Background: Ovodefensins are small peptides from eggs, related to avian antimicrobial defensins.
Results: The first three-dimensional structure of ovodefensins (gallin) is solved, and its antimicrobial properties are screened.
Conclusion: Gallin adopts a β-defensin fold, with significant variations. Its antibacterial spectrum was restricted to E. coli.
Significance: The first structural features may be related to E. coli specificity and/or other yet unknown functions.
Keywords: Antimicrobial Peptides, Defensins, Host Defense, NMR, Protein Structure, AvBD, SPPS, Ovodefensin
Abstract
Gallin is a 41-residue protein, first identified as a minor component of hen egg white and found to be antimicrobial against Escherichia coli. Gallin may participate in the protection of the embryo during its development in the egg. Its sequence is related to antimicrobial β-defensin peptides.
In the present study, gallin was chemically synthesized 1) to further investigate its antimicrobial spectrum and 2) to solve its three-dimensional NMR structure and thus gain insight into structure-function relationships, a prerequisite to understanding its mode(s) of action. Antibacterial assays confirmed that gallin was active against Escherichia coli, but no additional antibacterial activity was observed against the other Gram-positive or Gram-negative bacteria tested. The three-dimensional structure of gallin, which is the first ovodefensin structure to have been solved to date, displays a new five-stranded arrangement. The gallin three-dimensional fold contains the three-stranded antiparallel β-sheet and the disulfide bridge array typical of vertebrate β-defensins. Gallin can therefore be unambiguously classified as a β-defensin. However, an additional short two-stranded β-sheet reveals that gallin and presumably the other ovodefensins form a new structural subfamily of β-defensins. Moreover, gallin and the other ovodefensins calculated by homology modeling exhibit atypical hydrophobic surface properties, compared with the already known vertebrate β-defensins. These specific structural features of gallin might be related to its restricted activity against E. coli and/or to other yet unknown functions. This work provides initial understanding of a critical sequence-structure-function relationship for the ovodefensin family.
Introduction
There is currently a real explosion in antibiotic and multidrug resistance (1–4). A highly promising approach to overcome this issue is to explore and exploit the huge diversity of innovative bioactive-engineered molecules provided by nature to fight off microbes. Among these natural products involved in the defense systems of living organisms, antimicrobial peptides, also called host defense peptides, may represent new clues to investigate (5–10). These natural molecules of innate immunity ensure the defense of a multitude of organisms (plants, insects, and vertebrates) against the majority of pathogenic organisms. Most of them have a wide spectrum of activity, which covers the main bacterial and fungi species but also encapsulated viruses and protozoa (11). Among the host defense peptides, vertebrate defensins are widely studied (for a review, see Ref. 12). To date, these cysteine-rich cationic peptides have been classified in three groups, depending on the disulfide arrangement and the position of the six conserved cysteines: namely α-, β-, and cyclic θ-defensins (the last group being identified exclusively in some primates). Despite their divergence in disulfide pairing, α- and β-defensins share the same three-dimensional fold, composed of a three-stranded antiparallel β-sheet. Whereas both α- and β-defensins are found in mammals, only β-defensins have been to date identified in birds. It is presumed that α-defensins and β-defensins arose from a single ancestral β-defensin-like gene by gene duplication after the divergence of birds and mammals (13).
Avian defensins (14) are small cationic non-glycosylated peptides (<10 kDa) containing six conserved cysteines involved in three disulfide bonds, with the exception of chicken avian β-defensin 11 (AvBD11),3 a double size defensin with 12 cysteines that form six disulfide bridges (15). The consensus sequence XnCX4–6CX3–4CX9CX5–6CCXn corresponds to that of mammal β-defensins. The determination of the first tridimensional NMR structures of king penguin AvBD103b and chicken AvBD2 defensins showed the disulfide bridge motif typical of β-defensins (i.e. C1-C5/C2-C4/C3-C6) (16, 17), thus corroborating this terminology. Last, AvBDs generally possess antimicrobial activities against various microorganisms, including Gram-positive and Gram-negative bacteria as well as fungi (18–21).
The sequencing of the chicken (Gallus gallus) genome revealed the presence of a cluster of 14 different genes on chromosome 3 coding for avian defensins and designated as AvBD1 to -14 (22), with specific tissue distribution (13, 22–25). Proteomic analyses of the chicken egg revealed the presence of AvBDs, especially AvBD9, AvBD10, and AvBD11, identified in the egg yolk, eggshell, and eggshell/egg white/vitelline membrane, respectively (26–29). In addition to these AvBDs, another cysteine-rich and cationic peptide of 41 residues (4731.68 Da), named gallin, was also detected in the hen egg white and vitelline membrane (27, 30). This peptide was named gallin because of its sequence homology with meleagrin and cygnin, two peptides present in the egg whites of turkey (Meleagris) and swan (Cygnus), respectively (31, 32). Two other related peptides, dBPS1 and dBPS2, were also found in duck egg white (33). Their cysteine spacing (CX3–5CX3CX11C3–4CC) provides evidence that they are related to defensins, but the gallin gene does not belong to the cluster of AvBD genes, and its primary structure differs from typical AvBD sequences (34). This new subfamily of defensins was therefore termed ovodefensins. In view of the slight difference in cysteine spacing, it has been proposed that ovodefensins and avian defensins may have diverged from a common ancestor (34). Interestingly, gallin is potentially coded by three different genes (34), which are clustered on chromosome 3 and separated from the cluster of AvBD genes. To date, the ovodefensin subfamily counts seven members, including the two sequences, taeniopygin-1 and taeniopygin-2, predicted from the zebra finch genome (34). Besides the classical members of the ovodefensin subfamily, it is noteworthy that the cysteine spacings of dBPS1 and taeniopygin-2 are slightly different and were first described as “related to ovodefensins” (34).
From a functional point of view, recombinant gallin was shown to be able to inhibit the growth of Escherichia coli (34), thus suggesting that this defensin belongs to the antimicrobial innate immune system. However, its potency against other bacteria, especially Gram-positive bacteria, is still unknown.
The purpose of the present work is therefore to further characterize the functional and structural features of gallin. This study successively describes the chemical synthesis of gallin and then its use for the screening of its antibacterial spectrum and for the resolution of its three-dimensional structure.
EXPERIMENTAL PROCEDURES
Reversed Phase High Performance Liquid Chromatography and Mass Spectrometry
HPLC analyses were carried out on either an Elite LaChrom system, composed of a Hitachi L-2130 pump, a Hitachi L-2455 diode array detector, and a Hitachi L-2200 autosampler, or on a LaChrom 7000 system, composed of a Merck-Hitachi L-7100 pump, a Merck-Hitachi L-7455 diode array detector, and a Merck-Hitachi D-7000 interface, which was also used for semipreparative purification. The machines were equipped with C18 reversed phase columns (Nucleosil), 300 Å, 5 μm, 250 × 4.6 mm for the analytical separations or 250 × 10.5 mm for purification. Solvents A and B containing 0.1% TFA were H2O and MeCN, respectively.
MS analyses were performed on an Autoflex MALDI-TOF instrument (Bruker Daltonics, Bremen, Germany) equipped with a 337-nm nitrogen laser and a gridless delayed extraction ion source. The instrument was used in reflector positive ion mode with a 150-ns delay and an accelerating voltage of 19 kV. Instrument control and external calibration were accomplished using Flex-Control software (Bruker). The observed m/z values correspond to the monoisotopic ions. The sample was co-crystallized with a solution of α-cyano-4-hydroxycinnamic acid as a matrix, using the dry droplet method. Synthetic gallin was further analyzed by MS and MS/MS on a nanoESI-Q-TOF Ultima Global mass spectrometer (Waters, Manchester, UK), as described previously (15). A multicharged precursor ion with an m/z value of 788.42 (charge state 6 (+6)) was selected for the fragmentation (data not shown).
Gallin Synthesis and Oxidative Folding
Solid-phase peptide synthesis was run on an automated synthesizer 433A from Applied Biosystems using Fmoc/tBu chemistry at a 0.1-mmol scale with hexafluorophosphate salt O-(benzotriazol-lyl)-tetramethyl uronium/1-hydroxybenzotriazole as coupling reagents. Fmoc-Lys(Boc)-methylphenoxyproprionic acid (PolyPeptide) (122.37 mg, 0.25 mmol) was manually coupled onto the aminomethyl polyethylene glycol dimethylacrylamide resin (3 g, wet weight, 0.1 mmol) in the presence of hexafluorophosphate O-(7-azabenzotriazol-lyl)-tetramethyl uronium (95 mg, 0.25 mmol) and N,N-diisopropylethylamine (86 μl, 0.5 mmol) for 2 h. The elongation was then carried out automatically using a 10-fold excess of protected amino acids and coupling reagents. The side-chain protecting groups used were Asn(Trt), Cys(Trt), Gln(Trt), His(Trt), Lys(Boc), Ser(tBu), Thr(tBu), Trp(Boc), and Tyr(tBu). The 0.1-mmol scale program purchased from the manufacturer was used, and each coupling step was followed by capping with acetic anhydride. Fmoc deprotection was performed with 20% piperidine in N-methyl-2-pyrrolidone. The peptide resin was then treated for 3 h at room temperature with TFA/H2O/iPr3SiH/phenol (87.5:5:2.5:5). The released peptide was precipitated with ice-cold diethyl ether, recovered by centrifugation, washed three times with ether, dried under vacuum, and analyzed by HPLC and mass spectrometry. The main peak corresponds to the fully reduced form of gallin with a [MH]+ of 4729.5 Da. Calculated [MH+] for C213 H327 N54 O54 S7 was 4729.25 Da.
For purification, the crude reduced product was dissolved in distilled water containing 0.1% TFA, oxidized, and purified by semipreparative C18 reversed-phase HPLC. The oxidative folding was performed by mixing the crude reduced form of gallin at 500 μg/ml with reduced and oxidized glutathione at a molar ratio of 1:100:10 (peptide/GSH/GSSG) in 100 mm Tris-HCl buffer, pH 8.5, containing 1 mm EDTA. The gallin concentration was measured using UV spectrophotometry at 280 nm (ϵ = 21,345 m−1 cm−1).
The time course of an analytical oxidative folding reaction was determined by taking, at intervals, aliquots from the reaction mixture, quenching the oxidation by acidifying the sample using TFA (final concentration 2%), and then analyzing the sample by analytical C18 reversed phase HPLC. Oxidized gallin was eluted earlier than its reduced form, 12 min versus 24.6 min, respectively. Mass spectrometry analysis of the oxidized form of gallin was [MH]+ 4723.38 ± 38 Da in accordance with the calculated [MH+] for C213 H321 N54 O54 S7 (4723.1998 Da with the three theoretical disulfide bonds).
Antimicrobial Activity Assays
Eight different strains were used to assess the antimicrobial activity: Staphylococcus aureus ATCC 29740, Listeria monocytogenes strain EGD, Escherichia coli ATCC 25922, E. coli BEN3578, E. coli CFT073, Salmonella enterica serovar Enteritidis ATCC 13076, S. enterica serovar Enteritidis LA5, and S. enterica serovar Typhimurium ATCC 14028. S. enterica serovar Enteritidis LA5 is a wild-type strain (nalidixic acid-resistant at 20 μg/ml) isolated from natural chicken infections. S. enterica serovar Enteritidis ATCC 13076, and S. enterica serovar Typhimurium ATCC 14028 were purchased from the Centre de Ressources Biologiques de l'Institut Pasteur (Institut Pasteur, Paris, France). S. aureus ATCC 29740 was kindly provided by Dr. Pascal Rainard (INRA, UR1282, Nouzilly, France), L. monocytogenes strain EGD and S. enterica serovar Enteritidis LA5 were kindly provided by Dr. Philippe Velge (INRA, UR1282, Nouzilly, France). E. coli BEN3578, an avian pathogenic strain isolated from the oviduct of a hen with colibacillosis in France in 2010, was kindly provided by Dr. Catherine Schouler (INRA, UR1282, Nouzilly, France). E. coli CFT073 is a human uropathogenic strain whose genome has been sequenced (35).
Antibacterial activities of gallin were measured by a radial diffusion assay according to the method described by Lehrer et al. (36). Bacteria incubated overnight were diluted in trypticase soy broth or brain heart infusion at an absorbance of 0.02 and were incubated for 2.5–4 h, depending on the bacterial strain, at 37 °C to obtain a mid-logarithmic phase culture. Bacterial concentrations were determined by plating serial 10-fold dilutions of bacterial suspension on trypticase soy agar plates and by counting colony-forming units (cfu) after 24 h of incubation at 37 °C. A volume containing bacteria at 1 × 107 cfu was centrifuged at 900 × g for 5 min at 4 °C, and bacteria were washed once with cold 10 mm sodium phosphate buffer (pH 7.4), resuspended in a small volume of cold sodium phosphate buffer, and mixed with 25 ml of previously autoclaved, warm (42 °C) 10 mm phosphate buffer, containing 0.03% trypticase soy broth medium, 1% (w/v) low endosmosis-type agarose (Sigma-Aldrich), and 0.02% Tween 20. The agarose solution containing bacteria was poured into a Petri dish to form a 1-mm-deep uniform layer. A 2.5-mm-diameter gel punch was used to make 36 evenly spaced wells. In each well, 5 μl of peptide dilutions or control solutions were added. MSI-94 (a linear amphipathic magainin variant displaying a broad antimicrobial spectrum) was used as a positive control. MSI-94 was a kind gift from Dr. Philippe Bulet (BioPark, Archamps, France). The peptides were allowed to diffuse in the bacteria-containing gel while the plates were incubated for 3 h at 37 °C. The gel was then overlaid with 25 ml of agar consisting of a double strength (6% (w/v)) solution of trypticase soy broth containing 1% (w/v) agarose. After an overnight incubation at 37 °C, the diameter of the clearing zone surrounding each well was measured. For each bacterial strain, three identical independent measurements of antibacterial activity were performed. The minimum inhibitory concentration (MIC) of each peptide was determined as described by Lehrer et al. (36). The best fit straight line was determined using linear regression with Excel Microsoft 2003 software. The MIC was calculated by finding the intersection of the line with the x axis, indicating the lowest peptide concentration at which no clear zone was obtained.
NMR Experiments
A standard set of two-dimensional 1H NMR experiments (COSY, 80-ms TOCSY, and 160-ms NOESY) was performed on a 1 mm aqueous solution of the synthetic gallin (H2O/D2O (90:10) and 100% D2O) at pH 5.5 and at 293 K. All spectra were recorded on a 600-MHz VARIAN INOVA NMR spectrometer and processed with the NMRPipe/NMRDraw software package (37, 38). 1H chemical shifts were assigned according to classical procedures, and NOE cross-peaks were assigned within the NMRView software (37). 13C-1H HSQC spectra were acquired in natural abundance, and 13C chemical shifts were combined with 1H chemical shifts to deduce 31 (φ,ψ) angular restraints within the TALOS software (39).
Structure Calculations
Distance constraints were obtained from the volume integration of NOE correlations, using the NMRView software. The calculations were initiated using the default parameters of ARIA1.1 software (40) and a first set of easily assigned NOEs. At the end of each run, the new assignments proposed by ARIA were checked manually and introduced (or not) in the following calculation. Ambiguous intersulfur distances, an option assuming that a given half-cystine is part of a bridge without supposing a particular partner, were used in the first runs. With this “ambiguous disulfide bridges” protocol, each half-cystine is allowed to be linked to one of the five others, leading to 15 possibilities of pairing. During the calculations, each disulfide bridge floats freely, and the protocol is driven to the most compatible disulfide bridge array, under the influence of all the other NMR restraints. When enough experimental data had been introduced into the iterative ARIA process to converge to a specific disulfide bridge pairing, covalent bonds were added between sulfur atoms involved in each bridge. Final structures were displayed and analyzed using the MOLMOL program (41), and their quality was evaluated using the PROCHECK and PROMOTIF software packages (42, 43).
Comparative Modeling
Comparative modeling, or homology modeling, was performed with the MODELLER software (44), taking gallin as the template, to build the three-dimensional structures of the four closest ovodefensins (Fig. 1a). The level of sequence identity between targets and template is >60% for meleagrin, cygnin, and dBPS2 from the egg white of turkey (Meleagris), swan (Cygnus), and duck, respectively (31–33), and is 41% for taeniopygin-1 from zebra finch (Taeniopygia guttata) (45). This makes it possible to follow the standard MODELLER protocol. We first checked for each sequence that no other protein with a known related structure displayed a greater sequence similarity. Then each ovodefensin was aligned with the gallin sequence, without needing to introduce any gap. When gaps are absent in the sequence alignment, the three-dimensional model construction is straightforward, and the confidence in accuracy is high. Models were not built for dPBS1 and taeniopygin-2 (Fig. 1a), which were considered too far from the template, gallin, to obtain reliable models and to precisely analyze the resulting molecular surfaces.
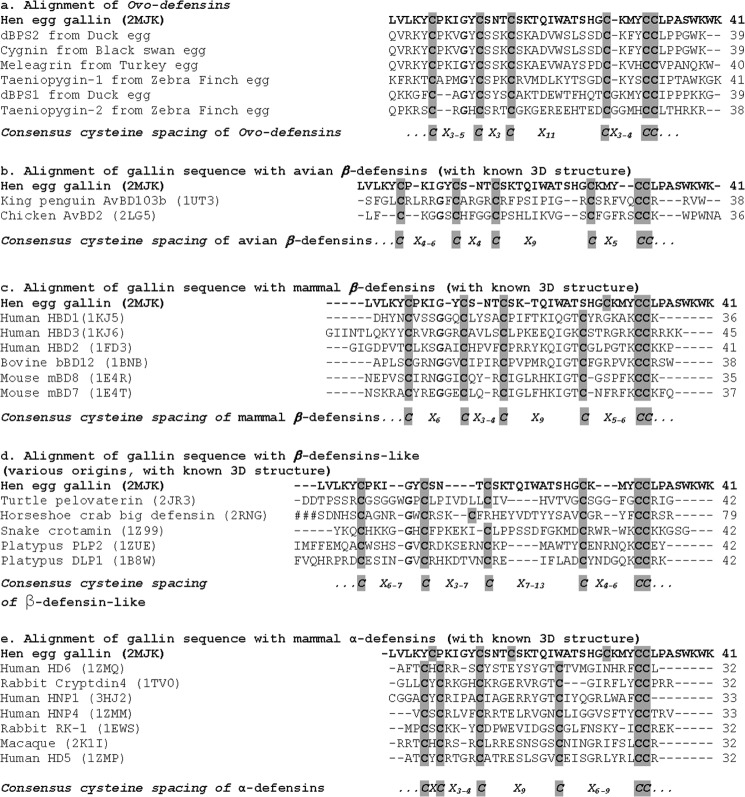
FIGURE 1.
a, sequence alignment of hen egg gallin versus ovodefensins. b–e, multiple alignments of hen egg gallin versus other vertebrate defensins with known three-dimensional structures: avian β-defensins (b), mammal β-defensins (c), β-defensins-like (d), and mammal α-defensins (e), respectively. Consensus cysteine spacings are given for each group. For clarity, the amino-terminal region of Horseshoe crab big defensin (79 residues) is not shown (###). Alignments were done with ClustalW2. The number of residues of each sequence is given at the end of each line.
RESULTS
Because of the difficulties of purifying gallin present at very low concentration in egg white (26), a prerequisite to the functional and structural study of gallin was its chemical synthesis.
Synthesis and Folding of Gallin
Peptide elongation of gallin was carried out by solid-phase peptide synthesis using the polyethylene glycol dimethylacrylamide resin as a polymeric matrix facilitating the synthesis of “difficult sequences” (46). A Wang-type linker was attached to the aminomethyl polyethylene glycol dimethylacrylamide resin by using the commercially available Fmoc-Lys(Boc)-methylphenoxyproprionic acid. The completion of the reaction was checked by Kaiser's test. Elongation was then carried out by the Fmoc/tBu strategy. After TFA treatment, the reduced form of gallin was precipitated using cold tert-butyldimethyl ether and analyzed by HPLC and mass spectrometry.
The oxidative folding was optimized on the fully reduced HPLC-purified gallin. The folding kinetics was followed by analytical HPLC (Fig. 2). At room temperature in the presence of a redox system (GSSG/GSH) at pH 8.6, the reaction was shown to be complete in less than 30 min. Finally, the synthetic gallin was purified by semipreparative HPLC and lyophilized.
FIGURE 2.
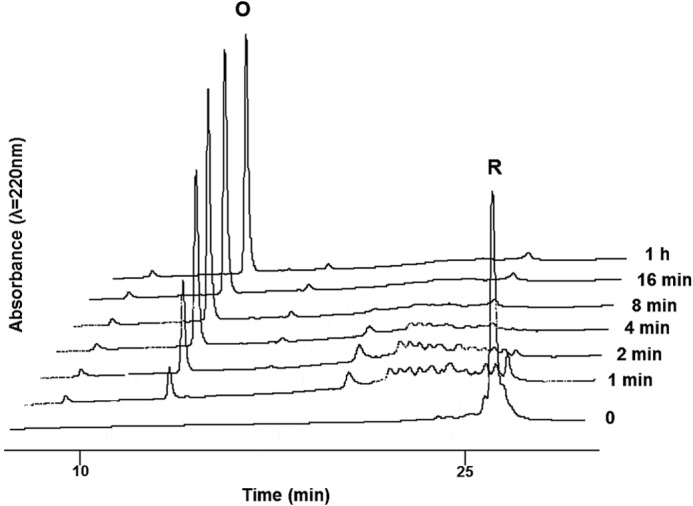
HPLC traces of gallin during oxidative folding. The reduced form of gallin (0 min) (R) was folded at room temperature, in 100 mm Tris-HCl buffer, containing 1 mm EDTA, pH 8.6, in the presence of GSH/GSSG. The oxidative folding (O) was monitored at 220 nm. The reaction was quenched by acidification before being analyzed by HPLC. C18 RP-HPLC gradient: 20–40% in solvent B over 30 min. The initial conditions (time 0) correspond to the time prior to the addition of the redox system.
Antibacterial Activity of Gallin
The antimicrobial activities of gallin was evaluated using a radial diffusion assay against a panel of eight different bacterial strains, mostly composed of pathogenic bacteria. Inhibition zones in units were plotted as a function of log10 (peptide concentration) for each bacterial strain, as illustrated on Fig. 3 for gallin against E. coli ATCC 25922. The MICs obtained from gallin and MSI-94, used as a positive control, are shown in μm in Table 1. No antibacterial activity was observed for gallin against the pathogenic bacteria S. enterica serovar Enteritidis ATCC 13076, S. enterica serovar Enteritidis LA5, S. enterica serovar Typhimurium, S. aureus, and L. monocytogenes. However, results showed that gallin inhibits all tested E. coli strains. MICs were 0.84, 1.23, and 2.00 μm, respectively, for the non-pathogenic ATCC 25922 and for the pathogenic CFT073 and BEN3578 E. coli strains.
FIGURE 3.
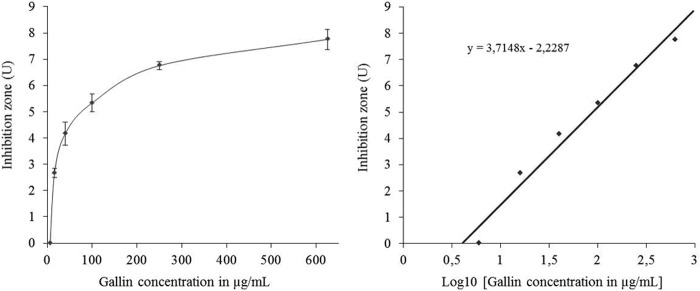
Antimicrobial activity of gallin against E. coli ATCC 25922. Left, dose response of gallin and its inhibitory effect on E. coli ATCC 25922, reflected by the diameter of the clear zone (inhibition zone is expressed in units = the diameter in mm of the clear zone on the plate minus the diameter in mm of the well). Each point represents the means ± S.E. (error bars) of three separate experiments. Right, the best fit straight line was determined by linear regression. MIC of gallin versus E. coli ATCC 25922 was calculated by finding the intersection of the line with the x axis, indicating the lowest peptide concentration at which no clear zone was detected.
TABLE 1.
MIC of synthesized gallin
| Bacterial group | MICa (S.E.) |
|
|---|---|---|
| Control MSI-94b | Gallin | |
| μm | μm | |
| Gram-negative | ||
| E. coli ATCC 25922 | 0.32 (0.04) | 0.84 (0.03) |
| E. coli CFT073 | NDc | 1.23 (0.07) |
| E. coli BEN3578 | ND | 2.00 (0.81) |
| S. enterica Enteritidis ATCC 13076 | 0.28 (0.03) | >53 |
| S. enterica Enteritidis LA5 | 0.31 (0.05) | >53 |
| S. enterica Typhimurium ATCC 14028 | 0.29 (0.01) | >53 |
| Gram-positive | ||
| S. aureus ATCC 29740 | 0.45 (0.07) | >53 |
| L. monocytogenes | 0.32 (0.04) | >53 |
a The MIC was determined by a radial diffusion assay for each bacterial strain.
b MSI-94 is a magainin variant used as a positive control.
c ND, not determined.
NMR Assignments
All protons were assigned except the NH of Ser13, Gly27, and Met30. Chemical shifts have been deposited in the BioMagResBank with the entry code 19729. Low field-shifted Hα chemical shifts and a characteristic set of interstrand connectivities delineate the β-sheet. An unusual chemical shift was observed for NH of Cys16 (4.38 ppm), outside of the already large range of chemical shifts listed in the BioMagResBank. In the three-dimensional structure, the NH of Cys16 points toward Trp22, which explains the unusual chemical shift observed for Cys16.
Structure Calculations
NOE assignments were progressively introduced during the iterative process of ARIA and manually validated between each run until complete assignment of the NOESY map. In the first runs, the “ambiguous disulfide bridges” option was used, to prevent any speculation about the disulfide bridge array (Cys6–Cys33, Cys12–Cys28, and Cys16–Cys32 for α-defensins; Cys6–Cys32, Cys12–Cys28, and Cys16–Cys33 for β-defensins; or any other possibility). Then an intermediate run (500 initial structures, 250 structures refined in water, 50 best structures kept for analysis) was used to unambiguously define the cysteine pairing before further refinement. Among this set of 50 structures (already displaying a root mean square deviation calculated on Cα atoms of 0.79Å), 40 structures (80%) unambiguously form the three disulfide bridges of β-defensins. In all of these 50 structures (50/50), the C16 with exponents C33 disulfide bridge typical of β-defensin is formed, whereas no structure (0/50) displays the C6 with exponents C33 or the C16 with exponents C32 disulfide bridge, both of which are expected in α-defensins. Further measurement of distances between the sulfur atoms of each half-cystine clearly discredited α-defensin pairing (Fig. 4). To switch β- to α-defensins, a completed reversal of the central strand of the sheet is required, which is totally inconceivable without numerous violations of distance constraints. The determined disulfide bridge array was introduced in final runs of calculation. The last run, performed with 1000 initial structures, used a final list of 782 NOE-derived distance restraints (Table 2), divided into 331.2 intraresidue, 174.9 sequential, 84 medium range (2 ≤ |i − j| ≤ 4), and 191.9 long range (|i − j| ≥ 5) restraints, with an average of 20 restraints/residue. Among these NOE restraints, 727 are non-ambiguous. In the last iteration, 200 structures were refined in a shell of water, 181 did not show any NOE violation greater than 0.1 Å, and the worst 19 structures only showed one violation above 0.1 Å. Among the best refined structures, 10 structures were selected, in agreement with all of the experimental data and the standard covalent geometry, and considered as representative of the solution structure of gallin. They were deposited in the Protein Data Bank with the entry code 2MJK. The Ramachandran plot exhibits 96.6% of the (φ,ψ) angles in the most favored and additional allowed regions according to the PROCHECK software nomenclature. Root mean square deviations testify that the three-dimensional structure is very well defined with a pairwise root mean square deviation on the Cα atoms of 0.47Å, and calculated energies are totally satisfactory (Table 2).
FIGURE 4.
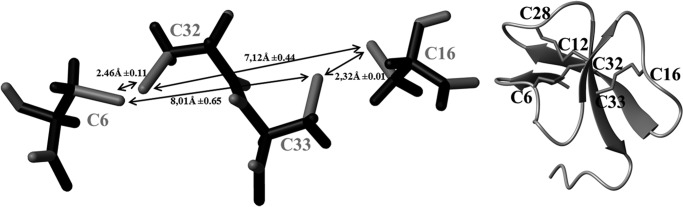
Analysis of an intermediate run of calculations. Left, measurements of distances between sulfur atoms (mean values of the 50 best structures) unambiguously define the disulfide pairing before further refinement. Right, ribbon representation of the protein in the same orientation, with the disulfide bridge array of β-defensins.
TABLE 2.
Structural statistics for the 10 models representative of the solution structure of gallin (1 mm H2O/D2O, pH 5.5, 293 K)
| NOE restraints | |
| Total | 782 |
| Intraresidue (|i | j| = 0) | 331.2 |
| Sequential (|i − j| = 1) | 174.9 |
| Medium range (2 ≤ |i − j| ≤ 4) | 84 |
| Long range (|i − j| ≥ 5) | 191.9 |
| Root mean square deviation on backbone Cα atoms (pairwise) (Å)a | |
| Global | 0.72 ± 0.10 |
| Secondary structures (residues 2–3, 11–13, 20–25, and 31–35) | 0.47 ± 0.11 |
| β1 (residues 2–3) | 0.09 ± 0.05 |
| β2 (residues 11–13) | 0.15 ± 0.07 |
| β3 (residues 20–23) | 0.22 ± 0.19 |
| β4 (residues 24–25) | 0.13 ± 0.07 |
| β5 (residues 31–35) | 0.28 ± 0.14 |
| Ramachandran plotb (%) | |
| Most favored regions | 87.2 |
| Additional allowed regions | 9.4 |
| Generously allowed regions | 3.4 |
| Disallowed regions | 0 |
| Energiesc (kcal·mol−1) | |
| Electrostatic | −1133 ± 32 |
| van der Waals | −152 ± 9 |
| ENOE | 9.3 ± 1.9 |
| Total energy | −1189 ± 47 |
a Fit on secondary structures (residues 2–3, 11–13, 20–25, and 31–35).
b Determined by PROCHECK.
c Calculated with the standard parameters of ARIA.
Functional Prediction Resulting from Comparison with Available Three-dimensional Structures
In the case of multifunctional proteins (47, 48), such as defensins or such as host defense peptides in general, it could be particularly instructive to explore whether a priori unrelated proteins, with very low sequence identity, could match with the protein of interest. Structural databanks were explored to determine if other proteins adopt the five-stranded β-sheet fold described for gallin. A comparison of the three-dimensional structure of gallin with about 75,000 experimentally determined coordinate files available in the Protein Data Bank (interactive service authored by E. Krissinel and K. Henrick) did not reveal any other protein with such a five-stranded sheet arrangement. Thus, the gallin three-dimensional fold can be considered a newly identified fold. The only proteins considered to be similar to gallin are β-defensins from humans, mice, and birds and one of the α-defensins (rabbit RK1). Comparable results were obtained by questioning the DaliLite version 3 software on the DALI server (49), where only α- and β-defensin three-dimensional structures were found to be similar to gallin, with a better fit with β-defensin folds than with α-defensins. Not surprisingly, β-defensin-like peptides (β-defensin disulfide bridges, but less structured) were not highlighted by our analysis. Thus, the gallin three-dimensional fold can be considered a newly described five-stranded fold and a new variation for the β-defensin fold.
DISCUSSION
Gallin is a chicken cysteine-rich cationic peptide previously identified in the chicken egg white and vitelline membrane by proteomic approaches (26, 27). Its antibacterial activities against the Gram-negative bacteria E. coli were recently demonstrated (34). Moreover, gallin was described as being related to the β-defensin family due to the presence of six conserved cysteine residues (34). However, the spacing between these conserved cysteines, which is characteristic of ovodefensins, a group of peptides isolated from the egg white of different avian species, is different from that observed for other known β-defensins. Moreover, the disulfide bond pairing of ovodefensins is not known and must be determined to unambiguously classify ovodefensins as members of the α- or β-defensin family. The lack of structural and functional data thus prompted us to define the three-dimensional structure of gallin in aqueous solution by NMR and to explore its antimicrobial activity against a series of Gram-positive and Gram-negative bacteria.
Structural Description
The gallin three-dimensional NMR structure is the first three-dimensional structure solved for an ovodefensin. The global fold of gallin contains a twisted three-stranded antiparallel β-sheet (11–13, 20–23, 31–35) with a (+2X, −1) topology, typical of vertebrate defensins (Fig. 5). This sheet is common to vertebrate α- and β-defensins, which differ only by the cysteine pairing. Molecular modeling of gallin under NMR restraints converged only with a C6 with exponents C32, C12 with exponents C28, C16 with exponents C33 pairing, attesting that gallin can unambiguously be classified as a β-defensin.
FIGURE 5.
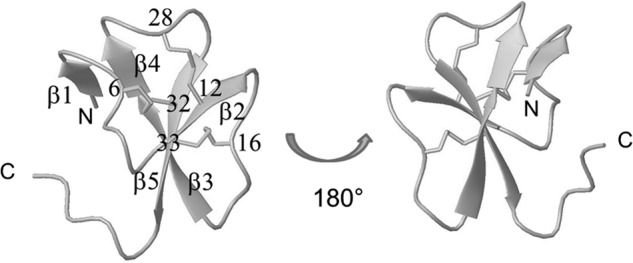
Schematic representation of gallin backbone and disulfide bridges (drawn with MOLMOL software (41)). The five strands of β-sheets are numbered from the amino to the carboxyl terminus (β1–β5), and cysteine residues are numbered to clearly identify the disulfide bridges.
Some variations of the β-defensin fold have already been described, mainly the presence of an amino-terminal α-helix or an α-helical propensity in several structures, such as human hBD1–3 (PDB codes 1IJV, 1FD4, and 1KJ6 respectively), mouse mBD7–8 (PDB codes 1E4T and 1E4R), and king penguin AvBD103b (PDB code 1UT3), whereas other β-defensin structures do not contain this additional amino-terminal helical propensity (such as chicken AvBD2 and bovine bBD12, PDB codes 2LG5 and 1BNB, respectively). In the present study, the global fold of gallin does not contain any amino-terminal helix but contains an additional two-stranded parallel β-sheet (residues 2–3 and 24–25) (Fig. 5). This observation reveals that the gallin structure adopts a new variation of the classical β-defensin fold. To our knowledge, this is the first structural proof that ovodefensins belong to the β-defensin family.
Structural Comparison with Avian Defensins
Only two three-dimensional structures of avian defensins are currently available (Fig. 1b): king penguin AvBD103b defensin, which our group determined in 2004 (PDB code 1UT3 (50)), and chicken AvBD2, which we recently solved (PDB code 2LG5 (16)). When these three structures are aligned, their three-stranded β-sheets can be easily superimposed (Fig. 6). However, they differ in their amino-terminal region; whereas gallin contains an additional two-stranded parallel β-sheet, AvBD103b has a high propensity to form an α-helix, as already observed for some mammal β-defensins. Chicken AvBD2 is shorter, lacking the amino-terminal residues, and consequently cannot form an additional amino-terminal structural element. The presence (or absence) of this amino-terminal helix cannot be related to antimicrobial properties, because both king penguin AvBD103b and chicken AvBD2 demonstrate large antimicrobial spectrum (21, 51). However, the amino-terminal region could be involved in other yet unknown functions.
FIGURE 6.
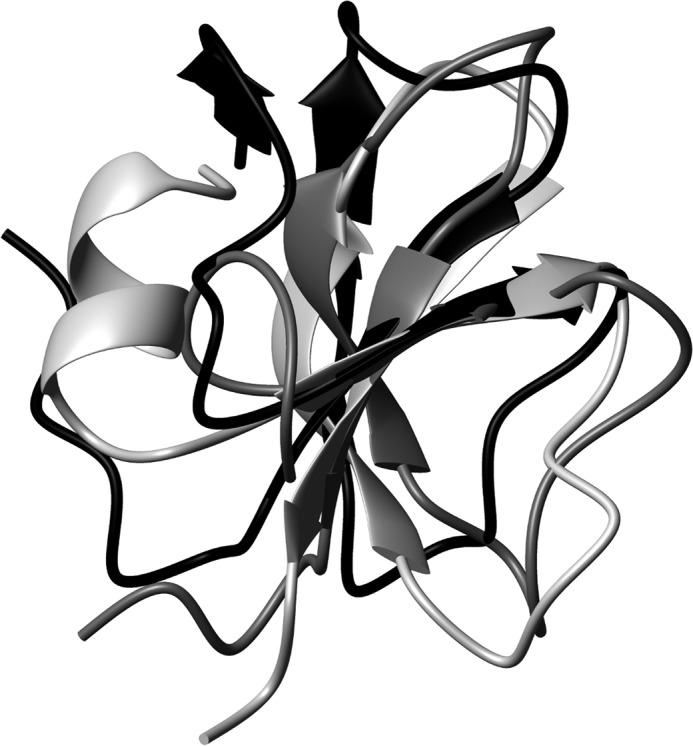
Superimposition of gallin, chicken AvBD2, and king penguin AvBD103b in black, dark gray, and light gray, respectively. Whereas the common three-stranded β-sheet can be easily superimposed, each defensin differs in the amino-terminal region.
Sequence-Structure Relationships
The consensus sequence of ovodefensins, CX3–5CX3CX11CX3–4CC, is different from that observed for other known β-defensins found in mammals or birds. In particular, the spacing between the third and fourth cysteine has been pointed out (34). Fig. 1, b–e, illustrates multiple alignments of gallin with vertebrate α- and β-defensin sequences with known three-dimensional structures. (i) Alignments with avian or mammal β-defensins clearly underline the particularities of the fourth cysteine; X11CX3–4 for ovodefensins, contrasting with X9CX5–6 for typical β-defensins, forces us to introduce gaps or to move this cysteine forward within the alignment (Fig. 1, b and c). This shift in sequence allows the two additional residues before the fourth cysteine to form an additional parallel sheet on the three-dimensional structure (Fig. 5) and is therefore directly related to the formation of the additional sheet. (ii) When the gallin sequence is compared with the five β-defensin-like peptides from various origins (Fig. 1d) (i.e. defensins with the typical disulfide bridges of β-defensins but lacking the complete three-dimensional structures), the consensus sequence almost fits (CX3–5CX3CX11CX3–4CC for the ovodefensin group versus CX6–7CX3–7CX7–13CX4–6CC for β-defensin-like peptides), particularly with regard to the spacing between the third and fourth cysteine. However, gallin is well structured and therefore does not belong to this β-defensin-like group. (iii) Finally, even if three-dimensional structures of α- and β-defensins are globally superimposable, the primary structure of gallin is clearly different from α-defensin sequences (Fig. 1e). We conclude that ovodefensins form a new subfamily of β-defensins, with a specific consensus sequence, accounting for a variation in the β-defensin three-dimensional fold.
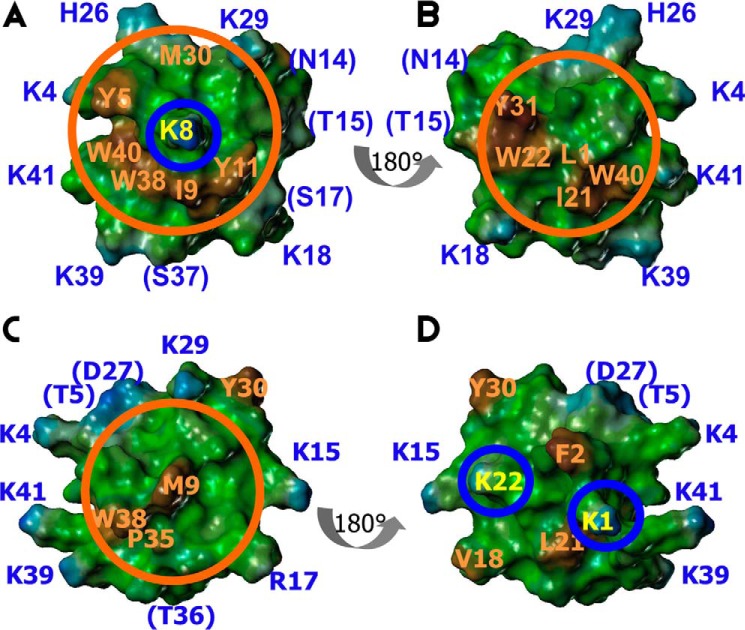
Surface Properties of Gallin
Gallin is a highly cationic molecule, with the six charged residues distributed over the surface of the molecule, resulting in an overall positive molecular surface (electrostatic potentials at the surface not shown). The representation of the hydrophobic properties at the molecular surface is more instructive; indeed, hydrophobic and hydrophilic residues are not randomly distributed at the surface. The gallin surface displays two hydrophobic sides (Fig. 7, A and B) surrounded by a crown of hydrophilic residues (Asn14, Thr15, Ser17, Lys18, Ser37, Lys39, Lys41, Lys4, His26, and Lys29). A basic residue, Lys8, points out of one face, in the middle of a rather hydrophobic region (Fig. 7A). This lysine is conserved in dBPS2, cygnin, and meleagrin sequences and could be essential to (yet unknown) biological function(s), and/or it could be essential to prevent oligomerization/aggregation.
FIGURE 7.
Surface properties of gallin from hen egg (A and B, with 180° rotation) and taeniopygin-1 from zebra finch (C and D, with 180° rotation). Hydrophobic and hydrophilic potential areas, calculated with the MOLCAD option of SYBYL software (TRIPOS Inc., St. Louis, MO) at the Connolly surfaces, are displayed in brown and blue, respectively (scale −0.20, +0.20). Green surfaces represent an intermediate hydrophobicity.
A comparison of gallin surface properties with those of other ovodefensins could give insight into structural properties of the ovodefensin subfamily. Thus, with gallin as a target, three-dimensional models were built for the four closest ovodefensins currently identified (dBPS2 from ducks, cygnin from black swans, and meleagrin from turkeys (displaying 62% of sequence identities with gallin)) as well as taeniopygin-1 predicted from the zebra finch genome and exhibiting 41% sequence identity with gallin (Fig. 1a).
The first three ovodefensins exhibit the same surface properties as gallin. In particular, the distribution of hydrophobic and hydrophilic residues at the surface, which could be related to function(s), is well conserved. On the contrary, the model built for the fourth ovodefensin, taeniopygin-1, leads to a mirror distribution of hydrophobic and hydrophilic residues at the surface of the three-dimensional models (Fig. 7, C and D). The crown of hydrophilic residues is preserved, ensuring a sort of “circular amphiphily.” However, the substitution of Lys8 (gallin numbering; Figs. 1a and 7A) by Pro in taeniopygin-1 makes this face hydrophobic (Fig. 7C). Two lysines, Lys1 and Lys22, point outward on the opposite face and replace Leu1 and Trp22 in gallin (Fig. 7D). They could compensate for gallin Lys8 for biological implications and/or for preventing aggregation of the protein.
Functional Data
In the present study, we observed that gallin has a potent antimicrobial activity against several E. coli strains, including an avian strain isolated from the oviduct of a hen with colibacillosis (BEN 3578). The observed anti-E. coli effect is in agreement with previous results showing that gallin at a concentration of 0.25 μm was able to inhibit the growth of an E. coli strain different from those used in our study (34). In contrast, no activities against the other tested bacteria, including Gram-negative (S. enterica serovar Enteritidis and S. enterica serovar Typhimurium) and Gram-positive (S. aureus and L. monocytogenes) bacteria, were observed for gallin at a maximal concentration of 53 μm. These results are unexpected because generally defensins possess a broad spectrum of antimicrobial activities, targeting both Gram-positive and Gram-negative bacteria (15, 21, 51). However, to our knowledge, no antimicrobial activity has been revealed for the other known members of the ovodefensin family to date. Indeed, when used at a concentration as high as 22 μm, turkey meleagrin displayed no anti-E. coli (LE392 strain) activity (32). Likewise, the duck ovodefensins dBPS1 and dBPS2 at a concentration of 56 μm possess no intrinsic antimicrobial properties, at least against the following Gram-positive and negative bacteria tested: S. aureus, B. subtilis, S. enterica serovar Enteritidis, and E. coli (52). The antimicrobial spectrum of the swan ovodefensin cygnin remains still undefined. Therefore, gallin is to date the only member of the ovodefensin family for which antimicrobial activity has been described, even if it is limited to E. coli. Gallin potentially participates in the antibacterial defense of the egg and the hen oviduct (reproductive organ involved in egg formation) against E. coli contamination. Nevertheless, this narrow antimicrobial activity raises questions, particularly whether this activity reflects the main function of this molecule. In fact, it is likely that gallin and, to a greater extent, all ovodefensins may have biological functions other than those related to antimicrobial defense. In this regard, potent anti-lipase activities have recently been found for the duck ovodefensins dBPS1 and dBPS2, suggesting that these molecules may play a role in the protection of lipids from lipase-mediated hydrolysis (52). Interestingly, dBPS1/dBPS2 proteins have been predominantly detected in the oviduct, where the synthesis of the egg takes place, and also to a lesser extent in the gall bladder, where they are believed to exert potentially their anti-lipase activities (33, 52). Similarly, maximum expression of the gallin genes is observed in the magnum (oviduct part responsible for the egg white protein synthesis) when compared with other oviduct parts (34). The biological activities of gallin need to be further investigated to highlight its function in the hen egg.
CONCLUSION
The gallin three-dimensional structure is the first ovodefensin structure to have been solved to date. No other protein with the same five-stranded arrangement could be found in structure databases. The gallin three-dimensional structure contains the typical β-sheet and the typical disulfide bridge array of β-defensins. Hence, gallin can be unambiguously classified as a β-defensin. However, gallin displays an additional small sheet, which can be directly related to the cysteine spacing in the sequence. Thus, gallin is the first member of a new structural subfamily of defensins. The apparent antibacterial selectivity of gallin toward E. coli strains, which may be related to the atypical hydrophobic properties of the gallin molecular surface, will need further investigation.
Finally, gallin is to date the only member of the ovodefensin family for which an antimicrobial activity has been described. The atypical structural features of gallin, compared with other defensins, could be related to other functions that remain to be characterized.
Acknowledgments
We thank Anne-Christine Lalmanach (Institut National de la Recherche Agronomique (INRA) Nouzilly) for help in MIC determination, Catherine Schouler (INRA Nouzilly) for valuable advice on the choice of pathogenic E. coli strains, Jean-Baptiste Madinier (CBM Orléans) for involvement in peptide synthesis, and Stéphane Bourg (CBM Orléans) for providing access to the SYBYL software. We thank the CBM mass spectrometry platform, and in particular Guillaume Gabant, for recording most of the MALDI-TOF spectra.
This work was supported by the French National Research Agency (OVO-mining, ANR-09-BLAN-0136).
The atomic coordinates and structure factors (code 2MJK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Chemical shifts have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) with the entry code 19729.
- AvBD
- avian β-defensin
- Boc
- t-butoxycarbonyl
- dBPS
- duck basic protein small
- Fmoc
- N-(9-fluorenyl)methyloxycarbonyl
- tBu
- tert-butyl
- Trt
- triphenylmethyl
- MIC
- minimum inhibitory concentration
- PDB
- Protein Data Bank.
REFERENCES
- 1. Bayer A. S., Schneider T., Sahl H. G. (2013) Mechanisms of daptomycin resistance in Staphylococcus aureus. Role of the cell membrane and cell wall. Ann. N.Y. Acad. Sci. 1277, 139–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breidenstein E. B., de la Fuente-Núñez C., Hancock R. E. (2011) Pseudomonas aeruginosa. All roads lead to resistance. Trends Microbiol. 19, 419–426 [DOI] [PubMed] [Google Scholar]
- 3. Master R. N., Deane J., Opiela C., Sahm D. F. (2013) Recent trends in resistance to cell envelope-active antibacterial agents among key bacterial pathogens. Ann. N.Y. Acad. Sci. 1277, 1–7 [DOI] [PubMed] [Google Scholar]
- 4. Smith P. A., Romesberg F. E. (2007) Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem. Biol. 3, 549–556 [DOI] [PubMed] [Google Scholar]
- 5. Brogden N. K., Brogden K. A. (2011) Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int. J. Antimicrob. Agents 38, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cederlund A., Gudmundsson G. H., Agerberth B. (2011) Antimicrobial peptides important in innate immunity. FEBS J. 278, 3942–3951 [DOI] [PubMed] [Google Scholar]
- 7. Guaní-Guerra E., Santos-Mendoza T., Lugo-Reyes S. O., Terán L. M. (2010) Antimicrobial peptides. General overview and clinical implications in human health and disease. Clin. Immunol. 135, 1–11 [DOI] [PubMed] [Google Scholar]
- 8. Yount N. Y., Yeaman M. R. (2013) Peptide antimicrobials. Cell wall as a bacterial target. Ann. N.Y. Acad. Sci. 1277, 127–138 [DOI] [PubMed] [Google Scholar]
- 9. Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 10. Alba A., López-Abarrategui C., Otero-González A. J. (2012) Host defense peptides. An alternative as antiinfective and immunomodulatory therapeutics. Biopolymers 98, 251–267 [DOI] [PubMed] [Google Scholar]
- 11. Jenssen H., Hamill P., Hancock R. E. (2006) Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Selsted M. E., Ouellette A. J. (2005) Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 13. Xiao Y., Hughes A. L., Ando J., Matsuda Y., Cheng J. F., Skinner-Noble D., Zhang G. (2004) A genome-wide screen identifies a single β-defensin gene cluster in the chicken. Implications for the origin and evolution of mammalian defensins. BMC Genomics 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Dijk A., Veldhuizen E. J., Haagsman H. P. (2008) Avian defensins. Vet. Immunol. Immunopathol. 124, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hervé-Grépinet V., Réhault-Godbert S., Labas V., Magallon T., Derache C., Lavergne M., Gautron J., Lalmanach A. C., Nys Y. (2010) Purification and characterization of avian β-defensin 11, an antimicrobial peptide of the hen egg. Antimicrob. Agents Chemother. 54, 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Derache C., Meudal H., Aucagne V., Mark K. J., Cadène M., Delmas A. F., Lalmanach A. C., Landon C. (2012) Initial insights into structure-activity relationships of avian defensins. J. Biol. Chem. 287, 7746–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landon C., Thouzeau C., Labbé H., Bulet P., Vovelle F. (2004) Solution structure of spheniscin, a β-defensin from the penguin stomach. J. Biol. Chem. 279, 30433–30439 [DOI] [PubMed] [Google Scholar]
- 18. Sugiarto H., Yu P. L. (2004) Avian antimicrobial peptides. The defense role of β-defensins. Biochem. Biophys. Res. Commun. 323, 721–727 [DOI] [PubMed] [Google Scholar]
- 19. Harmon B. G. (1998) Avian heterophils in inflammation and disease resistance. Poult. Sci. 77, 972–977 [DOI] [PubMed] [Google Scholar]
- 20. Evans E. W., Beach F. G., Moore K. M., Jackwood M. W., Glisson J. R., Harmon B. G. (1995) Antimicrobial activity of chicken and turkey heterophil peptides CHP1, CHP2, THP1, and THP3. Vet. Microbiol. 47, 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Derache C., Labas V., Aucagne V., Meudal H., Landon C., Delmas A. F., Magallon T., Lalmanach A. C. (2009) Primary structure and antibacterial activity of chicken bone marrow-derived β-defensins. Antimicrob. Agents Chemother. 53, 4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lynn D. J., Higgs R., Lloyd A. T., O'Farrelly C., Hervé-Grépinet V., Nys Y., Brinkman F. S., Yu P. L., Soulier A., Kaiser P., Zhang G., Lehrer R. I. (2007) Avian β-defensin nomenclature. A community proposed update. Immunol. Lett. 110, 86–89 [DOI] [PubMed] [Google Scholar]
- 23. Subedi K., Isobe N., Nishibori M., Yoshimura Y. (2007) Changes in the expression of gallinacins, antimicrobial peptides, in ovarian follicles during follicular growth and in response to lipopolysaccharide in laying hens (Gallus domesticus). Reproduction 133, 127–133 [DOI] [PubMed] [Google Scholar]
- 24. Meade K. G., Higgs R., Lloyd A. T., Giles S., O'Farrelly C. (2009) Differential antimicrobial peptide gene expression patterns during early chicken embryological development. Dev Comp. Immunol. 33, 516–524 [DOI] [PubMed] [Google Scholar]
- 25. Ebers K. L., Zhang C. Y., Zhang M. Z., Bailey R. H., Zhang S. (2009) Transcriptional profiling avian β-defensins in chicken oviduct epithelial cells before and after infection with Salmonella enterica serovar Enteritidis. BMC Microbiol. 9, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mann K. (2007) The chicken egg white proteome. Proteomics 7, 3558–3568 [DOI] [PubMed] [Google Scholar]
- 27. Mann K. (2008) Proteomic analysis of the chicken egg vitelline membrane. Proteomics 8, 2322–2332 [DOI] [PubMed] [Google Scholar]
- 28. Mann K., Macek B., Olsen J. V. (2006) Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics 6, 3801–3810 [DOI] [PubMed] [Google Scholar]
- 29. Mann K., Mann M. (2008) The chicken egg yolk plasma and granule proteomes. Proteomics 8, 178–191 [DOI] [PubMed] [Google Scholar]
- 30. Mann K., Olsen J. V., Macek B., Gnad F., Mann M. (2008) Identification of new chicken egg proteins by mass spectrometry-based proteomic analysis. Worlds Poult. Sci. J. 64, 209–218 [Google Scholar]
- 31. Simpson R. J., Morgan F. J. (1983) Isolation and complete amino acid sequence of a basic low molecular weight protein from black swan egg white. Int. J. Pept. Protein Res. 22, 476–481 [PubMed] [Google Scholar]
- 32. Odani S., Koide T., Ono T., Takahashi Y., Suzuki J. (1989) Covalent structure of a low-molecular-mass protein, meleagrin, present in a turkey (Meleagris gallopavo) ovomucoid preparation. J. Biochem. 105, 660–663 [DOI] [PubMed] [Google Scholar]
- 33. Naknukool S., Hayakawa S., Sun Y., Ogawa M. (2008) Structural and physicochemical characteristics of novel basic proteins isolated from duck egg white. Biosci. Biotechnol. Biochem. 72, 2082–2091 [DOI] [PubMed] [Google Scholar]
- 34. Gong D., Wilson P. W., Bain M. M., McDade K., Kalina J., Hervé-Grépinet V., Nys Y., Dunn I. C. (2010) Gallin. An antimicrobial peptide member of a new avian defensin family, the ovodefensins, has been subject to recent gene duplication. BMC Immunol. 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Welch R. A., Burland V., Plunkett G., 3rd, Redford P., Roesch P., Rasko D., Buckles E. L., Liou S. R., Boutin A., Hackett J., Stroud D., Mayhew G. F., Rose D. J., Zhou S., Schwartz D. C., Perna N. T., Mobley H. L., Donnenberg M. S., Blattner F. R. (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. (1991) Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137, 167–173 [DOI] [PubMed] [Google Scholar]
- 37. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 38. Johnson B. A., Blevins R. A. (1994) NMRview. A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 39. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 40. Linge J. P., O'Donoghue S. I., Nilges M. (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 41. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 42. Hutchinson E. G., Thornton J. M. (1996) PROMOTIF. A program to identify and analyze structural motifs in proteins. Protein Sci. 5, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 44. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 45. Hellgren O., Ekblom R. (2010) Evolution of a cluster of innate immune genes (β-defensins) along the ancestral lines of chicken and zebra finch. Immunome Res. 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cremer G. A., Tariq H., Delmas A. F. (2006) Combining a polar resin and a pseudo-proline to optimize the solid-phase synthesis of a “difficult sequence”. J. Pept. Sci. 12, 437–442 [DOI] [PubMed] [Google Scholar]
- 47. Yeung A. T., Gellatly S. L., Hancock R. E. (2011) Multifunctional cationic host defence peptides and their clinical applications. Cell Mol. Life Sci. 68, 2161–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hazlett L., Wu M. (2011) Defensins in innate immunity. Cell Tissue Res. 343, 175–188 [DOI] [PubMed] [Google Scholar]
- 49. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Landon C., Barbault F., Legrain M., Menin L., Guenneugues M., Schott V., Vovelle F., Dimarcq J.-L. (2004) Lead optimization of antifungal peptides with 3D NMR structures analysis. Protein Sci. 13, 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thouzeau C., Le Maho Y., Froget G., Sabatier L., Le Bohec C., Hoffmann J. A., Bulet P. (2003) Spheniscins, avian β-defensins in preserved stomach contents of the king penguin, Aptenodytes patagonicus. J. Biol. Chem. 278, 51053–51058 [DOI] [PubMed] [Google Scholar]
- 52. Naknukool S., Hayakawa S., Ogawa M. (2011) Multiple biological functions of novel basic proteins isolated from duck egg white. Duck basic protein small 1 (dBPS1) and 2 (dBPS2). J. Agric. Food Chem. 59, 5081–5086 [DOI] [PubMed] [Google Scholar]