker G. C. (2011) Efficient extension of slipped DNA intermediates by DinB is required to escape primer template realignment by DnaQ. J. Bacteriol. 193, 2637–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeLucia A. M., Grindley N. D., Joyce C. M. (2007) Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry 46, 10790–10803 [DOI] [PubMed] [Google Scholar]
- 26. Zhou B. L., Pata J. D., Steitz T. A. (2001) Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8, 427–437 [DOI] [PubMed] [Google Scholar]
- 27. Silvian L. F., Toth E. A., Pham P., Goodman M. F., Ellenberger T. (2001) Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 8, 984–989 [DOI] [PubMed] [Google Scholar]
- 28. Beard W. A., Shock D. D., Vande Berg B. J., Wilson S. H. (2002) Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 277, 47393–47398 [DOI] [PubMed] [Google Scholar]
- 29. Mukherjee P., Lahiri I., Pata J. D. (2013) Human polymerase κ uses a template-slippage deletion mechanism, but can realign the slipped strands to favour base substitution mutations over deletions. Nucleic Acids Res. 41, 5024–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Godoy V. G., Jarosz D. F., Simon S. M., Abyzov A., Ilyin V., Walker G. C. (2007) UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28, 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]