Background: Regulation of MutYH DNA glycosylase is essential for genomic integrity.
Results: MutYH is ubiquitinated by the E3 ligase Mule changing its protein levels and recruitment to chromatin. MutYH levels in cancer cells influence the mutagenic potential of reactive oxygen species.
Conclusion: Ubiquitination of MutYH is an important regulatory step.
Significance: We identified the residues that are targets for ubiquitination by Mule.
Keywords: Base Excision Repair, DNA Damage, E3 Ubiquitin Ligase, Oxidative Stress, Ubiquitination, 8-Oxo-G, Mule, MutYH
Abstract
Oxidation of DNA is a frequent and constantly occurring event. One of the best characterized oxidative DNA lesions is 7,8-dihydro-8-oxoguanine (8-oxo-G). It instructs most DNA polymerases to preferentially insert an adenine (A) opposite 8-oxo-G instead of the appropriate cytosine (C) thus showing miscoding potential. The MutY DNA glycosylase homologue (MutYH) recognizes A:8-oxo-G mispairs and removes the mispaired A giving way to the canonical base excision repair that ultimately restores undamaged guanine (G). Here we characterize for the first time in detail a posttranslational modification of the human MutYH DNA glycosylase. We show that MutYH is ubiquitinated in vitro and in vivo by the E3 ligase Mule between amino acids 475 and 535. Mutation of five lysine residues in this region significantly stabilizes MutYH, suggesting that these are the target sites for ubiquitination. The endogenous MutYH protein levels depend on the amount of expressed Mule. Furthermore, MutYH and Mule physically interact. We found that a ubiquitination-deficient MutYH mutant shows enhanced binding to chromatin. The mutation frequency of the ovarian cancer cell line A2780, analyzed at the HPRT locus can be increased upon oxidative stress and depends on the MutYH levels that are regulated by Mule. This reflects the importance of tightly regulated MutYH levels in the cell. In summary our data show that ubiquitination is an important regulatory mechanism for the essential MutYH DNA glycosylase in human cells.
Introduction
Every organism is exposed to high levels of oxidative stress every day from a variety of endogenous and exogenous sources. One of the most often observed consequences is the formation of the highly mutagenic lesion 7,8-dihydro-8-oxoguanine (8-oxo-G),3 which arises ∼103 times per cell and day in normal tissue and up to 105 times in cancer tissues (1). The 8-oxo-G lesion is potentially highly mutagenic, because replicative pols tend to incorporate the incorrect A rather than the correct C opposite the lesion, leading to the formation of an A:8-oxo-G mispair. If these mispairs are not repaired before the cell undergoes replication, GC→TA transversion mutations occur. Such mutations were found to be frequently present in different types of cancer tissues (2), demonstrating the paramount importance of a mechanism ensuring the correct repair of 8-oxo-G lesions. As we have shown previously (3, 4), such a mechanism indeed exists: a specialized base excision repair (BER) pathway coordinated by the DNA glycosylase MutYH and DNA pol λ (5). MutYH recognizes a mispaired A and excises it, resulting in the formation of a one nucleotide gap, still bearing the 8-oxo G lesion on the template strand, that is subsequently filled by pol λ. In contrast to other pols, pol λ and to some extent pol η, both in the presence of proliferating cell nuclear antigen and replication protein A, are very efficient in the correct bypass of an 8-oxo-G lesion thereby preventing the generation of mutations (4).
In agreement with the important role of MutYH in the repair of oxidative DNA damage, mouse embryonic fibroblasts as well as embryonic stem cells derived from MutYH−/− mice show an increase in mutation frequency (6, 7). Furthermore, knock-out mice have a higher incidence of tumor formation compared with the wild type mice. This effect can even be increased by exposing the mice to an oxidant showing that the absence of MutYH drastically impairs the repair of oxidative damage (8). Intriguingly, MutYH transcript levels are inversely correlated with the survival outcome of patients suffering from gastric cancer (9).
The BER machinery needs to be tightly controlled to ensure an efficient and correct repair of damaged DNA. Already small differences in expression of individual BER proteins can disturb the entire pathway and thus lead to a reduced repair capacity (10). Recent data provide evidence that regulation of BER is mainly achieved through various posttranslational modifications, such as phosphorylation and ubiquitination (11–14). Even if it was shown that the MutYH expression levels fluctuate during the cell cycle (15) and can be increased upon exposure of cells to reactive oxygen species (16), no mechanism elucidating the regulation of MutYH on protein level has been identified so far.
In the present paper we provide the first evidence that MutYH is a target for ubiquitination, and the executing E3 ligase was identified to be Mule. Ubiquitinated MutYH is marked for proteasomal degradation; thereby not only the protein levels are modulated but also its recruitment to chromatin. We furthermore show that a tight regulation of MutYH is of great importance, because altered protein levels lead to an increase in mutation frequency at the HPRT locus of the ovarian cancer cells A2780.
EXPERIMENTAL PROCEDURES
Chemicals
Oligonucleotides for site-directed mutagenesis were purchased from Microsynth (Balgach, Switzerland) and KBrO3 from Sigma. The Mule siRNA (Hs_HUWE2) was purchased from Qiagen.
Cells, Extracts, and Cell Fractionation
HeLa cells were purchased from American Type Culture Collection. HEK293T and A2780 (described in Ref. 17) cells were gifts from R. Santoro (University of Zürich, Switzerland) and J. Jiricny (University of Zürich, Switzerland), respectively. Cells were grown under standard conditions. Whole cell extracts were prepared by scraping cells into lysis buffer (10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 50 mm Hepes (pH 7.5), 150 mm NaCl, 1 mm EGTA, 100 mm NaF, 10 mm Na2P2O7, 1 mm Na3VO4, 1 mm PMSF, 1 μg/ml bestatin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) and incubated for 5 min on ice. The cell lysates were sonicated at 4 °C for 2.5 min and centrifuged for 10 min at 15,000 × g. The supernatants were collected and stored in aliquots at −80 °C. Cell fractionations into cytoplasmic, soluble nuclear, and chromatin-bound fractions were performed as described in Ref. 12.
Western Blot Analysis
Western blot analysis was performed according to standard protocols and visualized by the Odyssey image analysis system (LI-COR).
Antibodies and Proteins
The antibodies (ABs) against MutYH and GST were purchased from Santa Cruz Biotechnology. The Mule AB was from Bethyl Laboratories (Montgomery, TX), the tubulin AB from Sigma-Aldrich, and the histone H3 AB from Abcam (Cambridge, UK). The FLAG-octapeptide and the Actin AB were from Sigma, the HA AB from Covance (Princeton, NJ) and the His AB from Qiagen. Recombinant MutYH was expressed and purified as described in Ref. 16 and the homologous to the E6-AP carboxyl terminus (HECT) domain of Mule according to Ref. 14.
RNA Interference
Cells were transfected using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions (7.5 pmol of siRNA and 10 μl of RNAiMax/10-cm plate) and harvested 72 h after transfection.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using PfuTurbo pol from Stratagene according to the manufacturer's instructions. The primers (Microsynth) were designed using PrimerX: MutYH KK477RR, 5′-GTTTCCACCGCCATGAGAAGGGTTTTCCGTGTGTATC-3′ and 5′-GATACACACGGAAAACCCTTCTCATGGCGGTGGAAAC-3′; K495R, 5′-CCTGTATGGGTTCCAGAAGGTCCCAGGTGTC-3′ and 5′-GACACCTGGGACCTTCTGGAACCCATACAGG-3′; KK506RR, 5′-CCGTGCAGTCGGAGAAGGCCCCGCATGGGCC-3′ and 5′-GGCCCATGCGGGGCCTTCTCCGACTGCACGG-3′; HECT C326A, 5′-CTGCCTTCAGCTCACACAGCTTTTAATCAGCTGGATC-3′ and 5′-GATCCAGCTGATTAAAAGCTGTGTGAGCTGAAGGCAG-3′. The primers were used on the pcDNA3 HA-MutYH and the pcDNA3 FLAG-MuleΔN447 plasmids, and the mutagenesis was confirmed by sequencing.
Quantitative PCR
Total RNA was isolated from cells using the Nucleo Spin RNAII kit (Macherey & Nagel (Düren, Germany)), RT-PCR was performed using the Qiagen OneStep RT-PCR kit. Quantitative PCR was performed using SYBR Green (Qiagen), 150 ng of total RNA, and the oligonucleotides (Microsynth) as follows: HA-MutYH, 5′-GCCAGCAAGTCCTGGATAAT-3′ and 5′-ATGCGTAGTCAGGCACGTC-3′; MutYH, 5′-CCAGAGAGTGGAGCAGGAAC-3′ and 5′-TTTCTGGGGAAGTGGACCAC-3′; L28, 5′-GCAATTCCTTCCGCTACAAC-3′ and 5′-TGTTCTTGCGGATCATGTGT-3′. L28 was used as internal standard.
KBrO3 Treatment and Cell Based Mutagenesis Assay
The cell-based mutagenesis assays were performed using the A2780 cell line as described in Ref. 17. Briefly, the cells were transfected with plasmid or siRNA as indicated in Fig. 6; 24 h later the cells were seeded for treatment with KBrO3 (concentrations as indicated, 30 min). After culturing the treated cells for 7 days, they were reseeded in five plates/cell line (1 × 105 cells/10-cm plate), and cells harboring a mutation in the HPRT gene were selected by adding 6-thioguanine (5 μg/ml; Sigma). In parallel each cell line was plated (150 cells/6-well plate) in triplicate without the addition of 6-thioguanine to determine the seeding efficiency. The mutation rate was calculated after 14 days under selection, as the ratio of the number of mutant colonies to the number of clone-forming cells, as also described in Ref. 17. Three independent experiments including always the control, Mule knockdown (KD), and Mule overexpression cells were performed.
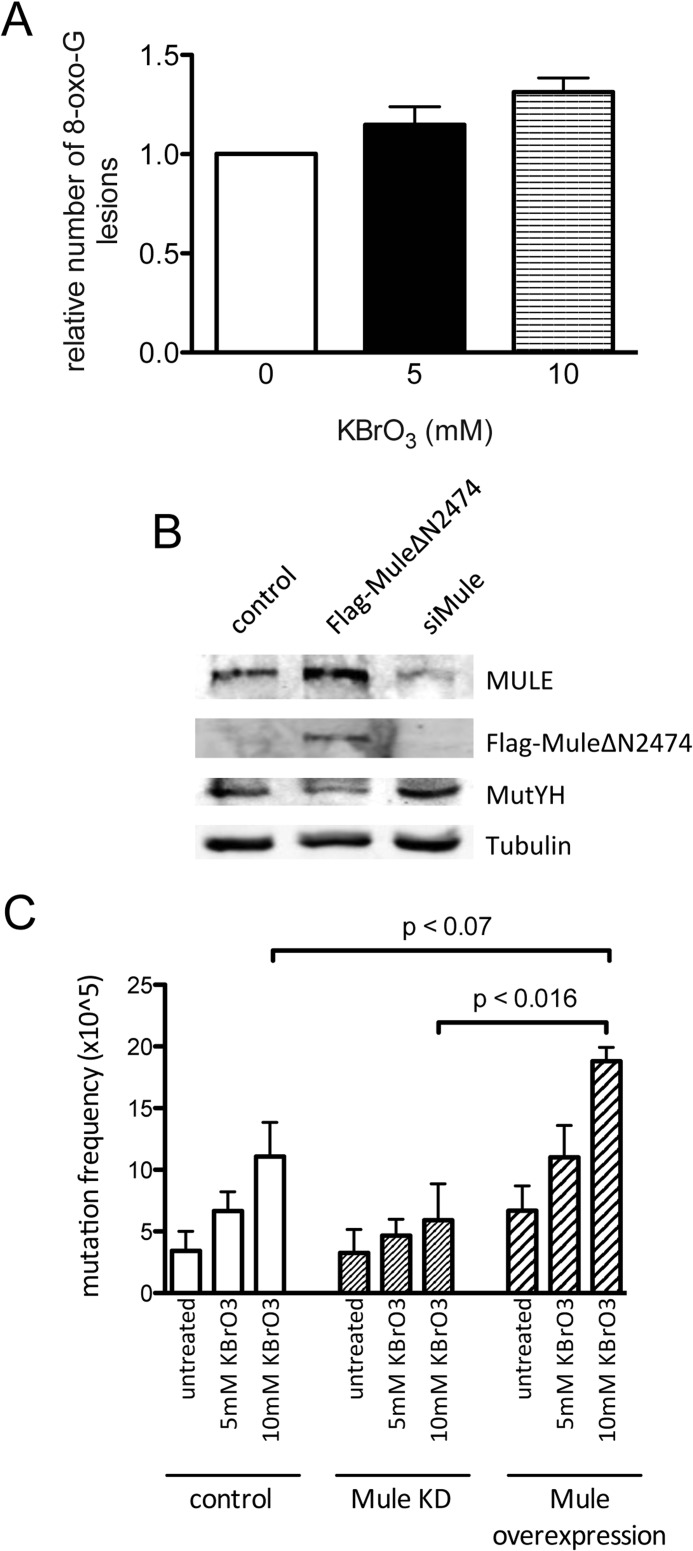
FIGURE 6.
Reactive oxygen species treatment increases the 8-oxo-G levels and the mutation frequency in A2780 ovarian cancer cells. A, A2780 cells were treated with increasing amounts of KBrO3, and the DNA was isolated and analyzed for 8-oxo-G as described under “Experimental Procedures.” The relative result of two independent experiments, normalized to the untreated cells, is shown. B, effect of Mule KD and FLAG-Mule ΔN2474 overexpression in A2780 cells, analyzed by Western blotting. C, mutation frequency at the HPRT locus in A2780 cells, either treated with Mule siRNA or transfected with FLAG-Mule ΔN2474 and, as indicated, incubated with increasing amounts of KBrO3. Error bars, S.D.
GST Pulldown Assay
The GST pulldown assay was performed using recombinant and purified GST-tagged MutYH (2 μg) coupled to GST-Sepharose beads (GE Healthcare) and incubated with 800 ng of recombinant HECT domain of Mule. The assay was performed as described in detail in Ref. 16.
Pulldown Assay
Cell extract containing overexpressed FLAG-tagged HECT domain of Mule was incubated with FLAG AB (1 μg) and 25 μl of protein G-Sepharose beads (GE Healthcare) for 2 h. After washing three times with a buffer containing PBS with 0.2% Triton X-100, 1 mm DTT, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml bestatin, and 1 mm PMSF 800 ng of recombinant purified protein was added to the beads. After washing three times, the beads were finally resuspended in Laemmli buffer and the samples analyzed by Western blotting.
Detection of Protein Ubiquitination
The protocol described in detail in Ref. 18 was used to eliminate all noncovalent protein binding.
Co-immunoprecipitation
Cell extracts were prepared as described, diluted 1:2 in HNTG buffer (20 mm Hepes (pH 7.5), 150 mm NaCl, 10% glycerol, 0.1% Triton-X-100, 10 mm NaF, 1 mm Na3VO4) and incubated with 2 μg of AB as indicated and 25 μl of protein G-Sepharose beads. After rotating for 2 h at 4 °C the beads were washed three times with HNTG buffer, resuspended in Laemmli buffer, and finally analyzed by Western blotting.
In Vitro Ubiquitination Assay
In vitro ubiquitination was performed as described in Ref. 14. Briefly, the assay was performed in a 15-μl reaction mixture containing E1 activating enzyme (0.7 pmol), E2 conjugating enzymes (9.5 pmol), and ubiquitin (0.6 nmol; Boston Chemicals). The reaction was carried out in buffer containing 25 mm Tris-HCl (pH 8.0), 4 mm ATP, 5 mm MgCl2, 200 μm CaCl2, 1 mm DTT, 10 mm MG132 for 1 h at 30 °C. Finally, Laemmli buffer was added, and the samples were analyzed by Western blotting.
DNA Isolation and 8-Oxo-G Determination
Approximately 2 × 107 cells were harvested and immediately stored at −80 °C. The DNA was isolated with the DNA Isolation kit for Cells and Tissues (Roche Applied Science) according to the manufacturer's instructions. The 8-oxo-G determination was carried out following the protocol depicted in Ref. 19.
Microscopy
HeLa cells were transfected with plasmids as indicated using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were seeded on glass coverslips and fixed using methanol for 20 min at −20 °C. After blocking with PBS/10% FCS the cells were incubated with the indicated primary AB (1:300 in PBS/10% FCS) for 2 h. The coverslips were washed three times with PBS/10% FCS (30 min), incubated with the secondary Cy3 (1:300 in PBS/10% FCS) for 1 h, washed three times, and fixed on object slides using Vectashield (Vector Laboratories). The pictures were taken using the Leica CTR 6000.
Statistical Analysis
For all statistical analysis the program GraphPad Prism was used. All experiments shown were repeated at least three times, if not stated differently in the figure legends. The shown error bars correspond to the S.D. Student's t test was used to evaluate the significance of at least three (otherwise as stated in the figure legends) independent experiments.
RESULTS
MutYH Is Ubiquitinated in Vitro and in Vivo by the E3 Ubiquitin Ligase Mule
Former studies identified the E3 ubiquitin ligase Mule as a regulator of the steady state levels of different proteins involved in BER (12, 14, 20). Here, we were aiming at the identification of the regulatory mechanisms controlling the MutYH protein levels in the cell. Using an in vitro ubiquitination approach we tested whether MutYH was a substrate for the E3 ubiquitin ligase Mule. Mule is a 482-kDa protein whose catalytic center lies within the HECT domain, mapped to the C-terminal 370 amino acids (21). Using the recombinant, purified HECT domain in an in vitro ubiquitination assay (Fig. 1A), we found that MutYH was ubiquitinated by the HECT domain in presence of the E2 conjugating enzymes H5b, H5c, and H7. To get more distinct bands a mutant ubiquitin, not able to form polyubiquitin chains, was used in all in vitro assays.
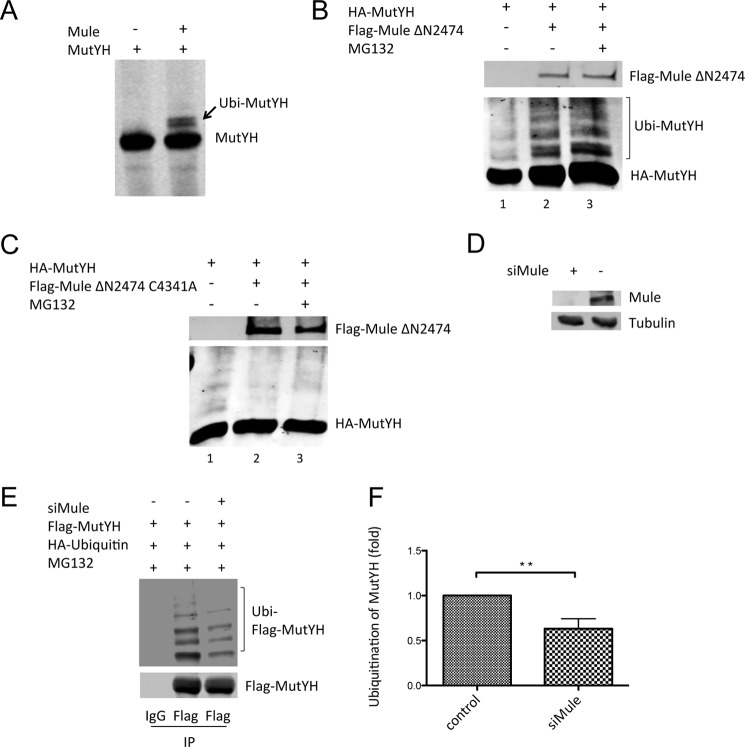
FIGURE 1.
MutYH is ubiquitinated in vitro and in vivo by the E3 ubiquitin ligase Mule. A, in vitro ubiquitination of MutYH by the HECT domain of Mule, using recombinant and purified proteins. B, co-transfection of FLAG-Mule ΔN2474 and HA-MutYH into HEK293T cells. The cells were treated with 10 μm MG132 for 16 h prior to harvest. C, co-transfection of FLAG-Mule ΔN2474 C4341A and HA-MutYH into HEK293T cells. The cells were treated with 10 μm MG132 for 16 h prior to harvest. D, Mule KD in HEK293T cells, analyzed by Western blotting. 5% of total cell extracts were loaded as input from E. E, immunoprecipitation (IP) of FLAG-MutYH, co-transfected with HA-ubiquitin. The cells were treated with Mule siRNA (efficiency confirmed in D) or scrambled (scr) siRNA as indicated, and the samples were analyzed by Western blotting. F, quantification of E. The amount of ubiquitinated FLAG-MutYH (upper panel) in every lane was normalized to the corresponding unmodified FLAG-MutYH bound to the beads (lower panel). The Mule KD sample was compared with the control (p ≤ 0.004). Error bar, S.D.
We next addressed the question of whether Mule also plays a role in the regulation of MutYH in vivo. We transfected an N-terminal deletion construct of Mule containing the full HECT domain (Mule ΔN2474) together with MutYH for overexpression into HEK293T cells. As shown in Fig. 1B the co-transfection led to the appearance of higher migrating MutYH bands (lanes 2 and 3) in contrast to the control (lane 1) in which just one form of MutYH was present. The fact that addition of the proteasomal inhibitor MG132 (lane 3) to the transfected cells prior to harvest resulted in an increased intensity in the higher migrating bands of MutYH further suggested that these bands represented an ubiquitinated form of the protein. To ensure that the catalytic activity of Mule was required for the modification of MutYH and to strengthen the result shown in Fig. 1B, an additional experiment was performed in parallel using a catalytic dead mutant of Mule (Mule C4341A). In contrast to the experiment using the Mule WT, the co-transfection of the Mule C4341A mutant together with MutYH (lanes 2 and 3) did not result in any higher migrating forms of MutYH (Fig. 1C). These results clearly showed that MutYH is modified by the catalytic activity of Mule.
We performed a transient KD of Mule in HEK293T cells (Fig. 1D) that overexpressed MutYH and ubiquitin. After inhibition of the proteasomal degradation by adding MG132 we purified MutYH by binding to beads and analyzed the amount of ubiquitinated MutYH (Fig. 1E). The KD of Mule resulted in a 40% reduction of modified MutYH, if normalized to the amount of unmodified MutYH bound to the beads (Fig. 1F), indicating that Mule is the enzyme responsible for ubiquitination of MutYH. The observed decrease in ubiquitinated MutYH upon KD of Mule further supports its role as the E3 ligase for MutYH ubiquitination.
MutYH Protein Levels Depend on the Amount of the E3 Ubiquitin Ligase Mule
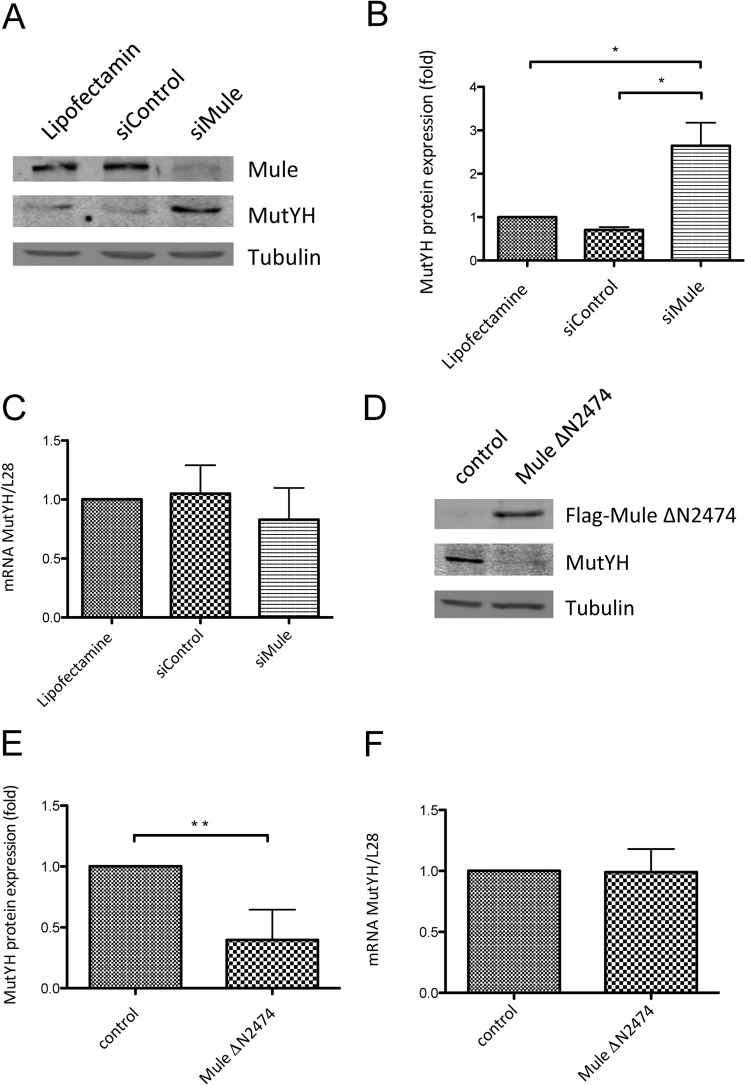
To address the relevance of regulation by Mule in vivo, we performed a transient KD of Mule using siRNA to see an effect on the endogenous protein levels of MutYH. As shown in Fig. 2A and quantified in Fig. 2B the KD of Mule led to a significant increase in MutYH protein levels, whereas neither the treatment with Lipofectamine nor with scrambled siRNA showed this effect. Unchanged mRNA levels of MutYH in these cells (Fig. 2C) were indicative that the regulation of MutYH (Fig. 1, A and B) took place at the protein level. Consistently we observed the opposite effect upon Mule overexpression. The transfection of HEK293T cells with a Mule construct caused a dramatic decrease in the endogenous protein levels of MutYH compared with the empty vector control, as shown in Fig. 2D and quantified in Fig. 2E. As expected, the MutYH mRNA levels did not change in these cells upon overexpression of Mule (Fig. 2F). The results showing that the amount of Mule in the cells directly influences the protein levels of MutYH led us to conclude that indeed Mule is the E3 ubiquitin ligase responsible for the regulation of MutYH on protein level in vivo.
FIGURE 2.
MutYH protein levels depend on the amount of the E3 ubiquitin ligase Mule. A, effect of Mule KD in HEK293T cells, analyzed by Western blotting. B, quantification of protein levels shown in A (p < 0.048 and p < 0.035). The protein levels of MutYH were normalized to tubulin. C, quantification of MutYH mRNA extracted from the samples shown in A. The MutYH mRNA was normalized to the levels of L28 mRNA. D, effect of FLAG-Mule ΔN2474 overexpression in HEK293T cells, analyzed by Western blotting. E, quantification of protein levels shown in D (p ≤ 0.01). The protein levels of MutYH were normalized to tubulin. F, quantification of the MutYH mRNA extracted from the samples shown in D. The MutYH mRNA was normalized to the levels of L28 mRNA. Error bars, S.D.
MutYH and Mule Physically Interact
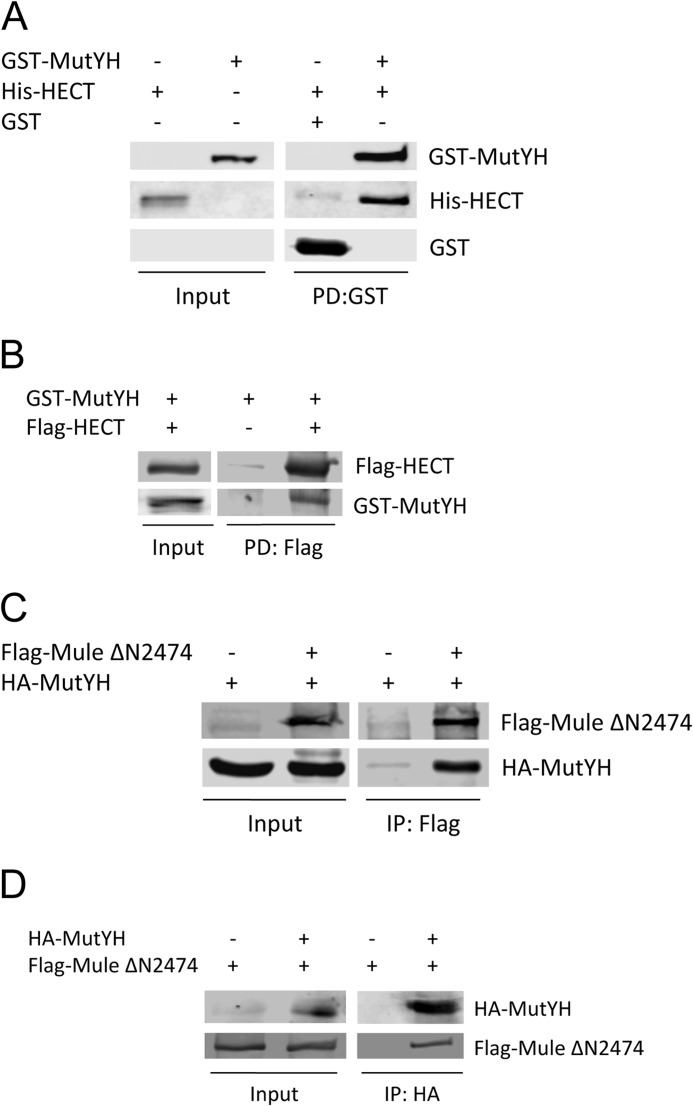
We questioned whether MutYH and Mule directly interact. In the first approach we could confirm the physical interaction between the two proteins by performing pulldown assays using tagged, recombinant and purified proteins. The pulldown of GST-tagged MutYH also enriched the HECT domain, demonstrating its direct interaction with MutYH (Fig. 3A). Moreover, the pulldown of the HECT domain using FLAG-beads confirmed the interaction between the two proteins (Fig. 3B).
FIGURE 3.
MutYH and Mule physically interact. A, GST pulldown of His-HECT with GST-MutYH. The recombinant and purified proteins were incubated with GST beads. The input corresponds to 20% of the used proteins. B, pulldown of GST-MutYH with FLAG-HECT. FLAG AB-coupled beads were incubated with cell extracts containing FLAG-HECT or vector control, washed, and incubated with the recombinant and purified GST-MutYH. The input corresponds to 10% of the used proteins. C, co-immunoprecipitation of HA-MutYH with FLAG-Mule ΔN2474. Whole cell extracts containing the overexpressed proteins or vector control were incubated with FLAG AB coupled to beads. The input corresponds to 20% of the used proteins. D, co-immunoprecipitation of FLAG-Mule ΔN2474 with HA-MutYH. Whole cell extracts containing the overexpressed proteins or vector control were incubated with HA AB coupled to beads. The input corresponds to 10% of the used proteins. PD, pulldown; IP, immunoprecipitation.
Finally, we showed that the interaction could be also recapitulated in cells. We performed co-immunoprecipitation experiments using whole cell extract containing overexpressed FLAG-tagged Mule and HA-tagged MutYH. By binding of Mule to FLAG-beads we could show that MutYH co-immunoprecipitated (Fig. 3C). The interaction could be also confirmed vice versa by coupling MutYH to HA-beads and analyzing the samples for co-immunoprecipitated Mule (Fig. 3D), strengthening the point that Mule directly interacts with MutYH in a cellular context.
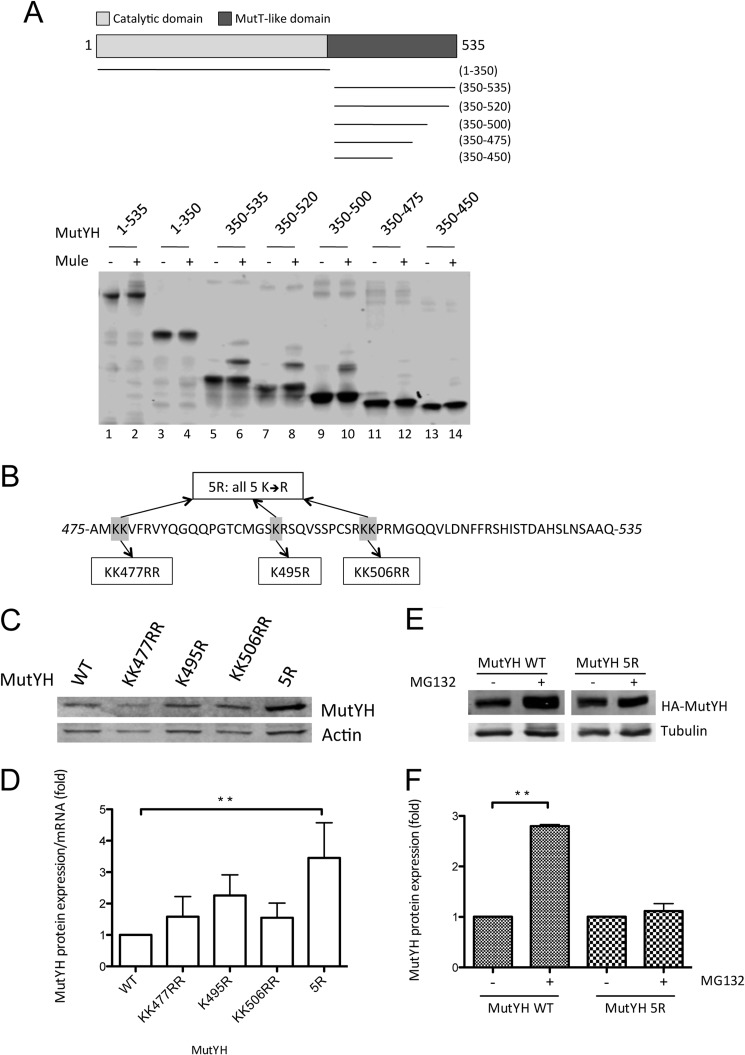
MutYH Is Ubiquitinated between Amino Acids 475 and 535, and Mutation of the Five Lysine Residues Located in this Part Stabilizes MutYH on the Protein Level
Ubiquitin chains are covalently attached to lysine residues on the target protein. The sequence of MutYH reveals a total of 17 lysine residues distributed all over the protein, with an accumulation on the C- and N-terminal sites. To identify the lysine residues that act as targets for ubiquitination by Mule we generated deletion constructs of MutYH (Fig. 4A): one construct spanning the N-terminal part (amino acids 1–350) containing the glycosylase domain of MutYH, the other construct consisting of the C-terminal MutT-like domain (amino acids 350–535). Using these constructs in an in vitro ubiquitination approach we could identify the MutT like domain of MutYH (lane 6) to be ubiquitinated by Mule, whereas the N-terminal part of MutYH was not modified (lane 4, Fig. 4A). To narrow the region we generated further deletion constructs of the MutT-like domain. Once the amino acids 475–535 were deleted the MutYH construct was not ubiquitinated any more (lanes 12 and 14) leading finally to the conclusion that the C-terminal 60 amino acids of MutYH contain the ubiquitination sites. The sequence of MutYH (Fig. 4B) contains five lysine residues within this region as potential targets for ubiquitination (Fig. 4B). To determine which function these lysine residues have for the regulation of MutYH we generated ubiquitination-deficient, lysine to arginine point mutants: KK477RR, K495R, KK506RR, and one construct containing all five lysine residues mutated to arginine (5R) (Fig. 4B). Consistent with the idea that ubiquitination of MutYH affects the proteasomal degradation of MutYH all lysine point mutants showed stabilization on the protein level if transfected in HEK293T cells (Fig. 4C). The mRNA amount of the transfected mutants was analyzed, and the protein levels were normalized to that (Fig. 4D) to show that the increase in MutYH was caused by a stabilization of the mutants on protein level rather than by differences in the transfection efficiency. The 5R mutant showed a more pronounced increase in protein level (4-fold compared with WT) than the single mutants (1.5–2.5-fold) (Fig. 4, C and D), leading to the assumption that all five lysine residues appear to contribute to the ubiquitination of MutYH. Strikingly, the treatment of cells with the proteasome inhibitor MG132 caused a dramatic increase in the protein level of the MutYH WT compared with the ubiquitination-deficient mutant 5R that is just slightly stabilized (Fig. 4, E and F). This result further underlines the fact that the MutYH 5R mutant is protected against ubiquitination and proteasomal degradation.
FIGURE 4.
MutYH is ubiquitinated between amino acids 475 and 535, and mutation of the five lysine residues located in this part stabilizes MutYH on the protein level. A, in vitro ubiquitination of different recombinant and purified MutYH deletion constructs, as depicted in the upper part. B, amino acid sequence of MutYH between positions 475 and 535. Highlighted are the lysine residues that were mutated into arginine. C, transfection of the MutYH point mutant constructs (as indicated) into HEK293T cells, analyzed by Western blotting. D, normalization of the mutant MutYH protein level as shown in C to the mRNA level of the respective expression construct (p < 0.004). The protein levels were normalized to actin and the mRNA levels to L28 mRNA. E, transfection of the MutYH WT and 5R construct into HEK293T cells. The cells were treated with 10 mm MG132 as indicated, 16 h prior to harvest. The samples were analyzed by Western blotting. F, quantification of the protein levels shown in E (p < 0.01). The MutYH levels were normalized to tubulin. Presented are the results of two independent experiments.
Ubiquitination-deficient MutYH Is Preferentially Bound to Chromatin
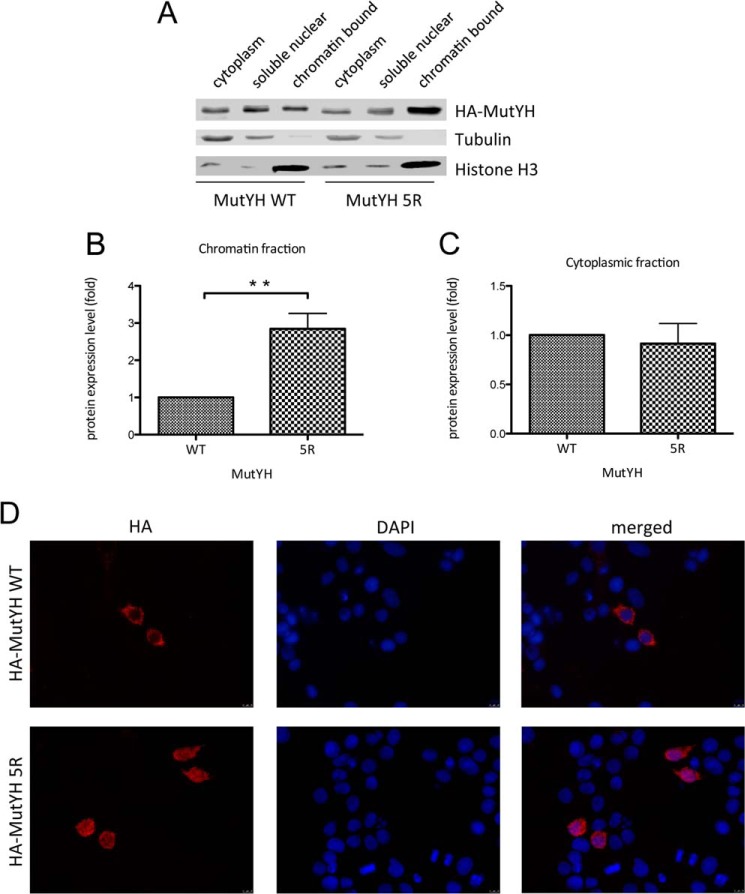
Besides protein stability, ubiquitination can also influence the intracellular localization of a protein (22). To figure out whether ubiquitination of MutYH determines its localization within the cell, cell fractionation experiments were performed (Fig. 5A). HEK293T cells transfected with either the MutYH WT or the 5R mutant were fractionated into cytoplasmic, soluble nuclear and chromatin-bound fractions. We observed an enrichment of the ubiquitination-deficient MutYH mutant in the chromatin-bound fraction with respect to the WT (Fig. 5B), whereas the amount of MutYH in the cytoplasm remained comparable (Fig. 5C). In addition to the cellular fractionation, this effect could be further confirmed by using an immunocytochemistry approach. HeLa cells were transfected with the HA-tagged MutYH WT or the 5R mutant and stained for HA and the nuclear marker DAPI. The result clearly showed that the MutYH WT was exclusively localized in the cytoplasm whereas the 5R mutant was also present in the nucleus to a substantial amount (Fig. 5D). Taken together, these results indicated that ubiquitination of MutYH not only influences its turnover but also changes its recruitment to chromatin.
FIGURE 5.
Ubiquitination-deficient MutYH is preferentially bound to chromatin. A, transfection of MutYH WT or 5R mutant into HEK293T cells. The cells were fractionated and analyzed by Western blotting. B, quantification of the MutYH protein levels in the chromatin-bound fraction shown in A (p < 0.002). The protein levels were normalized to the histone H3. C, quantification of the MutYH protein level in the cytoplasmic fraction shown in A. The protein levels were normalized to tubulin. D, fluorescence microscope images of HeLa cells transfected with MutYH WT or the 5R mutant. The cells were fixed and stained with HA AB and DAPI.
Reactive Oxygen Species Treatment Increases the 8-Oxo-G Levels and the Mutation Frequency in A2780 Ovarian Cancer Cells
MutYH plays an important role in the repair of 8-oxo-G thereby preventing the onset of deleterious mutations (23, 24). Consequently, we expected changes in the mutation frequency according to the amount of MutYH present in the cells. A2780 cells, either transfected with a Mule construct (Mule ΔN2474) or Mule siRNA, were treated with KBrO3 to stimulate the formation of 8-oxo-G lesions. From Fig. 6A it is evident that the 8-oxo-G levels in the cells increased upon treatment with increasing amounts of KBrO3, as determined in a mass spectrometry-based assay. The observed levels of 8-oxo-G are in the range of 1–5.5 8-oxo-G/106 nucleotides for the untreated cells (data not shown) and are in accordance with the numbers reported in literature (25). The results indicate a clear tendency of a higher oxidative burden for the cell upon treatment with KBrO3.
We have already shown that the MutYH levels depend on the amount of Mule (Fig. 2). Such a relationship was also confirmed in the A2780 cell system. Fig. 6B clearly shows an increase of endogenous MutYH protein levels upon KD of Mule and, conversely, a decrease upon Mule overexpression. The mutation frequency at the HPRT locus in these cells was analyzed by selection with 6-thioguanine, a toxic guanine analogue that is not incorporated in the DNA by cells harboring a mutation at the HPRT locus. Mule KD cells had a higher MutYH level (Fig. 6B) and a lower incidence of mutations in the HPRT locus (Fig. 6C). The opposite was observed for the Mule (Mule ΔN2474)-overexpressing cells, showing a decrease in MutYH level with respect to the control (Fig. 6, B and C).
These data confirmed that Mule-dependent regulation of MutYH influences the susceptibility of cells to mutagenesis caused by exposure to oxidative stress. Thus, the regulation of MutYH has a direct impact on the mutation frequency in cells, underlining again the importance of a tight and controlled regulation of the BER machinery.
DISCUSSION
In this work we provide evidence that ubiquitination regulates not only the steady state levels of MutYH, but also its subcellular localization. We identified Mule to be the E3 ubiquitin ligase responsible for the modification of MutYH. Mule mainly appears to monoubiquitinate MutYH. Whether other E3 ubiquitin ligases are involved in the polyubiquitination and proteasomal degradation remains so far elusive. Consistent with the predicted role of Mule in the proteasomal degradation of MutYH, we find the MutYH protein levels to be reversely correlated with the levels of Mule present in HEK293T cells. Five lysine residues could be identified as targets for ubiquitination by Mule. The clear stabilization of the ubiquitination-deficient MutYH mutants confirms their role in the proteasomal degradation of the protein.
The enzymatic activity of MutYH needs to be directed to the nucleus to act on the newly synthesized strand immediately after replication, to prevent the formation of deleterious G:C→T:A transversion mutations (15, 26). MutYH was already shown to be localized in different cellular compartments (27). Our data implicate that the ubiquitination of MutYH plays an important role in enrichment of MutYH on chromatin. We observed that mutation of the ubiquitination sites results in strong accumulation of MutYH in the chromatin-bound cell fraction, respectively, in the nucleus as shown by immunofluorescence. We previously found that the recruitment of pol λ to chromatin is mediated by interplay of ubiquitination and phosphorylation as well as by formation of active repair complexes on chromatin (12, 28, 29). Comprising the recent data it seems very likely that MutYH might coordinate the MutYH/pol λ-dependent LP-BER pathway (16). Whether it is an orchestration of different posttranslational modification adjusting the subcellular localization and protein levels of MutYH remains to be elucidated.
As noted in the literature, a cell contains 1000–7000 8-oxo-G as a steady state level that can be repaired any time (25). MutYH has an important role in the repair of these lesions, thus misregulation or mutations of the enzyme cause an increased mutation frequency (10, 30). In accordance we see differences in the mutation frequency depending on the levels of MutYH. In general, the mutation frequency at the HPRT locus increases upon KBrO3 treatment of A2780 cells. Further, the data suggest that cells with elevated MutYH levels, due to KD of Mule, are better able to cope with oxidative stress than cells with a lower amount of MutYH, upon Mule overexpression. These results are underlined by a very recent publication showing that the amount of MutYH influences directly the susceptibility of cells to oxidative damage as well as the amount of 8-oxo-G upon treatment with hydrogen peroxide (31). To elucidate whether this regulation of MutYH has an impact in the context of cancer requires additional research.
In conclusion, the data presented here underline the importance of a tight and controlled regulation of BER, because already small differences in protein levels can have a pivotal effect on the genome integrity. In line with previous results, showing BER (12–14) enzymes to be regulated by posttranslational modification to ensure a faithful repair of 8-oxo-G lesions, we observed for the first time a similar regulation for the essential MutYH DNA glycosylase.
Acknowledgments
We thank R. Santoro and J. Jiricny for cells used in this study and Q. Zhong for the Mule expression construct; G. Dianov and S. Khoronenkova for valuable input and expertise; M. Ghodgaonkar for support in the experimental setup of the cell-based mutagenesis assay; and G. Maga for critical reading of the manuscript and suggestions.
Footnotes
- 8-oxo-G
- 7,8-dihydro-8-oxoguanine
- AB
- antibody
- BER
- base excision repair
- HECT
- homologous to the E6-AP C terminus
- KD
- knockdown
- pol
- polymerase.
REFERENCES
- 1. Collins A. R. (1999) Oxidative DNA damage, antioxidants, and cancer. BioEssays 21, 238–246 [DOI] [PubMed] [Google Scholar]
- 2. Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maga G., Crespan E., Wimmer U., van Loon B., Amoroso A., Mondello C., Belgiovine C., Ferrari E., Locatelli G., Villani G., Hübscher U. (2008) Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc. Natl. Acad. Sci. U.S.A. 105, 20689–20694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U. (2007) 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447, 606–608 [DOI] [PubMed] [Google Scholar]
- 5. Braithwaite E. K., Kedar P. S., Lan L., Polosina Y. Y., Asagoshi K., Poltoratsky V. P., Horton J. K., Miller H., Teebor G. W., Yasui A., Wilson S. H. (2005) DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 280, 31641–31647 [DOI] [PubMed] [Google Scholar]
- 6. Hirano S., Tominaga Y., Ichinoe A., Ushijima Y., Tsuchimoto D., Honda-Ohnishi Y., Ohtsubo T., Sakumi K., Nakabeppu Y. (2003) Mutator phenotype of MUTYH-null mouse embryonic stem cells. J. Biol. Chem. 278, 38121–38124 [DOI] [PubMed] [Google Scholar]
- 7. Russo M. T., De Luca G., Casorelli I., Degan P., Molatore S., Barone F., Mazzei F., Pannellini T., Musiani P., Bignami M. (2009) Role of MUTYH and MSH2 in the control of oxidative DNA damage, genetic instability, and tumorigenesis. Cancer Res. 69, 4372–4379 [DOI] [PubMed] [Google Scholar]
- 8. Sakamoto K., Tominaga Y., Yamauchi K., Nakatsu Y., Sakumi K., Yoshiyama K., Egashira A., Kura S., Yao T., Tsuneyoshi M., Maki H., Nakabeppu Y., Tsuzuki T. (2007) MUTYH-null mice are susceptible to spontaneous and oxidative stress-induced intestinal tumorigenesis. Cancer Res. 67, 6599–6604 [DOI] [PubMed] [Google Scholar]
- 9. Shinmura K., Goto M., Suzuki M., Tao H., Yamada H., Igarashi H., Matsuura S., Maeda M., Konno H., Matsuda T., Sugimura H. (2011) Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J. Pathol. 225, 414–423 [DOI] [PubMed] [Google Scholar]
- 10. Parker A. R., O'Meally R. N., Oliver D. H., Hua L., Nelson W. G., DeWeese T. L., Eshleman J. R. (2002) 8-Hydroxyguanosine repair is defective in some microsatellite stable colorectal cancer cells. Cancer Res. 62, 7230–7233 [PubMed] [Google Scholar]
- 11. Khoronenkova S. V., Dianov G. L. (2013) USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res. 41, 1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Markkanen E., van Loon B., Ferrari E., Parsons J. L., Dianov G. L., Hübscher U. (2012) Regulation of oxidative DNA damage repair by DNA polymerase λ and MutYH by cross-talk of phosphorylation and ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 109, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parsons J. L., Tait P. S., Finch D., Dianova I. I., Allinson S. L., Dianov G. L. (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell 29, 477–487 [DOI] [PubMed] [Google Scholar]
- 14. Parsons J. L., Tait P. S., Finch D., Dianova I. I., Edelmann M. J., Khoronenkova S. V., Kessler B. M., Sharma R. A., McKenna W. G., Dianov G. L. (2009) Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 28, 3207–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boldogh I., Milligan D., Lee M. S., Bassett H., Lloyd R. S., McCullough A. K. (2001) hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 29, 2802–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Loon B., Hübscher U. (2009) An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc. Natl. Acad. Sci. U.S.A. 106, 18201–18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blasi M. F., Ventura I., Aquilina G., Degan P., Bertario L., Bassi C., Radice P., Bignami M. (2006) A human cell-based assay to evaluate the effects of alterations in the MLH1 mismatch repair gene. Cancer Res. 66, 9036–9044 [DOI] [PubMed] [Google Scholar]
- 18. El-Shemerly M., Janscak P., Hess D., Jiricny J., Ferrari S. (2005) Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 65, 3604–3609 [DOI] [PubMed] [Google Scholar]
- 19. Taghizadeh K., McFaline J. L., Pang B., Sullivan M., Dong M., Plummer E., Dedon P. C. (2008) Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 3, 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khoronenkova S. V., Dianova I. I., Ternette N., Kessler B. M., Parsons J. L., Dianov G. L. (2012) ATM-dependent down-regulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol. Cell 45, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 22. Laine A., Ronai Z. (2007) Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 26, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Russo M. T., De Luca G., Degan P., Parlanti E., Dogliotti E., Barnes D. E., Lindahl T., Yang H., Miller J. H., Bignami M. (2004) Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 64, 4411–4414 [DOI] [PubMed] [Google Scholar]
- 24. Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D. E., Lindahl T., McIlhatton M., Fishel R., Miller J. H. (2004) Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 64, 3096–3102 [DOI] [PubMed] [Google Scholar]
- 25. Friedberg E., Walker G., Siede W. (2006) DNA Repair and Mutagenesis, 2nd Ed., p. 16, ASM Press, Washington, D. C [Google Scholar]
- 26. Markkanen E., Dorn J., Hübscher U. (2013) MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA. Front. Genet. 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takao M., Zhang Q. M., Yonei S., Yasui A. (1999) Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res. 27, 3638–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wimmer U., Ferrari E., Hunziker P., Hübscher U. (2008) Control of DNA polymerase λ stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep. 9, 1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frouin I., Toueille M., Ferrari E., Shevelev I., Hübscher U. (2005) Phosphorylation of human DNA polymerase λ by the cyclin-dependent kinase Cdk2/cyclin A complex is modulated by its association with proliferating cell nuclear antigen. Nucleic Acids Res. 33, 5354–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruggieri V., Pin E., Russo M. T., Barone F., Degan P., Sanchez M., Quaia M., Minoprio A., Turco E., Mazzei F., Viel A., Bignami M. (2013) Loss of MUTYH function in human cells leads to accumulation of oxidative damage and genetic instability. Oncogene 32, 4500–4508 [DOI] [PubMed] [Google Scholar]
- 31. Hwang B. J., Shi G., Lu A. L. (2014) Mammalian MutY homolog (MYH or MUTYH) protects cells from oxidative DNA damage. DNA Repair 13, 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]