Background: Fut7 gene expression is tightly regulated and controls leukocyte traffic.
Results: A 12.7 kb genomic interval controls Fut7 gene expression in all cell types.
Conclusion: This interval therefore represents the functional boundaries of the Fut7 locus.
Significance: All cis-acting genetic elements that control Fut7 expression are within this genomic interval.
Keywords: Cell Adhesion, Gene Expression, Glycobiology, Glycosyltransferases, Immunology, Leukocyte, Transcription Regulation
Abstract
Fut7 encodes an α1,3-fucosyltransferase critical for biosynthesis of glycan ligands for all three selectins. Consistent with this function, Fut7 expression is limited to hematopoietic cells and high endothelial cells which express selectin ligands. Mechanisms that govern Fut7 expression are poorly defined. To begin to understand the molecular genetic basis for transcriptional regulation of Fut7, a transgenic, gain-of-function, genetic complementation approach in mice was used to define the “functional boundaries” of the murine Fut7 locus, defined here as any uninterupted stretch of genomic DNA that contains all cis-acting genetic elements essential for accurate physiologic expression. A 12.7-kb contiguous genomic interval, which lies completely between the highly conserved flanking Npdc1 and Abca2 loci on chromosome 2 and which contains the complete transcriptional unit plus ∼7.4 kb upstream of the transcriptional start site and ∼2 kb downstream of the transcriptional termination and polyadenylation sites, was used as a transgene (Tg) on a Fut7 null background. Tg+ mice exhibited restoration of Fut7 gene expression and physiologic levels of selectin ligand expression and function on neutrophils, activated T cells, and high endothelial cells and corrected the functional defects in these cells found in Fut7 null mice without leading to detectable expression of Fut7 in normally non-expressing tissues. These results demonstrate that all genetic information essential for appropriate and selective expression of Fut7 in diverse cell types and in response to distinct developmental signals is contained within this comparatively small genetic region.
Introduction
The selectins are a family of three carbohydrate binding adhesion molecules that play critical roles in leukocyte traffic both during inflammation and under normal homeostasis. Work by numerous investigators has definitively shown that L-selectin is essential for normal lymphocyte recirculation by mediating binding of blood-borne lymphocytes to high endothelial cells (HEC)2 of postcapillary venules of secondary lymphoid organs (1–4). E- and P-selectin are crucial for homing of hematopoietic stem cells to their niches in bone marrow (5, 6) as well as migration of dendritic cells into skin (7). All three selectins participate in recruitment of neutrophils, monocytes, and dendritic and T cells to sites of inflammation (8–15).
The α1,3 fucosyltransferase FucT-VII, encoded by Fut7, is well established as critical for the biosynthesis of ligands for all three selectins on all cell types on which these ligands are expressed, including L-selectin ligands on HEC and E- and P-selectin ligands on all classes of leukocytes (16–18). Fut7 gene expression is found in all types of blood-borne mature myeloid cells, including monocytes, dendritic cells, and neutrophils, in activated but not naive T cells (19, 20), in IgG plasma cells (21), and in HEC of lymph nodes and Peyer's patches (18), mirroring expression of selectin ligands on these cell types. Fut7 expression is not detected in normal non-malignant epithelial or mesenchymal cells or tissues. The restricted expression of Fut7 to cell types that express glycan ligands for one or more selectins and the absence in Fut7 null mice of any phenotypes unrelated to defects in selectin ligand expression strongly suggests that this enzyme is specialized for selectin ligand formation.
Despite the critical importance of FucT-VII, little is known regarding transcriptional regulation of Fut7. In murine CD4+ T cells, T-bet expression is essential for maximal, IL-12-driven expression of Fut7 (22). Experiments in Jurkat T cells indicated that Fut7 promoter activity is positively regulated by a complex of T-bet, Sp1, and p300 (23). Fut7 is also transcriptionally induced by the HTLV-1 tax protein (24) via a mechanism involving CREB (cAMP-response element (CRE)-binding protein)/ATF proteins, possibly CREB1 and/or CREMα, binding with tax and CBP (CREB-binding protein) to a CRE element, which is close to the T-bet and Sp1 binding sites (23, 25). In contrast, Gata3, whose expression is driven by IL-4, repressed Fut7 promoter activity via recruitment of HDACs and interference with T-bet binding (23), which may account for the ability of IL-4 to inhibit Fut7 expression (26). However, the activities of these transcription factors cannot fully explain the pattern of expression of Fut7 even in T cells, and no transcription factors that control Fut7 expression in myeloid cells or in HEC have been identified. Moreover, no cis-acting genetic elements that govern Fut7 expression, apart from the promoter, have been characterized.
A detailed understanding of transcriptional mechanisms that govern Fut7 expression requires, among other things, comprehensive identification of cis-acting elements that are responsible for expression or repression of Fut7 in different cell types. The most stringent and definitive test to determine if all essential cis-acting regulatory regions of a gene are contained within a given sequence of genomic DNA is the ability of that DNA, in the form of a transgene (Tg), to restore physiologically accurate expression and appropriate function in mice which harbor homozygous null mutations in the endogenous gene of interest. Therefore, a transgenic, gain-of-function, genetic complementation approach in mice was used to define the functional boundaries of the murine Fut7 locus. The results show that a remarkably small contiguous genomic interval contains all genetic information essential for the correct physiologic pattern of expression of Fut7.
EXPERIMENTAL PROCEDURES
Mice
C57BL6/J WT mice were obtained from Jackson Laboratory and maintained and bred in a barrier facility at Northwestern University according to standard protocols. Mice with homozygous null mutations in Fut7 (16) were obtained from Dr. John Lowe, University of Michigan, Ann Arbor, MI, and were backcrossed >10 generations to C57BL6/J. Tg mice were produced using standard approaches by microinjecting SN1 Tg DNA (described below) into Fut7 null fertilized eggs. This approach maintains the isogenic strain background and obviated any need for intercrossing SN1 Tg+ mice with Fut7 null mice. Founder pups were identified by PCR using three different primer pairs. The first pair (fut7 Ben primers) is contained completely within the single large exon encoding the catalytic domain (see Fig. 1), which is largely absent in the fut7 null allele (16) and gives a band of 476 bp on WT or Tg+ but not Fut7 null mice. The second primer pair (SN1 primers) is unique to the Tg as a result of non-genomic plasmid sequence contained within the sense primer, with the antisense primer within the Fut7 locus; this primer pair gives a band of 434 bp only from Tg+ mice but not WT or Fut7 null mice. The third primer pair (neo primers) is completely within the neo cassette originally used to disrupt the Fut7 locus to produce Fut7 null mice (16) and gives a 248-bp band from Tg+ and Fut7 null mice but not from WT mice. All primer sequences for this entire study are given in Table 1, and their locations (for those that amplify Fut7 sequences) are depicted in Fig. 1. Tg+ founders, which by definition are homozygous for the endogenous Fut7 null allele (because they were produced with fut7 null eggs), were mated with (non-Tg) Fut7 null mice, and Tg+ pups were identified as above. Mice derived from two independently derived founders, designated SN1.1 and SN1.2, were produced and analyzed in this study.
FIGURE 1.
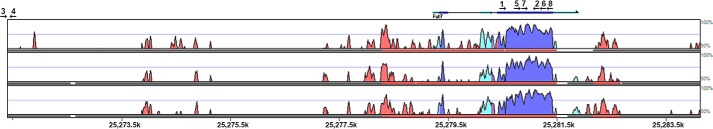
Structure and conservation of the Fut7 locus and the SN1 Tg. Shown is a Vista-derived diagram of the genomic interval used as the SN1 Tg. Using the mouse as the base genome (not pictured), the conservation of the mouse with three other mammalian species (human, dog, horse, top to bottom) is depicted. The left (5′) edge of the figure represents the location of the SpeI site, and the right (3′) edge of the figure represents the location of the NotI site, initially used to excise this genomic interval from the original BAC clone. The major Fut7 transcriptional unit in the mouse is depicted at the top, aligned with the other species. The location of the major transcriptional start site (TSS) in the mouse is indicated as the left edge of the transcript at bp 7401 of the 12,729-bp SN1 Tg. Evolutionary conservation of the mouse with each of the indicated species is indicated by peaks, with colors indicating the nature of the sequence: dark blue/purple, coding; light blue, 5′- or 3′-untranslated sequence; pink, non-coding, i.e. conserved non-coding sequence. The scale goes from 50 to 100%, with 100% representing complete identity at the nucleotide level. Note that both coding and non-coding sequences of the murine mRNA are highly conserved in the other species, as are nearly all of the conserved noncoding sequences. Genomic position numbers for the mouse on chromosome 2 are indicated at the bottom. Oligonucleotides used for genotyping and other analysis are indicated by arrows, which give their orientation and numbers, which are in Table 1. Oligonucleotide 3, the SN1 sense oligo, is not part of native genomic DNA and is therefore “off scale” left. See “Results” for further details.
TABLE 1.
Oligonucleotide primers used in this study
| Numbera | Name | Sequence | Use |
|---|---|---|---|
| 1 | fut7 Ben sense | GTCTGGGCCTCCATGGAATCGCCCAG | Genotyping, Fig. 6 |
| 2 | fut7 Ben antisense | TCCACGTGTACAAAGGCATCTGGTGGC | |
| 3 | SN1 sense | ATAAGCTTGATATCGAATTCCTGCAGC | Genotyping |
| 4 | SN1 antisense | ACATGCAACTTGTCTCAAAGAAAACTGA | |
| Neo sense | CTTGGGTGGAGAGGCTATTC | Genotyping | |
| Neo antisense | AGGTGAGATGACAGGAGATC | ||
| 5 | fut7-1 sense | GTACCGCTTCTACCTGGCCTTTGAG | Figs. 2, 3B, and 4B |
| 6 | fut7-1 antisense | ATAACGACTCTCATTCATGCTGAC | |
| 7 | fut7-2 sense | AACTCACAGCATCGGGACTACATC | Fig. 2 |
| 8 | fut7-2 antisense | CACGATAACGACTCTCATTCATGC | |
| hprt sense | GGATATGCCCTTGACTATAATGAG | Figs. 3B, 4B, and 6 | |
| hprt antisense | GCCACAGGACTAGAACACC | ||
| pgk2 sense | CTCTTTCTGCTAAGTTGACTCTGGAC | Fig. 2 | |
| pgk2 antisense | CTCTAATGAATACTTGTCTGGCATAGG |
a Numbers are used in Fig. 1. Primers corresponding to genes other than Fut7 are not numbered.
Construction of the SN1 Tg
The BAC clone RP23–273F21, which contains ∼197 kb of genomic DNA from mouse chromosome 2 with the Fut7 locus approximately centered, was obtained from Invitrogen. A ∼12.7-kb SpeI/NotI fragment (see Fig. 1) containing virtually all genomic sequence between the nearest upstream (Npdc1) and downstream (Abca2) loci was subcloned into the SpeI/NotI sites of pBS/SK (Promega, Madison, WI). The insert, designated SN1, was excised with ClaI and SacII, gel-purified, and used for microinjection into Fut7 null fertilized eggs.
Flow Cytometry
Expression of E- and P-selectin ligands was quantitated using supernatants of COS cells transfected with expression plasmids expressing E-selectin/IgM and P-selectin/IgM fusion proteins (16), exactly as described (19, 22), using Cy5- or AF647-coupled goat anti-human IgM (BIOSOURCE) as the second step. FITC- or phosphatidylethanolamine-conjugated anti-Gr1 or CD4 were obtained from eBioscience. Data were gathered on a FACSCalibur or FACSCanto (BD Biosciences) and analyzed by FloJo software.
WBC Counts and Differential
Approximately 50 μl of blood was collected from the tail vein in EDTA-coated tubes and analyzed on a Hemavet 950 (Drew Scientific, Oxford, CT) complete blood counter.
Peritonitis Assay
Mice were injected intraperitoneally with 1 ml of 4% thioglycollate in PBS. After 4 h, mice were sacrificed, and cells were collected from the peritoneum by thorough lavage after injection of ∼10 ml of PBS/EDTA and counted. In all cases, recovered cells consisted of ∼90% neutrophils, as assessed by flow cytometry.
Genomic qPCR
DNA from tail snips for four mice of each genotype was analyzed by SYBR Green qPCR according to the manufacturer's instructions using a Bio-Rad iQ5 thermal cycler and two different oligonucleotide pairs within the catalytic domain of Fut7. Oligonucleotides for pgk2 were used as the normalization controls. Gene copy number was calculated by the standard 2−ΔΔCt method. Results were normalized by first standardizing the quantified PCR product from fut7-specific primers to that of the pgk2 gene for each individual mouse, deriving a mean ± S.D. for each genotype and primer and then normalizing the mean of each group for each primer pair to WT mice = 2. Results are reported as means ± S.D. for each primer set and each genotype.
qRT-PCR
Total RNA was isolated from various cells and tissues using TRIzol (Invitrogen). 1 μg of total RNA was treated with DNase (Roche Applied Science) for 1 h at 37 ºC, precipitated, and reverse-transcribed using the SuperScript II kit (Invitrogen). Equal volumes of cDNA were then amplified using SYBR Green qPCR as above for genomic DNA. Oligonucleotides for Hprt were used to assess RNA and cDNA integrity and as normalization controls. Normalized fold values were calculated as above and were expressed relative to the mean for WT (for neutrophils) or WT without IL-12 or TGFβ1 (for activated CD4 cells).
End Point RT-PCR
RNA and cDNA from various tissues were prepared as above, and conventional end point PCR was carried out using standard conditions and primers for Fut7 and Hprt and cycle numbers that gave robust bands for the positive controls. The entire PCR reaction was run on standard 1% agarose gels and visualized with ethidium bromide.
T Cell Activation Cultures
CD4+ T cells were isolated from spleens of mice by positive selection using CD4 magnetic beads and columns (Miltenyi). T cells were activated by plate-bound anti-CD3/CD28 (eBioscience) for ∼40 h either with no added cytokine or with 10 ng/ml IL-12 (Peprotech) or 5 ng/ml TGFβ1 (R&D) and subsequently cultured with 20 ng/ml IL-2 (BIOSOURCE) either alone or with IL-12 or TGFβ1 as described previously by us (19, 22, 26). Media were changed every 2 days with the addition of fresh cytokines. Cells were analyzed by flow cytometry at the indicated time points.
Induction of Selectin Ligands and T Cell Homing in Vivo
Mice were infected with the ME49 strain of Toxoplasma gondii, and brain lysates were prepared 3 weeks later as described (27, 28). Brain lysates containing ∼20 T. gondii cysts were injected intraperitoneally, and mice were sacrificed 7 days later. Splenic CD4+ T cells were analyzed for expression of CD44 and E- and P-selectin ligands, and the fraction of activated (CD44-hi) cells that expressed E- and P-selectin ligands was determined. Total peritoneal exudate cells (PEC) were harvested and counted and analyzed by FACS for CD4, and the total number of CD4 cells harvested from the peritoneum was calculated.
Lymphocyte Homing Assay
Spleen cells were isolated from WT mice, labeled with 1 μm CMFDA, and washed, and 2 × 107 cells were injected intravenously into recipients. After 1 h, spleens and inguinal, brachial, and axillary LNs were harvested, and single cell suspensions were prepared and analyzed by flow cytometry. For all LNs, the entire cell sample of individual LNs was analyzed. Results are expressed as % of total cells, which are CMFDA+.
Immunohistology
Brachial lymph nodes were frozen in OCT (Tissue-Tek), and 10-μm sections were cut in a Leica cryostat. The sections on SuperFrost Plus slides (Thermo Fisher) were fixed for 10 min in ice-cold acetone, air-dried for 1 h, and incubated with Block (3% BSA in Dulbecco's CMF-PBS with 5% normal goat serum (Sigma)) for 10 min. The Block was removed, and either 5 μg/ml recombinant mouse L-selectin/human IgG chimera (R&D Systems) ± 10 mm EDTA in Block or 1 μg/ml of MECA-79 (BioLegend) or rat IgM isotype control (eBioscience) in Block was applied to the sections for 1 h at room temperature. After washing in CMF, 1.5 μg/ml Cy3-conjugated goat anti-human IgG or 2 μg/ml biotinylated goat anti-rat IgM (Jackson ImmunoResearch) in Block was added and incubated for 45 min at room temperature. The sections stained with the biotinylated secondary were washed in CMF, and 1.7 μg/ml Cy2-conjugated streptavidin (Jackson ImmunoResearch) in Block was added for 30 min at room temperature. Finally, all sections were washed in CMF, lightly counterstained with Harris hematoxylin (Sigma), and coverslipped with Fluoro-Gel (Electron Microscopy Sciences). Pictures were obtained with a Zeiss Axiocam attached to a Nikon Optiphot using a Fluor 20× objective. The exposure time for bright field was 3 ms, for Cy2 was 800 ms, and for Cy3 was 5000 ms.
In Silico Analysis
Analysis of evolutionary conservation of Fut7 and surrounding loci were performed using the Vista Browser (available at genome.lbl.gov).
Statistical Analysis
All statistical analyses were carried out using the Mann-Whitney non-parametric test within the Prizm program. p values for pairwise comparisons are given in the figure legends.
RESULTS
Production of Mice Expressing the SN1 Tg
A 12.7-kb contiguous genomic segment flanked by locally unique SpeI and NotI restriction sites, encompassing nucleotides 25,271,404–25,284,140 (from the Vista Browser, mouse July 2007 build) on mouse chromosome 2, was subcloned into pBS from the BAC clone RP23-273F21. This DNA segment, which we have designated SN1, encompasses the entire transcriptional unit, including the major transcriptional start site, transcriptional termination site, and polyadenylation site along with ∼7.4 kb of upstream sequence and ∼2 kb of downstream sequence (Fig. 1) and represents essentially all of the sequence between the immediate upstream (Npdc1) and downstream (Abca2) genes. This genomic segment also contains a number of conserved noncoding sequences both upstream and downstream of the transcriptional unit (Fig. 1), suggesting their possible involvement in gene regulation.
The SN1 DNA was used as a Tg for pronuclear injection into fertilized eggs from Fut7 null mice. One founder was identified among pups born from each of two independent microinjections, and these two founder lines were designated SN1.1 and SN1.2. For both lines, ∼50% of both male and female progeny were Tg+, indicating normal Mendelian inheritance. Analysis of Tg copy number using qPCR with two independent primer sets within the single large exon, which encodes the glycosyltransferase domain, revealed that SN1.1 mice harbored ∼3-fold higher gene copy numbers of the Tg compared with WT, whereas SN1.2 mice harbored ∼2.5 copies fold higher gene copy numbers (Fig. 2). These Tg mice, therefore, harbor ∼6 and ∼5 copies of the SN1 Tg, respectively.
FIGURE 2.
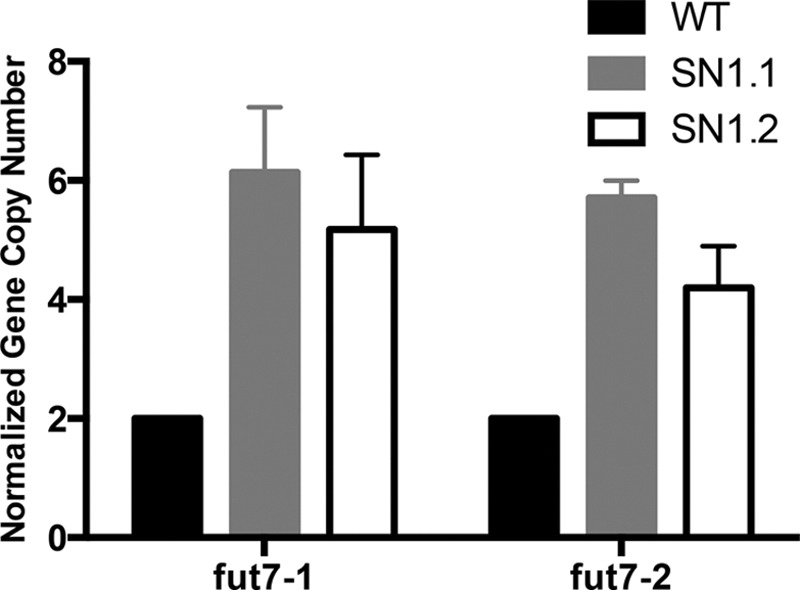
Gene copy number of the SN1 Fut7 Tg in SN1 Tg mice. Tg copy number in SN1 Tg mice was determined by qPCR of genomic DNA using two different pairs of primers, both within the catalytic domain of Fut7 (see Table 1). n = 4 for WT, and SN1.2 and n = 3 for SN1.1. Results for each mouse were normalized first against pgk2 primers for that mouse. The mean empirically determined gene copy number for each genotype for each primer was then determined. Finally, the normalized gene copy number was then calculated by normalizing for the WT = 2.
Fut7 Gene Expression and Function in Neutrophils in Tg Mice
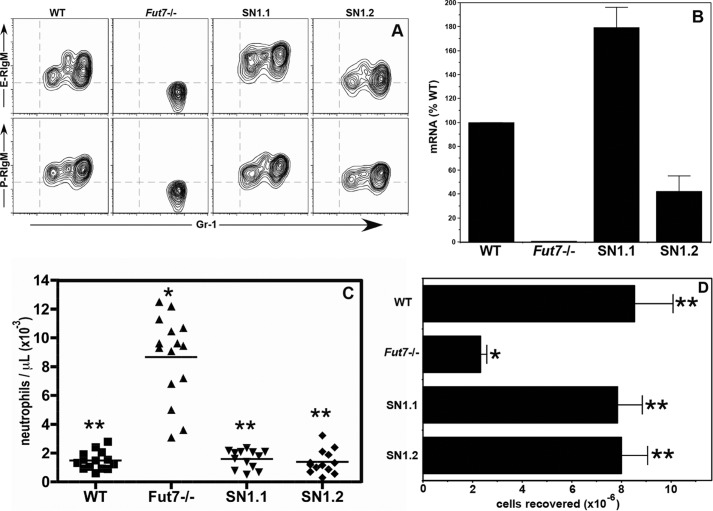
Fut7 null mice display a spectrum of abnormalities in hematopoietic and lymphoid tissues. Within the myeloid compartment, these include a nearly complete loss of both E- and P-selectin ligands, significantly higher blood neutrophil counts, and sharply impaired neutrophil recruitment in acute peritonitis models (16). Expression of selectin ligands can be reliably quantitated on a per cell basis by flow cytometry with selectin-IgM chimeras (16, 19). We found that the SN1 Tg restored E- and P-selectin ligands to neutrophils in the blood (Fig. 3A) and bone marrow (data not shown). Levels of selectin ligands were slightly higher than WT on neutrophils from SN1.1 mice and slightly lower than WT on neutrophils from SN1.2 mice (Fig. 3A), and these levels of selectin ligands correlated roughly with Fut7 mRNA levels in these cells (Fig. 3B). Critically, the neutrophilia found in Fut7 null mice was eliminated in both Tg lines (Fig. 3C). Similarly, neutrophil recruitment into the peritoneum in response to thioglycollate was also restored to WT levels (Fig. 3D). Taken together, these data show that Fut7 gene expression in neutrophils was restored to physiologic levels by the SN1 Tg, and this level of Fut7 gene expression led to expression of functional selectin ligands on these cells.
FIGURE 3.
The SN1 Tg restores Fut7 and functional selectin ligand expression in neutrophils. A, flow cytometry analysis of E- and P-selectin ligand expression on blood neutrophils. Neutrophils were identified by characteristic forward and side scatter properties (gating not shown). Total blood cells were stained with anti-Gr-1, which identifies neutrophils, and E- or P-selectin/IgM chimera. B, qRT-PCR of Fut7 mRNA levels in neutrophils. For each mouse, results were first normalized to hprt for that mouse and then to the WT (=100%). One of two similar experiments is shown. C, absolute neutrophil numbers in peripheral blood of each genotype of mice. Each symbol represents a single mouse. D, total PEC recovered from the peritoneum of mice 4 h after intraperitoneal injection of 4% thioglycollate. Depicted is the mean ± S.D. for n = 12–16 for each genotype. For C and D: *, different (p < 0.01) from all other groups; **, no difference between groups.
Expression of Fut7 in Activated T Cells
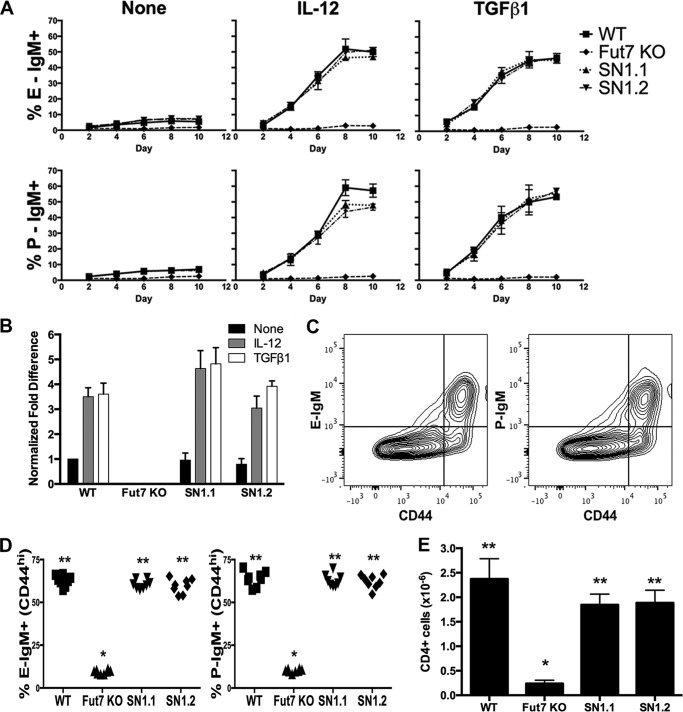
CD4 T cells activated via the TCR and cultured with IL-2 alone show low levels of E- and P-selectin ligands, whereas CD4 T cells activated via the TCR and cultured with IL-2 plus either IL-12 or TGFβ1 show significantly higher levels of selectin ligands (19, 29). CD4 T cells from WT, Fut7 null, SN1.1, and SN1.2 mice were isolated, activated by plate-bound anti-CD3/CD28 mAb, and cultured with IL-2 alone, with IL-2 plus IL-12, or with IL-2 plus TGFβ1 to determine if the SN1 Tg could restore the ability of these cells to express Fut7 and selectin ligands in response to signals emanating from the TCR, IL-12 receptor, and/or TGFβ receptor. Cells were analyzed every 2 days to ensure that any shift in the kinetics of expression would be detected. We found that CD4 T cells from both the SN1.1 and SN1.2 mice expressed WT levels of E- and P-selectin ligands in response to activation and culture in IL-2 alone or with inclusion of either IL-12 or TGFβ1 (Fig. 4A). For each cytokine condition, levels of Fut7 mRNA were similar to WT in both SN1.1 and SN1.2 CD4 cells (Fig. 4B). These data show that the SN1 Tg contains all cis-acting elements essential for induction of Fut7 gene expression in response to signals originating from the TCR, IL-12 receptor, and TGFβ receptor.
FIGURE 4.
Fut7 and functional selectin ligand expression in activated CD4 T cells in SN1 Tg mice. A, CD4 T cells were isolated, activated with plate-bound anti-CD3/CD28 without any added cytokine (none), with IL-12, or with TGFβ1, and subsequently cultured with IL-2 alone, IL-12 plus IL-2, or TGFβ1 plus IL-2 and analyzed by flow cytometry with selectin chimeras every 2 days. Depicted are the fractions that are positive for E-selectin ligands (top row) and P-selectin ligands (bottom row) for each genotype at each time point. Mean values ± S.D. from three experiments. B, qRT-PCR of Fut7 mRNA levels in activated CD4 T cells cultured with cytokines as above. Cells were harvested on day 6. As for neutrophils, values were normalized first to hprt levels. These values were then normalized to WT IL-2 only (=1) and expressed as -fold difference over WT IL-2 only. One of two similar experiments is shown. C, induction of E- and P-selectin ligands on splenic CD4 T cells in response to T. gondii infection. Spleens were harvested 7 days after intraperitoneal injection of T. gondii cysts. Plots are gated on CD4+ cells (gating not shown). Representative FACS plots of WT cells show that E- and P-selectin ligand-expressing cells are largely confined to the CD44-hi (activated) population. D, the fraction of CD44-hi CD4+ cells expressing ligands for either E- or P-selectin was calculated. Each symbol represents an individual mouse. E, total CD4+ PEC recovered on day 7 from each of the 4 genotypes of mice. For D and E: *, different (p < 0.01) from all other groups; **, no difference between groups.
To determine whether restoration of Fut7 expression was restored on activated CD4 T cells in vivo, we injected each genotype of mice intraperitoneally with brain lysate prepared from WT mice chronically infected with T gondii, which triggers a robust Th1 response (27, 28), and analyzed induction of selectin ligands on activated (CD44-hi) splenic CD4 T cells 7 days after infection. Selectin ligands are induced predominantly on the activated (CD44-hi) subset of cells (Fig. 4C). Induction of E- and P-selectin ligands on SN1.1 and SN1.2 CD4+CD44-hi cells was equivalent to that of WT (Fig. 4D). To determine if recruitment of T cells to the peritoneum in response to infection with T. gondii was selectin-dependent, we analyzed CD4+ PEC at day 7 in WT and Fut7 null mice. These data showed an ∼90% reduction of accumulated CD4+ PEC in the Fut7 null mice (Fig. 4E), indicating a strong requirement for selectins in T cell migration to the peritoneum, paralleling that of neutrophils (9–12). We, therefore, analyzed recruitment of CD4 T cells to the peritoneum in the SN1 Tg mice infected with T. gondii. We found that the numbers of CD4 T cell recruited to the peritoneum of SN1 Tg mice were equivalent to WT (Fig. 4E). Taken together, these data demonstrate that the SN1 Tg restored Fut7 expression and functional selectin ligands on activated CD4 T cells in response to infection with T. gondii.
Restoration of Fut7 Expression in HEC of Tg Mice
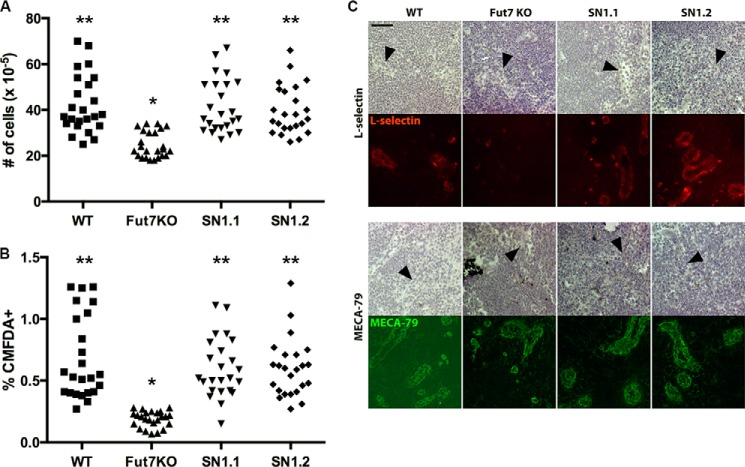
Lymph nodes from Fut7 null mice display significantly lower total lymphocyte numbers compared with WT, concomitant with immunohistochemically undetectable L-selectin ligands, and support significantly lower levels of lymphocyte homing (16). We examined each of these aspects of Fut7 expression in our SN1 mice. Total cell numbers in peripheral LNs were restored to WT levels in both SN1.1 and SN1.2 mice (Fig. 5A), and these LNs contained normal ratios of T and B cells (data not shown). HEC in SN1 mice were fully able to support short term homing of CMFDA-labeled lymphocytes (Fig. 5B), consistent with restoration of immunohistochemically detectable L-selectin ligands on HEC (Fig. 5C). Direct analysis of Fut7 mRNA levels in HEC was not possible due to the inability to purify sufficient numbers of HEC but was clearly sufficient to restore expression and function of L-selectin ligands on these cells. These data demonstrate that physiologic levels of Fut7 gene expression are restored to HEC in SN1 Tg mice.
FIGURE 5.
Restoration of Fut7 expression and L-selectin ligands in HEC. A, total lymphocyte numbers isolated from individual axillary, brachial, and inguinal LNs of each genotype. LNs were harvested, single cell suspensions were prepared, and cells were counted. Each symbol represents a single LN in one mouse, with axillary, brachial, and inguinal LNs combined for each genotype. B, short term homing of labeled lymphocytes in mice of each genotype. Cells were labeled with CMFDA, washed, and injected into the tail vein. One hour later mice were sacrificed, and the axillary, brachial, and inguinal LNs were harvested, and single cell suspensions were prepared and analyzed by flow cytometry. As in A, each symbol represents a single LN in one mouse, with axillary, brachial, and inguinal LNs combined for each genotype. For A and B: *, different (p < 0.01) from all other groups; **, no difference between groups. C, expression of L-selectin ligands on HEC. Top, staining with L-RIgM; bottom, staining with MECA-79, which recognizes HEC regardless of Fut7 expression. The top set of panels in each case is brightfield, with HEC indicated by black arrowheads; the bottom set of panels is staining with either L-selectin or MECA-79. Each vertical pair (brightfield, fluorescence) depicts the same field. There was no detectable L-selectin binding in the presence of EDTA (not shown).
Absence of Fut7 Expression in Inappropriate Tissues in Tg Mice
A striking aspect of Fut7 expression is its highly restricted pattern of expression, which is in contrast to most other glycosyltransferases, including those involved in selectin ligand biosynthesis. Therefore, to determine whether any silencers or other regulatory elements essential to tissue-specific expression of Fut7 were absent from the SN1 Tg, we analyzed Fut7 mRNA expression in a panel of non-hematopoietic tissues that do not normally express Fut7. The results showed that Fut7 mRNA was undetectable in any of the tissues examined, including brain, lung, liver, kidney, skeletal muscle, and small intestine (Fig. 6). Fut7 expression is, therefore, absent from inappropriate cell types in these Tg mice. Taken together with the data presented above, the lack of detectable Fut7 expression in cell types that do not normally express this gene demonstrates that all essential cis-acting genetic elements involved in Fut7 gene expression, both positive and negative, are contained within the 12.7-kb SN1 Tg.
FIGURE 6.
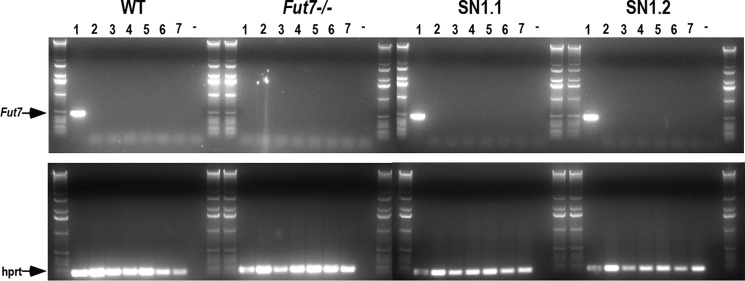
Absence of Fut7 expression in inappropriate cell types. Mice were anesthetized, perfused with PBS, sacrificed, and total RNA was isolated and reverse-transcribed, and PCR was performed on organs from WT, Fut7 KO, SN1.1, and SN1.2 mice. Lanes represent RT-PCR from bone marrow (1), brain (2), lung (3), liver (4), kidney (5), skeletal muscle (6), and small intestine (7). The dash (−) represents negative control of no cDNA input to the PCR reaction. Primers for hprt were used as a positive control on all samples (lower panels). The entire PCR reaction was run on 1% agarose gels, with each set of eight samples flanked by DNA size ladders.
DISCUSSION
Complex and highly restricted patterns of gene expression are determined in part by an array of functionally distinct cis-acting genetic elements, including enhancers, silencers, locus control regions, and insulators. An essential prerequisite for comprehensive and unbiased identification of cis-acting elements that control expression of any particular gene is the delineation of the functional boundaries of the locus under study, defined as any contiguous stretch of genomic DNA that contains all cis-acting genetic elements essential for accurate physiologic expression. Here, we have used a genetic complementation/gain-of-function approach in Tg mice to define the functional boundaries of the murine Fut7 locus. We found that a 12.7-kb contiguous genomic region, which lies completely between the flanking genes Npdc1 and Abca2, fully restores normal levels of Fut7 expression and selectin ligands in neutrophils, activated CD4 T cells, and HEC when analyzed as a Tg on a Fut7 null background. In addition, no aberrant expression of Fut7 was observed in a panel of tissues that do not normally express this gene. These findings, obtained in mice derived from two independently generated Tg founder mice, convincingly demonstrate that this 12.7-kb Tg incorporates all essential cis-acting genetic elements required for accurate physiologic expression of Fut7.
This 12.7-kb genomic segment, therefore, encompasses genetic elements required to respond to an array of distinct signals in diverse cell types. In HEC, Fut7 expression may be controlled by dendritic cell-triggered LTβR signaling (30), which classically involves NF-κB and TRAF (TNR receptor-associated factor) adaptor proteins (31, 32). In myeloid cells, G-CSF is capable of inducing E-selectin ligands on mobilized peripheral blood progenitors (33), but no signaling pathways or transcription factors that control Fut7 expression or selectin ligand expression in response to G-CSF or other cytokines involved in myelopoiesis have thus far to our knowledge been identified. In CD4 T cells, signals initiated by the TCR evoke modest levels of Fut7 expression and selectin ligands, possibly mediated by Ras (34, 35), and these lower levels of Fut7 and selectin ligands are significantly enhanced by IL-12 or TGFβ1 (19, 29). The signaling and transcriptional pathways downstream of the TCR, IL-12 receptor, and TGFβ receptor, which are responsible for induction and/or up-regulation of Fut7 and selectin ligands, remain incompletely characterized. We previously uncovered a requirement for the Th1 transcription factor T-bet in IL-12-driven Fut7 and selectin ligand expression (22) and a requirement for p38 MAPK activity in TGFβ1-induced Fut7 and selectin ligand expression (29). Whether T-bet and/or p38 MAPK directly or indirectly control Fut7 expression and selectin ligand expression remains unclear. Despite this limited information, it nonetheless seems likely that T lymphocytes, myeloid cells, and HEC use distinct transcriptional mechanisms for induction or maintenance of Fut7 gene expression. Our results show that all cis-acting genetic elements required for each of these signals to act are present within the 12.7-kb SN1 Tg. Thus, complex inputs from multiple distinct pathways converge in a relatively small genomic space.
Expression of Fut7 is not detected in a wide variety of cells and tissues that do not express selectin ligands. Crucially, we also found that Fut7 expression in our Tg mice was undetectable in tissues that do not normally express Fut7. The absence of Fut7 expression in epithelial and mesenchymal tissues could be due to the absence of appropriate transcription factors, the presence of negative cis-acting genetic elements, silencing epigenetic chromatin modifications, or a combination of these. Our results indicate that if any negative regulatory elements such as silencers, boundary elements, or insulators are required to maintain the absence of Fut7 expression in epithelial and mesenchymal tissues, these regulatory elements must also be present in the 12.7-kb SN1 Tg.
As analyzed using the Vista Browser, the Fut7 locus is nestled within a large, evolutionarily conserved array of genes whose pattern of expression is quite distinct both from that of Fut7 and from each other. In particular, the presence, order, sequence, genomic spacing, and overall arrangement of all of the genes surrounding Fut7 are evolutionarily highly conserved among human, dog, horse, and mouse for at least 466 kb upstream and 278 kb downstream of Fut7, indicating strong evolutionary pressure to maintain this higher order genomic arrangement. This large region contains at least 46 genes, each of which is found and conserved in its exon/intron organization in all four of the species examined. This finding makes it likely that mechanisms to prevent inappropriate expression or silencing of neighboring loci, including insulators and boundary elements, are also conserved within this overall genomic arrangement and is consistent with our results in this report that no essential cis-acting elements required for physiologically accurate Fut7 expression lie within the boundaries of the upstream or downstream genes.
Other investigators have carried out similar experiments to define the functional boundaries of other mammalian genes. Engel and co-workers (37) have shown that the minimal functional boundaries of the Gata2 gene required for mouse viability and normal development was 413 kb (36). Lichtenheld and co-workers (37) identified an ∼150 kb interval of the human PRF1 gene that conferred accurate lineage- and stage-specific expression of perforin in primary cytotoxic T and NK cells. Weaver and co-workers (38) utilized an ∼160-kb “BAC-in” reporter gene strategy to delineate Ifng expression in Tg mice and achieved faithful expression of the reporter in T cells (i.e. expression of the reporter gene exclusively in cells that also expressed the endogenous Ifng gene). Each of these studies identified a genomic interval of at least 150 kb that conferred physiologically accurate expression. In contrast, the current study shows that physiologically accurate expression of Fut7 required a much smaller genomic region.
The comparatively small size of the Fut7 locus, coupled to its sharply restricted expression, should facilitate studies aimed at exploring how distinct classes of cis-acting genetic elements function to control Fut7 gene expression in distinct cell types and in response to distinct developmental signals. The work presented here, therefore, provides a platform for mechanistic studies of cis-acting genetic elements that control leukocyte traffic and suggests that further studies of this gene could yield insights into fundamental questions of gene regulation.
Acknowledgments
We gratefully acknowledge the efforts of Mark Singer and Dr. Steven D. Rosen, University of California, San Francisco, for performing immunohistochemistry for L-selectin ligands and MECA79 on peripheral lymph nodes (Fig. 5C) and for helpful advice and Dr. Dragana Jankovic, Laboratory of Parasitic Diseases, NIAID, National Institutes of Health for provision of T. gondii-containing brain lysates and critical advice. The genetically engineered mice were generated with the assistance of Northwestern University Transgenic and Targeted Mutagenesis Laboratory. The Northwestern University Transgenic and Targeted Mutagenesis Laboratory is supported in part by National Institutes of Health Grant CA60553 (to the Robert H. Lurie Comprehensive Cancer Center at Northwestern University).
This work was supported, in whole or in part, by National Institutes of Health Grant 1 R21 HL092357-01A1 (to G. S. K.). This work was also supported in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to G. S. K.).
- HEC
- high endothelial cell(s)
- PEC
- peritoneal exudate cell(s)
- Tg
- transgenic
- LN
- lymph node
- qPCR
- quantitative PCR
- CMFDA
- 5-chloromethylfluorescein (CMF) diacetate
- TCR
- T cell receptor.
REFERENCES
- 1. Gallatin W. M., Weissman I. L., Butcher E. C. (1983) A cell-surface molecule involved in organ-specific homing of lymphocytes. Nature 304, 30–34 [DOI] [PubMed] [Google Scholar]
- 2. Lasky L. A., Singer M. S., Yednock T. A., Dowbenko D., Fennie C., Rodriguez H., Nguyen T., Stachel S., Rosen S. D. (1989) Cloning of a lymphocyte homing receptor reveals a lectin domain. Cell 56, 1045–1055 [DOI] [PubMed] [Google Scholar]
- 3. Stoolman L. M., Yednock T. A., Rosen S. D. (1987) Homing receptors on human and rodent lymphocytes- evidence for a conserved carbohydrate-binding specificity. Blood 70, 1842–1850 [PubMed] [Google Scholar]
- 4. Kishimoto T. K., Jutila M. A., Butcher E. C. (1990) Identification of a human peripheral lymph node homing receptor. A rapidly down-regulated adhesion molecule. Proc. Natl. Acad. Sci. U.S.A. 87, 2244–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frenette P. S., Subbarao S., Mazo I. B., von Andrian U. H., Wagner D. D. (1998) Endothelial selectins and vascular adhesion molecule-1 promote hematopoietic progenitor homing to bone marrow. Proc. Natl. Acad. Sci. U.S.A. 95, 14423–14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nabors L. K., Wang L. D., Wagers A. J., Kansas G. S. (2013) Overlapping roles for endothelial selectins in murine hematopoietic stem/progenitor cell homing to bone marrow. Exp. Hematol. 41, 588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robert C., Fuhlbrigge R. C., Kieffer J. D., Ayehunie S., Hynes R. O., Cheng G., Grabbe S., von Andrian U. H., Kupper T. S. (1999) Interaction of dendritic cells with skin endothelium. A new perspective on immunosurveillance. J. Exp. Med. 189, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arbonés M. L., Ord D. C., Ley K., Ratech H., Maynard-Curry C., Otten G., Capon D. J., Tedder T. F. (1994) Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin (CD62L) deficient mice. Immunity 1, 247–260 [DOI] [PubMed] [Google Scholar]
- 9. Bullard D. C., Kunkel E. J., Kubo H., Hicks M. J., Lorenzo I., Doyle N. A., Doerschuk C. M., Ley K., Beaudet A. L. (1996) Infectious susceptibility and severe deficiency of leukocyte rolling and recruitment in E-selectin and P-selectin double mutant mice. J. Exp. Med. 183, 2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frenette P. S., Mayadas T. N., Rayburn H., Hynes R. O., Wagner D. D. (1996) Susceptibility to infection and altered hematopoiesis in mice deficient in both P- and E-selectins. Cell 84, 563–574 [DOI] [PubMed] [Google Scholar]
- 11. Mayadas T. N., Johnson R. C., Rayburn H., Hynes R. O., Wagner D. D. (1993) Leukocyte rolling and extravasation are severely compromised in P-selectin-deficient mice. Cell 74, 541–554 [DOI] [PubMed] [Google Scholar]
- 12. Labow M. A., Norton C. R., Rumberger J. M., Lombard-Gillooly K. M., Shuster D. J., Hubbard J., Bertko R., Knaack P. A., Terry R. W., Harbison M. L. (1994) Characterization of E-selectin-deficient mice. Demonstration of overlapping function of the endothelial selectins. Immunity 1, 709–720 [DOI] [PubMed] [Google Scholar]
- 13. Robinson S. D., Frenette P. S., Rayburn H., Cummiskey M., Ullman-Culleré M., Wagner D. D., Hynes R. O. (1999) Multiple, targeted deficiencies in selectins reveal a predominant role for P-selectin in leukocyte recruitment. Proc. Natl. Acad. Sci. U.S.A. 96, 11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ley K., Kansas G. S. (2004) Selectins in T cell recruitment to non-lymphoid tissues and sites of inflammation. Nat. Rev. Immunol. 4, 325–335 [DOI] [PubMed] [Google Scholar]
- 15. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Getting to the site of inflammation. The leukocyte adhesion cascade updated. Nat. Rev. Immunol 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 16. Malý P., Thall A., Petryniak B., Rogers C. E., Smith P. L., Marks R. M., Kelly R. J., Gersten K. M., Cheng G., Saunders T. L., Camper S. A., Camphausen R. T., Sullivan F. X., Isogai Y., Hindsgaul O., von Andrian U. H., Lowe J. B. (1996) The α(1,3)-fucosyltransferase FucT-VII controls leukocyte trafficking through an essential role in L-, E-, and P-selectin ligand biosynthesis. Cell 86, 643–653 [DOI] [PubMed] [Google Scholar]
- 17. Homeister J. W., Thall A. D., Petryniak B., Malý P., Rogers C. E., Smith P. L., Kelly R. J., Gersten K. M., Askari S. W., Cheng G., Smithson G., Marks R. M., Misra A. K., Hindsgaul O., von Andrian U. H., Lowe J. B. (2001) The alpha(1,3)fucosyltransferases FucT-IV and FucT-VII exert collaborative control over selectin-dependent leukocyte recruitment and lymphocyte homing. Immunity 15, 115–126 [DOI] [PubMed] [Google Scholar]
- 18. Smith P. L., Gersten K. M., Petryniak B., Kelly R. J., Rogers C., Natsuka Y., Alford J. A., 3rd, Scheidegger E. P., Natsuka S., Lowe J. B. (1996) Expression of the α1,3-fucosyltransferase Fuc-TVII in lymphoid aggregate high endothelial venules correlates with expression of L-selectin ligands. J. Biol. Chem. 271, 8250–8259 [DOI] [PubMed] [Google Scholar]
- 19. White S. J., Underhill G. H., Kaplan M. H., Kansas G. S. (2001) Cutting edge. Differential requirements for Stat4 in expression of glycosyltransferases responsible for selectin ligand formation in Th1 cells. J. Immunol. 167, 628–631 [DOI] [PubMed] [Google Scholar]
- 20. Blander J. M., Visintin I., Janeway C. A., Jr., Medzhitov R. (1999) α(1,3)-fucosyltransferase VII and α(2,3) sialyltransferase IV are upregulated in activated CD4 T cells and maintained after their differentiation into Th1 and migration into inflammatory sites. J. Immunol. 163, 3746–3752 [PubMed] [Google Scholar]
- 21. Underhill G. H., Minges Wols H. A., Fornek J. L., Witte P. L., Kansas G. S. (2002) IgG plasma cells display a unique spectrum of leukocyte adhesion and homing molecules. Blood 99, 2905–2912 [DOI] [PubMed] [Google Scholar]
- 22. Underhill G. H., Zisoulis D. G., Kolli K. P., Ellies L. G., Marth J. D., Kansas G. S. (2005) A crucial role for T-bet in selectin ligand expression in T helper 1 (Th1) cells. Blood 106, 3867–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen G. Y., Osada H., Santamaria-Babi L. F., Kannagi R. (2006) Interaction of GATA-3/T-bet transcription factors regulates expression of sialyl Lewis X homing receptors on Th1/Th2 lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 16894–16899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hiraiwa N., Hiraiwa M., Kannagi R. (1997) Human T cell leukemia virus-1 encoded tax protein transactivates α1,3-fucosyltransferase FucT-VII, which synthesizes sialyl Lewis X, a selectin ligand expressed on adult T cell leukemia cells. Biochem. Biophys. Res. Comm. 231, 183–186 [DOI] [PubMed] [Google Scholar]
- 25. Hiraiwa N., Yabuta T., Yoritomi K., Hiraiwa M., Tanaka Y., Suzuki T., Yoshida M., Kannagi R. (2003) Transactivation of the fucosyltransferase VII gene by human T-cell leukemia virus type 1 Tax through a variant cAMP-responsive element. Blood 101, 3615–3621 [DOI] [PubMed] [Google Scholar]
- 26. Wagers A. J., Waters C. M., Stoolman L. M., Kansas G. S. (1998) IL-12 and IL-4 control T cell adhesion to endothelial selectins through opposite effects on FucT-VII gene expression. J. Exp. Med. 188, 2225–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu J., Jankovic D., Oler A. J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., Feigenbaum L., Zhao K., Paul W. E. (2012) The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37, 660–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jankovic D., Kullberg M. C., Feng C. G., Goldszmid R. S., Collazo C. M., Wilson M., Wynn T. A., Kamanaka M., Flavell R. A., Sher A. (2007) Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med. 204, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagers A. J., Kansas G. S. (2000) Potent induction of FucT-VII by TGF-β1 through a p38 MAP kinase-dependent pathway. J. Immunol. 165, 5011–5016 [DOI] [PubMed] [Google Scholar]
- 30. Moussion C., Girard J. P. (2011) Dendritic cells control lymphocyte entry to lymph nodes through high endothelial venules. Nature 479, 542–546 [DOI] [PubMed] [Google Scholar]
- 31. VanArsdale T. L., VanArsdale S. L., Force W. R., Walter B. N., Mosialos G., Kieff E., Reed J. C., Ware C. F. (1997) Lymphotoxin-β receptor signaling complex. Role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor κB. Proc. Natl. Acad. Sci. U.S.A. 94, 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. (2002) The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 33. Dagia N. M., Gadhoum S. Z., Knoblauch C. A., Spencer J. A., Zamiri P., Lin C. P., Sackstein R. (2006) G-CSF induces E-selectin ligand expression on human myeloid cells. Nature medicine 12, 1185–1190 [DOI] [PubMed] [Google Scholar]
- 34. Barry S. M., Zisoulis D. G., Neal J. W., Clipstone N. A., Kansas G. S. (2003) Induction of FucT-VII by the Ras/MAP kinase cascade in Jurkat T cells. Blood 102, 1771–1778 [DOI] [PubMed] [Google Scholar]
- 35. Zisoulis D. G., Kansas G. S. (2004) H-Ras and PI3K cooperate to induce FucT-VII expression in Jurkat T cells. J. Biol. Chem. 279, 39495–39504 [DOI] [PubMed] [Google Scholar]
- 36. Brandt W., Khandekar M., Suzuki N., Yamamoto M., Lim K. C., Engel J. D. (2008) Defining the functional boundaries of the Gata2 locus by rescue with a linked bacterial artificial chromosome transgene. J. Biol. Chem. 283, 8976–8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pipkin M. E., Ljutic B., Cruz-Guilloty F., Nouzova M., Rao A., Zúñiga-Pflücker J. C., Lichtenheld M. G. (2007) Chromosome transfer activates and delineates a locus control region for perforin. Immunity 26, 29–41 [DOI] [PubMed] [Google Scholar]
- 38. Hatton R. D., Harrington L. E., Luther R. J., Wakefield T., Janowski K. M., Oliver J. R., Lallone R. L., Murphy K. M., Weaver C. T. (2006) A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729 [DOI] [PubMed] [Google Scholar]