Abstract
The p53 gene encodes 12 distinct isoforms some of which can alter p53 activity in the absence of genomic alteration. Endogenous p53 isoforms have been identified in cancers; however, the function of these isoforms remains unclear. In melanoma, the frequency of p53 mutations is relatively low compared to other cancers suggesting that these isoforms may play a larger role in regulating p53 activity. We hypothesized that p53 function and therefore cell fate might be altered by the presence of Δ40p53, an embryonic isoform missing the first forty N-terminal amino acids of the full-length protein including the transactivation and Mdm2 binding domains. To test this hypothesis, we transduced tumor and normal cells with a lentivirus encoding Δ40p53. We found that exogenous Δ40p53 caused apoptosis and increased levels of endogenous, activated p53 in both cancerous and non-cancerous cells, which led to significant levels of cell death, particularly in cancer cells. Activated p53 molecules formed nuclear hetero-tetramers with Δ40p53 and altered downstream p53 transcription target levels including p53-induced protein with death domain (PIDD) and cyclin dependent kinase inhibitor, p21. Δ40p53 altered promoter occupancy of these downstream p53 target genes in such a way that shifted cell fate toward apoptosis and away from cell cycle arrest. We show that tumor suppression by p53 can occur via an alternate route that relies on its interaction with Δ40p53.
Introduction
In order to understand the initiation and progression of cancers, numerous tumor suppressors have been screened for the presence of mutations and changes in protein expression (Cheok et al., 2011; Machado-Silva et al., 2010; Robles and Harris, 2010). p53 has been shown to orchestrate an appropriate tumor suppressor function by trans-activating or -suppressing cell cycle and apoptosis genes in response to a particular dose and quality of cellular stress (Beckerman and Prives, 2010; Belyi et al., 2010; Lane and Levine, 2010; Vousden and Prives, 2009). The importance of proper p53 function is emphasized by its high mutation frequency among human cancers (Hollstein et al., 1991; Levine et al., 1991; Petitjean et al., 2007) and the overexpression of ‘mutant’ p53 in certain tumors suggests that some mutations may have a dominant-negative effect on wildtype p53 (Goldstein et al., 2011; Oren and Rotter, 2010). Certain cancers such as melanomas harbor wildtype TP53, however, these tumors bypass the regulatory functions of p53 and continue to proliferate and metastasize (Albino et al., 1994; Gwosdz et al., 2006; Li et al., 2006; Montano et al., 1994; Soto et al., 2005; Weiss et al., 1995; Zerp et al., 1999). This poses the question of how melanoma cells continue to proliferate in the presence of wildtype TP53.
The TP53 gene encodes 12 protein isoforms that are missing specific regions of full-length p53 (Marcel et al., 2011) and are capable of altering p53 function (Courtois et al., 2002; Ghosh et al., 2004; Khoury and Bourdon, 2010). Specific p53 isoforms have been identified in both cancer (Anensen et al., 2006; Avery-Kiejda et al., 2008; Boldrup et al., 2007; Bourdon et al., 2005b; Marcel et al., 2010; Takahashi et al., 2012) and non-cancerous tissues (Ungewitter and Scrable, 2010b). One of these isoforms, Δ40p53, is missing the first 40 amino acids encoding the first transactivation domain and can be synthesized primarily by alternative translation initiation in exon 4 (Courtois et al., 2002; Grover et al., 2009; Grover et al., 2008; Grover et al., 2011; Yin et al., 2002) and by alternative splicing of intron 2 (Ghosh et al., 2004). Δ40p53 is uniquely expressed during early embryogenesis (Maier et al., 2004a; Ungewitter and Scrable, 2010b), is associated with less differentiated, proliferative cells, and cannot be found in corresponding adult tissues (Maier et al., 2004a; Medrano et al., 2009b; Takahashi et al., 2012). The overall frequency of Δ40p53 protein expression in cancer, including melanoma, remains unknown.
We previously showed that Δ40p53 is highly and consistently expressed at the protein level in both glioblastoma and gliosis tissues but not in normal brain cortex (Takahashi et al., 2012). Given that Δ40p53 is uniquely found in stem cell populations, the discovery of its endogenous expression in glioblastoma tumors raises the question of whether this isoform profile reflects a subpopulation of less differentiated, proliferative cells within a tumor or an attempt by the host to activate p53 and curb tumor growth. To distinguish between these possibilities, we generated a lentiviral vector that could deliver Δ40p53 to melanoma cells with high efficiency. We hypothesized that, if high expression of Δ40p53 were associated with proliferative, less differentiated cell types, then transduced cells would take on the growth characteristics of rapidly-dividing cells, resulting in increased cell number. Alternatively, if higher than normal expression of Δ40p53 reflected an attempt by the cells to slow down proliferation, then transduced cells would be expected to exhibit characteristics of tumor suppression, such as increased apoptosis or cell cycle arrest, resulting in reduced numbers of cells. We asked if Δ40p53 could re-activate endogenous p53 function in melanoma cells to favor tumor suppression. Our results are consistent with a model where p53 activation can occur via an alternate route that relies on its interaction with Δ40p53 to modify downstream targets and promote cell death over cell cycle arrest.
Results
Excess Δ40p53 increases the fraction of dead cells in cultures of tumor and normal cells
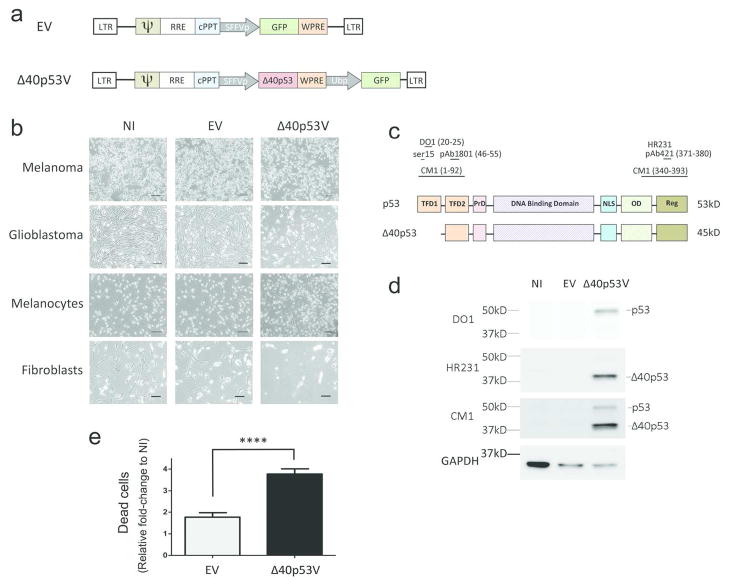
We used a lentivirus encoding Δ40p53 (Δ40p53V) driven by the spleen focus-forming virus promoter (SFFVp) (Fig. 1A) to overexpress Δ40p53 in four different cell types: A375 melanoma cells, primary glioblastoma xenograft cells, human melanocytes and mouse embryonic fibroblasts (Fig. 1B). A previously described lentivirus encoding GFP (EV, Fig. 1A) or no infection (NI) was used as controls (Demaison et al., 2002). We achieved transduction efficiencies of >95% as determined by expression of a GFP reporter included in both the empty vector and the Δ40p53-encoding lentivirus (Fig. 1A, Supplemental Fig. S1). Using a panel of p53 antibodies that bind specific domains of the p53 protein (Fig. 1C), which we previously developed to detect full-length p53 and its isoforms (Takahashi et al., 2012), we demonstrated that exogenous Δ40p53 was expressed in transduced cells (Fig. 1D). p53 antibodies HR231 and CM1 detected Δ40p53 in A375 melanoma cells transduced with Δ40p53V, but not in uninfected cells (NI) or cells transduced with the empty vector (EV). The N-terminal antibody DO1 detected endogenous p53 expression, but only in Δ40p53V-transduced cells. HR231 also detected endogenous p53 upon higher exposure (data not shown). Approximately five days after infection, there were fewer adherent A375 melanoma and primary glioblastoma cells in wells treated with Δ40p53 lentivirus compared to controls (Fig. 1B, top two panels). To determine if this was due to decreased proliferation or increased cell death, we incubated A375 melanoma cells with ethidium homodimer, a DNA binding molecule that is impermeable to cells with intact cell membranes. The relative number of dead cells was significantly increased in Δ40p53-infected cultures compared to empty vector controls (Fig. 1E). Using trypan blue exclusion, we did not find a significant difference in the number of viable cells between Δ40p53-infected cells and controls (data not shown). We also found decreased numbers of adherent cells in melanocytes and mouse embryonic fibroblasts, but at ten days rather than five days after infection (Fig. 1B, bottom two panels). Thus, Δ40p53 did appear to affect the growth of cultures of both tumor and normal cells by decreasing cell viability.
Figure 1. Δ40p53 increases the relative number of dead cells.
(A) Lentiviral constructs encoding Δ40p53 and green fluorescence protein (GFP) (Δ40p53V) or GFP alone (EV). Spleen Focus-Forming Virus promoter (SFFVp); Ubiquitin promoter (Ubp); Long terminal repeat (LTR); packaging signal (ψ); Rev-responsive element (RRE); HIV central polypurine tract (cPPT); Woodchuck hepatitis virus post-transcriptional regulatory element (WPRE). (B) Representative images of cells infected with EV, Δ40p53V, or no lentivirus (NI). Scale bar represents 1μM. (C) p53 and Δ40p53 domain structures and antibody epitopes. Δ40p53 is missing transcription factor domain 1 (TFD1), but retains transcription factor domain 2 (TFD2), proline-rich domain (PrD), DNA binding domain, nuclear localization signal (NLS), oligomerization domain (OD), and regulatory domain (Reg). (D) Western blot analysis of A375 melanoma cells infected with EV, Δ40p53V, or NI. Δ40p53V was detected by HR231 and CM1 but not DO1. (E) Relative fold-change of dead cells in EV- and Δ40p53V-infected cells normalized to NI control. ****p<0.0001.
Δ40p53 causes apoptosis
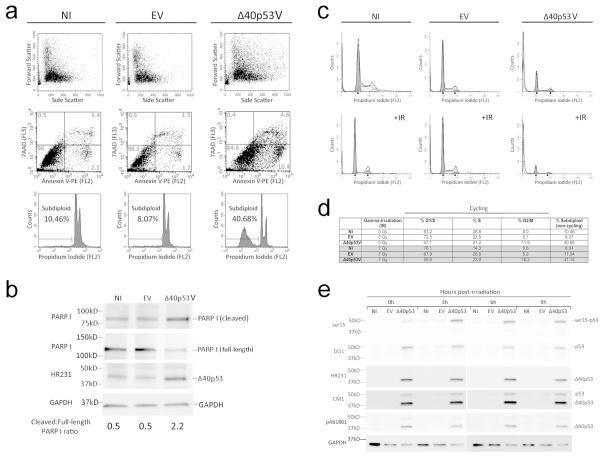
p53 activates pathways that can result in cell cycle arrest or apoptosis in response to different cellular stressors and damage. To determine if the visible decrease in viable cells in the presence of increased Δ40p53 expression was due to apoptosis or to prolonged cell cycle arrest, we analyzed apoptosis and membrane integrity in Δ40p53-infected cells with PE-conjugated Annexin V and a DNA binding dye, 7AAD, respectively (Fig. 2A, middle row). We found an approximately 3-fold increase in double-positive (late apoptotic) cells, a 2.7-fold increase in Annexin V single-positive (early apoptotic) cells, and a 4.5-fold increase in total Annexin V-positive cells with Δ40p53 infection compared to controls. Consistent with the apoptosis results, we observed a 4.4-fold increase in the proportion of subdiploid cells in Δ40p53-infected cultures compared to controls, as determined by propidium iodide staining and flow cytometric analysis (Fig. 2A, bottom row). We found a similar increase in apoptosis in primary glioblastoma xenograft cells infected with Δ40p53 (Supplemental Fig. S2). We further confirmed our findings with western blot analysis using antibodies that detect cleaved or full length poly-(ADP-ribose)-polymerase (PARP I), a caspase target that is cleaved during the late phase of apoptosis (Oliver et al., 1998). As shown in Fig. 2B, we found an increase in cleaved PARP I product in Δ40p53-infected lysates, a 2.2-fold increase relative to full-length PARP I, compared to 0.5-fold in both non-infected (NI) and empty vector (EV) controls. We carried out similar experiments in p53-deficient cells and did not observe an increase in the proportion of apoptotic cells in Δ40p53-infected cells compared to EV controls (Supplemental Fig. S3). These results indicate that Δ40p53 reduces cell viability by inducing apoptosis, but only in cells that express full-length p53.
Figure 2. Δ40p53 causes apoptosis, not cell cycle arrest.
(A) Flow cytometric analysis of apoptotic and subdiploid populations in A375 cells infected with EV, Δ40p53V, or NI. Percentages within each quadrant represent fraction of total cells. Bottom panel percentages indicate subdiploid fraction of cells. At least 20,000 events were collected per experimental sample. Representative plots shown. (B) Western blot analysis of poly-(ADP-ribose) polymerase (PARP I) cleavage. (C) Cell cycle profiles of infected A375 cells in the presence of propidium iodide following 0 or 7Gy of γ-irradiation (+IR). (D) Cell cycle distribution of infected vs. uninfected cells with (shaded area) or without γ-irradiation (non-shaded area). Subdiploid (non-cycling) percentages are shown separately. (E) Western blot analysis of p53 expression in infected A375 cells with γ-irradiation (7Gy). Cells were harvested at 0, 3, 6, and 9-hours post-irradiation.
Δ40p53 does not cause cell cycle arrest
To determine if Δ40p53 affected cell cycle progression, we infected A375 melanoma cells with Δ40p53 or empty vector and analyzed the cells by flow cytometry in the presence of propidium iodide. Consistent with our apoptosis results (Fig. 2A and 2B), we observed a 4.5-fold increase in the percentage of subdiploid cells infected with the Δ40p53 lentivirus (Fig. 2C, top row). However, we did not find significant differences in the cell cycle distributions of cycling Δ40p53-infected and empty vector-infected or non-infected A375 cells (Fig. 2D, unshaded area).
Next, we induced cell cycle arrest and determined cell cycle profiles at 0, 3, 6, and 9-hours following 7Gy of γ-irradiation. Representative plots and quantitation at the 3-hour time point are shown in Fig. 2C (bottom row) and Fig. 2D (shaded area), respectively. We did not find differences in cell cycle profiles with Δ40p53 compared to empty vector controls at any timepoints. By 9-hours post-irradiation, we observed a decrease in the percentage of S phase cells across all conditions indicating that both infected and uninfected cells were able to respond to γ-irradiation (data not shown). We obtained similar results in normal melanocytes, where the cell cycle distributions were the same with or without γ-irradiation and in both infected and non-infected cells (Supplemental Fig. S4). We also did not find an increase in the percentage of subdiploid cells in γ-irradiated samples compared to non-irradiated samples (Fig. 2D, % subdiploid). Collectively, these results suggested that Δ40p53 might selectively affect cell death while leaving cell cycle arrest unchanged.
One of the canonical pathways of tumor suppression by p53 involves ATM/ATR phosphorylation of p53 at serine 15 by ionizing radiation (Banin et al., 1998; Canman et al., 1998; Khanna et al., 1998; Siliciano et al., 1997). The choice between cell cycle arrest and apoptosis is affected by the level of p53 and post-translational modifications at residues such as serine 15 (Hollstein and Hainaut, 2010; Vousden and Prives, 2009). We compared the level of serine 15 phosphorylation in non-infected cells to cells infected with Δ40p53V or EV by western blot analysis at 0, 3, 6, and 9-hours after irradiation (Fig. 2E, ser15). Exogenous Δ40p53 expression was confirmed using antibodies HR231, CM1, and pAb1801 (Fig. 2E, bottom panels). We found that serine 15 phosphorylation was increased in cells infected with Δ40p53V even in the absence of ionizing radiation (Fig. 2E, 0h). Serine 15 phosphorylation was increased as early as 3-hours and sustained to 9-hours post irradiation in both uninfected and infected cells, but to significantly higher levels in Δ40p53 expressing cells (Fig. 2E, ser15). Total full-length p53 as detected by DO1 was similarly affected (Fig. 2E, DO1). Although there were increased levels of phospho-p53 in Δ40p53 expressing cells compared to EV control, there was no significant difference within each group over time (Supplemental Fig. S5). We observed similar effects of Δ40p53 and γ-irradiation in a second melanoma cell line (WM266) and in human melanocytes (Supplemental Fig. S6). These results suggested that Δ40p53 enhanced the activation of p53 in response to genotoxic stress through ATM/ATR-dependent phosphorylation of serine 15.
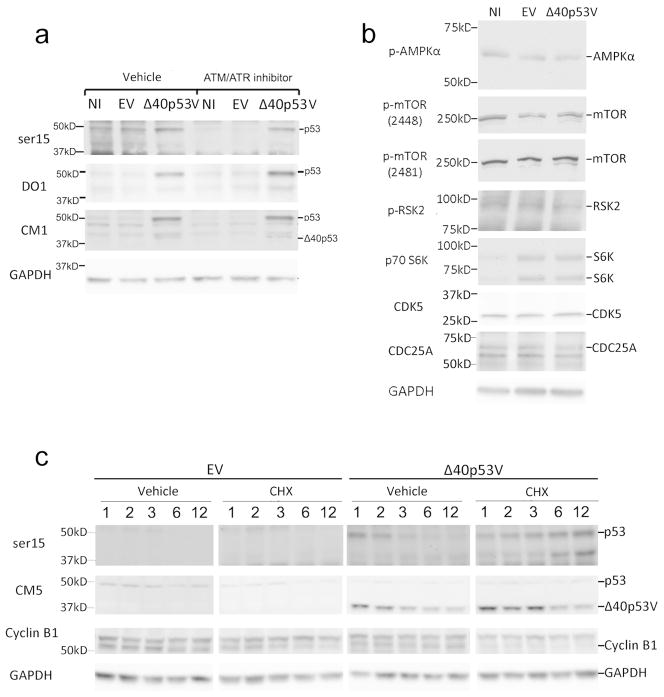
Given that p53 can be activated by γ-irradiation through the canonical ATM/ATR-serine15 phosphorylation pathway, we hypothesized that, Δ40p53, similar to γ-irradiation, might act through ATM/ATR to increase serine-15 phosphorylated p53 levels. To test the dependence of serine 15 phosphorylation on the activity of the ATM/ATR pathway, we again infected cells with Δ40p53V, but this time in the presence of CGK733, an ATM/ATR inhibitor (Bhattacharya et al., 2009; Crescenzi et al., 2008). Inhibition of ATM/ATR kinase activity prevented serine 15 phosphorylation in EV and NI controls, but had no effect on the level of serine 15 phosphorylation in Δ40p53-transduced cells (Fig. 3A). This suggests that Δ40p53V acts through a mechanism that is different from that of ATM/ATR activation seen with γ-irradiation. We also compared the levels of other kinases and phosphatases known to affect serine 15 phosphorylation, but did not find any significant changes that could account for the increased phosphorylation seen in Δ40p53-infected cells (Fig. 3B). What we did find, however, was a strong, Δ40p53-dependent increase in serine 15 phosphorylation under conditions that induce ER stress and the unfolded protein response (UPR) (Ito et al., 2006; Shenkman et al., 2007). As shown in Fig. 3C, blocking protein translation with cycloheximide led to a gradual increase in the level of serine 15 phosphorylated p53 concurrent with the decline of control proteins, such as cyclin B1, and virally-encoded Δ40p53 (Fig. 3C, CM5). Together with previous results, these data suggest that activation of p53 by Δ40p53, which is reflected in the phosphorylation of serine 15, is primarily in response to proteotoxic rather than genotoxic stress brought on by high levels of Δ40p53 relative to full-length p53.
Figure 3. Δ40p53 activates p53 under proteotoxic, but not genotoxic, conditions.
(A) Serine 15 phosphorylated p53 in A375 cells infected with EV, Δ40p53V, or NI treated with ATM/ATR inhibitor CGK733 (10μM). (B) Kinases and phosphatases affecting p53 serine 15 phosphorylation in Δ40p53V, EV, and NI in A375 cells. Phosphorylated AMP-activated protein kinase alpha (p-AMPKα); phosphorylated mammalian target of rapamycin (p-mTOR); phosphorylated ribosomal protein S6 kinase 2 (p-RSK2); p70 ribosomal S6 kinase (p70 RSK2); cyclin dependent kinase 5 (CDK5) and cell division cycle 25 homolog A (CDC25A). (C) Serine 15 phosphorylated p53 in Δ40p53-infected A375 cells treated with cycloheximide (CHX). Cells were infected with EV or Δ40p53V and treated with 0 or 100μg/mL cycloheximide for 1, 2, 3, 6, or 12-hours. p53 and Δ40p53V detected by ser15 and CM5. Cyclin B1 levels shown to determine effectiveness of cycloheximide treatment.
Δ40p53 increases the expression of apoptotic gene, PIDD, and suppresses expression of cell cycle arrest gene, p21
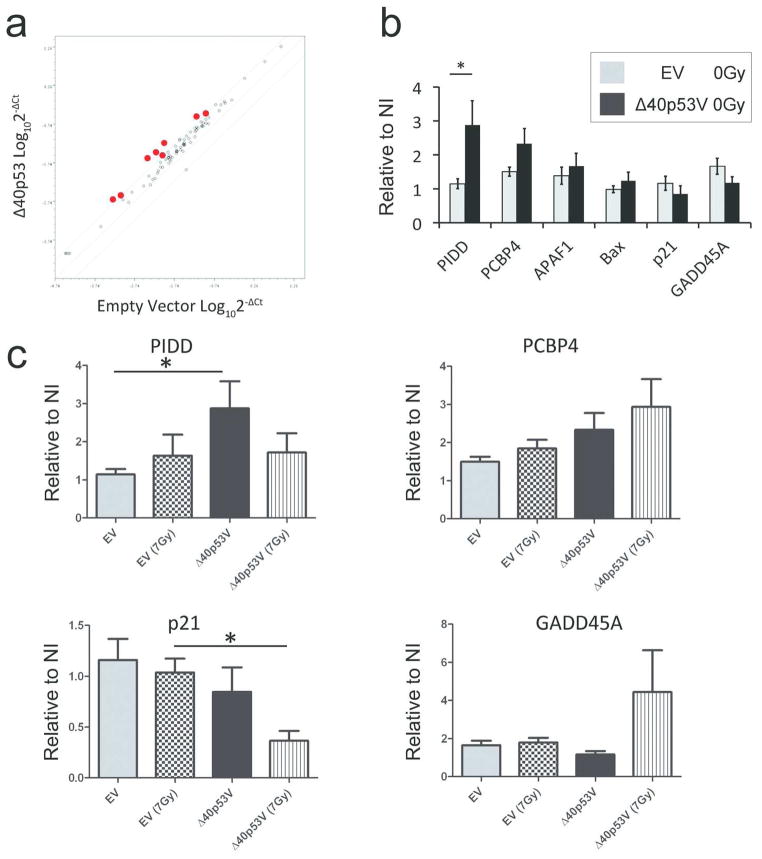
p53 exerts its tumor suppressor activity and cellular response to damage primarily as a transcription factor. We therefore asked if the presence of excess Δ40p53 might alter the levels of p53 transcription target gene expression. To discover candidate p53 targets altered by the introduction of Δ40p53, we used a PCR array containing 84 known p53 targets to compare fold changes in Δ40p53-transduced cells relative to EV infected cells (Supplemental Figure S7A, upper panel 0Gy). When comparing Δ40p53V samples to EV, we found that the majority of transcript levels relative to EV were within a 3-fold dynamic range based on a log10-scale (Fig. 4A, black circles). However, there were eight target genes outside this dynamic range (Fig. 4A, red circles), and 5 of these are involved in apoptosis (Supplemental Figure S7B). Based on these data, we focused on the expression of individual genes involved in apoptosis (PIDD, PCBP4, APAF1, and Bax) and on two cell cycle arrest genes (p21 and GADD45), which exhibited reduced expression in the array. Single gene transcript levels determined by qPCR revealed an increase in the apoptotic gene, PIDD, in Δ40p53 infected cells compared to EV controls (Fig. 4B). These data were consistent with the increased apoptosis observed in the presence of Δ40p53V (Fig. 2A and 2B).
Figure 4. Δ40p53 increases expression of apoptotic gene, PIDD, and suppresses expression of cell cycle arrest gene, p21.
(A) Potential p53 gene targets altered by Δ40p53V. Log-scale dot plot of gene target levels in Δ40p53V versus EV. Eight genes (red circles) fell outside of the 3-fold dynamic range (delineated by parallel lines); see Supplemental Fig. S7. (B) Relative expression of p53 gene targets. Quantitative PCR analysis of selected p53 targets in A375 cells infected with EV (grey bars) and Δ40p53V (black bars). p53-induced protein with a death domain (PIDD); Poly(rC)-binding protein 4 (PCBP4); Apoptotic protease activating factor 1 (APAF1); Bcl-2– associated X protein (Bax); Cyclin dependent kinase 1 (p21); Growth arrest and DNA-damage-inducible protein alpha (GADD45A). *p<0.05. (C) Relative expression of p53 targets (PIDD, PCBP4, p21, and GADD45A) in Δ40p53-infected, γ-irradiated A375 cells. *p<0.05.
Next, we screened 84 known p53 targets by PCR array 3-hours after exposure to 7Gy γ-irradiation in A375 melanoma cells infected with Δ40p53V or EV (Supplemental Figure S7A, bottom panel 7Gy). Changes in p21 and PIDD were verified by single-gene qPCR (Fig. 4C).
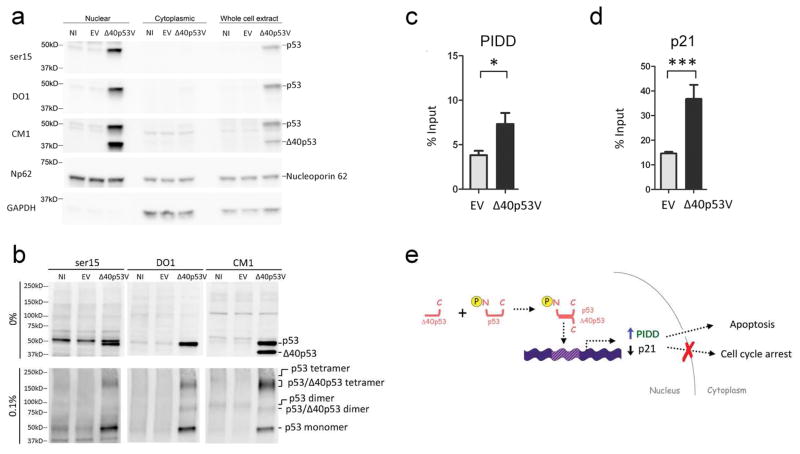
Δ40p53 oligomerizes with endogenous activated p53 in the nucleus
To determine the mechanism by which Δ40p53 could alter p53 target gene expression and increase apoptosis, we first asked if both activated p53 and Δ40p53 localized to the nucleus. We fractionated Δ40p53V or EV-infected A375 melanoma cells and determined the subcellular localization of Δ40p53 and full-length p53 by western blot analysis (Fig. 5A). Δ40p53, serine 15 phosphorylated p53, and total full-length p53 were exclusively found in the nucleus. The small amount of endogenous p53 detected in the cytoplasmic fraction is most likely due to carry-over from the nuclear fraction, as indicated by the presence of NP62, a nuclear protein. We could not detect any Δ40p53 in the cytoplasmic fraction. To determine if Δ40p53 formed hetero-tetramers with endogenous activated p53, we treated nuclear fractions of Δ40p53-infected cells with glutaraldehyde to cross link proteins and looked for higher molecular weight oligomers by western blot analysis as previously described (Powell et al., 2008; Ungewitter and Scrable, 2010a). Using antibodies that detect activated p53 (serine 15 phosphorylated p53), full length p53 (DO1 and CM1), and Δ40p53 (CM1), we found bands at molecular weights corresponding to p53 tetramers, p53/Δ40p53 tetramers, p53 dimers, and p53/Δ40p53 dimers in glutaraldehyde-treated, but not untreated, nuclear extracts (Fig. 5B; compare upper and lower panels). p53-specific antibodies that recognize epitopes in the N-terminus of the protein (ser15 and DO1) detected two bands, one migrating above the 150kD marker in glutaraldehyde treated samples and the other at 50kD corresponding to p53 monomers.
Figure 5. Δ40p53 oligomerizes with endogenous activated p53 in the nucleus and increases promoter occupancy of PIDD and p21.
(A) Nuclear and cytoplasmic fractions of A375 cells infected with Δ40p53V. Nucleoporin 62 (Np62) and GAPDH were used as nuclear and cytoplasmic markers, respectively. (B) Higher order oligomers in A375 melanoma cells infected with Δ40p53V. Nuclear fractions of cells infected were treated with glutaraldehyde (0.1%). p53 promoter occupancy at PIDD (C) and p21 (D) promoters. p53 antibodies were used to immunoprecipitate p53, Δ40p53, and activated p53 from Δ40p53V-infected A375 cells for ChIP analysis (see Supplemental Fig. S8). *p<0.05; ***p<0.001. (E) A model for how Δ40p53 alters cell fate. Endogenous p53 can be activated and directed by Δ40p53 to favor apoptosis over cell cycle arrest (even with γ-irradiation) in tumor cells. Our results are consistent with a role for Δ40p53 in the reactivation of p53-dependent tumor suppression.
Using CM1, which recognizes epitopes present in both p53 and Δ40p53, we found that all detectable Δ40p53 (migrating above the 37kD marker in 0% glutaraldehyde samples) was bound in tetramers with p53. These tetramers migrated above the 150kD marker in glutaraldehyde-treated samples and were detected by all 3 antibodies. We could detect p53 monomers but no Δ40p53 monomers in glutaraldehyde treated samples. A band migrating at 200kD above the p53/Δ40p53 tetramer band (best visualized with CM1 in NI and EV controls) indicated the apparent migration of endogenous p53 tetramers and was used as a reference point. Likewise in the CM1 lanes, a band at approximately 100kD in control lanes was used as a reference point for endogenous p53 dimers. The band migrating below 100kD in the Δ40p53V lane, therefore, must correspond to p53/Δ40p53 dimers. From these data, it is clear that Δ40p53 oligomerizes with full-length p53 and, more importantly, forms tetramers with activated p53 in the nucleus. This suggested a molecular model by which the presence of Δ40p53 might affect the expression of p53 target genes (identified by PCR array and qPCR analysis) by modifying the ability of activated tetramers to bind DNA.
Δ40p53 increases promoter occupancy of PIDD and p21
p53 isoforms, including Δ40p53, have been shown to modulate promoter occupancy of p53 gene targets by full-length p53, a known transcription factor, and subsequently alter the expression of downstream targets (Bourdon et al., 2005a; Mills, 2005; Ungewitter and Scrable, 2010a). We hypothesized that the increase in PIDD and decrease in p21 transcript levels could be due to altered promoter occupancy by Δ40p53/p53 complexes. We tested this hypothesis by infecting A375 melanoma cells with either EV or Δ40p53V and used chromatin immunoprecipitation (ChIP) assays to determine promoter occupancy at the LRDD and CDKN1A genes encoding PIDD and p21, respectively. p53 was immunoprecipitated from chromatin complexes using serine 15 phosphorylated p53, pAb421, and 9282 antibodies. Rabbit IgG was used as a control. At the PIDD promoter (Fig. 5C), immunoprecipitating polyclonal p53 antibody 9282 revealed significantly increased occupancy in Δ40p53-infected cells compared to cells infected with the empty virus. Similarly, analysis of p21 promoter occupancy demonstrated a significant increase in p53 molecules bound in the presence of Δ40p53V compared to EV with p53 antibodies pAb421 (Fig. 5D) and 9282 (Supplemental Fig. S8). We did not find a significant difference in promoter occupancy using the serine 15 phosphorylated p53 antibody (Supplemental Fig. S8).
In summary, we found that exogenous Δ40p53 increases p53-dependent cell death by apoptosis in both cancer and normal cells without altering cell cycle arrest. Consistent with previous studies, γ-irradiation did not induce cell cycle arrest (Kaufmann et al., 2008), nor did it change the fraction of dead cells. These data suggest that the decision to undergo apoptosis is favored in the presence of Δ40p53 and cannot be amended by exogenous stimuli such as γ-irradiation. Δ40p53 increased levels of endogenous, activated (serine 15 phosphorylated) p53 in A375 melanoma cells, which could be enhanced by proteotoxic agents, such as cycloheximide. Increased levels of endogenous p53 were not due to phosphorylation by ATM/ATR as seen with γ-irradiation. Δ40p53 increased transcript levels of apoptotic targets, such as PIDD, while suppressing the expression of cell cycle arrest genes, such as p21. We found that Δ40p53 formed nuclear hetero-tetramers with activated p53 and increased promoter occupancy at both PIDD and p21 genes.
Discussion
Δ40p53 is an isoform of p53 that is normally expressed only during embryogenesis and in the stem cell compartment of adult tissues (Medrano et al., 2009a; Ungewitter and Scrable, 2010b). Recently, we have also identified Δ40p53 as the only consistently expressed p53 isoform in glioblastoma multiforme (Takahashi et al., 2012), the most common brain tumor in adults. To gain insight into the role of Δ40p53 in cancer, we utilized a lentiviral system to overexpress Δ40p53 in tumors, such as melanoma, and normal counterparts (melanocytes) with or without functional p53. We chose an overexpression model to parallel the p53 expression profiles previously described in glioblastoma multiforme (Takahashi et al., 2012). Our results are summarized in the model presented in Fig. 5E. We found that Δ40p53 shifted melanoma cell fate in a p53-dependent manner to favor apoptosis over cell cycle arrest, even in the presence of γ-irradiation, a known inducer of DNA damage and cell cycle arrest. Δ40p53 increased endogenous, activated p53 levels in melanoma cells and increased expression of downstream p53 targets such as PIDD and suppressed others such as p21. Δ40p53 formed nuclear tetramers with endogenous, serine 15 phosphorylated p53 and directly altered promoter occupancy of PIDD and p21. Endogenous p53 phosphorylation by Δ40p53 was dependent on proteotoxic and not genotoxic damage, which led to significant levels of cell death. Our results are consistent with a role for Δ40p53 in the reactivation of p53-dependent tumor suppression.
Although p53 is the most frequently inactivated tumor suppressor in human tumors, certain types of cancer, such as melanoma, have relatively few TP53 mutations (Albino et al., 1994; Gwosdz et al., 2006; Li et al., 2006; Montano et al., 1994; Soto et al., 2005; Weiss et al., 1995; Zerp et al., 1999). Despite high levels of wildtype protein expression, studies have showed that p53 fails to execute downstream functions such as cell cycle arrest and apoptosis in melanoma primarily due to a failure in transactivating target genes (Avery-Kiejda et al., 2011; Houben et al., 2011; Knopf et al., 2011). Consequently, there have been a number of studies that have focused on the idea of re-activating or ‘rescuing’ p53 function in cells with wildtype TP53 alleles by introducing exogenous p53 (Kichina et al., 2003; Lane et al., 2010) or by inhibiting Mdm, which has been found to be overexpressed in melanomas (Danovi et al., 2004; Gembarska et al., 2012; Ji et al., 2012; Muthusamy et al., 2006; Terzian et al., 2010). Our results reveal a way in which endogenous p53 can be activated and directed to increase apoptosis in tumor cells. Validation of these results using in vivo tumor models would be necessary to determine any therapeutic utility of these findings.
Materials and Methods
Lentiviral vectors and transduction
A375 melanoma cells (ATCC, Manassas) and melanocytes (Life Technologies, Grand Island) were cultured according to manufacturer’s protocols. Primary glioblastoma cells and mouse embryonic fibroblasts (MEFs) were cultured as previously described (Carlson et al., 2011; Maier et al., 2004b). The Δ40p53 fragment was PCR amplified and cloned into the pSIN construct (gift of Dr. Yasuhiro Ikeda, Mayo Clinic) and lentivirus produced as previously described (Demaison et al., 2002). Transduction efficiency >95% (based on GFP fluorescence) was achieved in all infected cell types prior to carrying out downstream assays (approximately five days post infection for cancer cells and ten days for non-transformed cells).
Western blot analysis
Western blot analyses as previously described (Ungewitter and Scrable, 2010b). p53 antibody include: DO1 and HR231 (Santa Cruz Technologies, Inc., Santa Cruz), pAb421 and pAb1801 (Calbiochem/EMD Chemicals, Gibbstown), phospho-p53 (ser15) (Cell Signaling, Danvers), CM1 (Vector Labs, Burlingame), and GAPDH (Ambion, Foster City). PARP I antibodies (Promega, Madison) (Budihardjo et al., 1998). Serine 15 phosphorylation kinase and phosphatase antibodies: p-AMPKα, p-mTOR, p-RSK2, p70 S6K, CDK5, CDC25A (Cell Signaling, Danvers). Cyclin B1 (Cell Signaling, Danvers); Np62 (BD Transduction Laboratories, San Jose).
ATM/ATR inhibition and cycloheximide treatment
Infected A375 cells were treated with ATM/ATR inhibitor, CGK733 (10μM), or vehicle for approximately 60 hours and harvested for western blot analysis. Infected cells were treated with cycloheximide (100μg/mL) or vehicle and harvested at 1, 2, 3, 6, and 12 hours.
Subcellular fractionation
Infected A375 cells were harvested for nuclear and cytoplasmic fractionation using the Qproteome Cell Compartment Kit (Qiagen, Germantown). For tetramer detection, nuclear fractions were treated with 0.1% glutaraldehyde and analyzed as previously described (Ungewitter and Scrable, 2010b).
Cell cycle and apoptosis assays
The fraction of dead cells was determined by ethidium homodimer binding (Life Technologies, Grand Island), percentage of apoptotic cells by Annexin V: PE Apoptosis Detection Kit I (BD Pharmingen, San Jose), and cell cycle profiles by propidium iodide staining. PARP I products were detected by western blot analysis. Cells were γ-irradiated with 7Gy and harvested at 3, 6, or 9-hours post irradiation. Flow data were analyzed with ModFit and CellQuest.
Quantitative PCR and Chromatin Immunoprecipitation Assays
RNA harvested using a miRNeasy Mini Kit (Qiagen, Valencia) was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Grand Island). iQ SYBR Green Supermix was used for single gene PCR (Biorad, Hercules). The RT2 Profiler PCR array was used (SABiosciences, Valencia) for candidate p53 target discovery. Chromatin immunoprecipitation analysis was carried out using SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell Signaling, Danvers). Immunoprecipitating antibodies: serine 15 phosphorylated p53, 9282, and rabbit IgG (Cell Signaling, Danvers); pAb421 (Calbiochem/EMD Chemicals, Gibbstown).
Supplementary Material
Acknowledgments
We thank Dr. Luigi Puglielli (University of Wisconsin, Madison) for helpful comments on the UPR; Dr. Yasuhiro Ikeda for the original lentivirus construct and expert advice on lentiviral systems; Dr. Scott Kaufmann for PARP I antibodies and expert advice on apoptosis assays; Wendy Nevala and the Flow Cytometry Core facility for help with apoptosis and cell cycle analysis; Dr. Adrienne Grzenda for PCR array expertise.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Albino AP, Vidal MJ, McNutt NS, Shea CR, Prieto VG, Nanus DM, et al. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4:35–45. doi: 10.1097/00008390-199402000-00006. [DOI] [PubMed] [Google Scholar]
- Anensen N, Oyan AM, Bourdon JC, Kalland KH, Bruserud O, Gjertsen BT. A distinct p53 protein isoform signature reflects the onset of induction chemotherapy for acute myeloid leukemia. Clin Cancer Res. 2006;12:3985–92. doi: 10.1158/1078-0432.CCR-05-1970. [DOI] [PubMed] [Google Scholar]
- Avery-Kiejda KA, Bowden NA, Croft AJ, Scurr LL, Kairupan CF, Ashton KA, et al. P53 in human melanoma fails to regulate target genes associated with apoptosis and the cell cycle and may contribute to proliferation. BMC Cancer. 2011;11:203. doi: 10.1186/1471-2407-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery-Kiejda KA, Zhang XD, Adams LJ, Scott RJ, Vojtesek B, Lane DP, et al. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin Cancer Res. 2008;14:1659–68. doi: 10.1158/1078-0432.CCR-07-1422. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harbor perspectives in biology. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2:a001198. doi: 10.1101/cshperspect.a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Ray RM, Johnson LR. Role of polyamines in p53-dependent apoptosis of intestinal epithelial cells. Cell Signal. 2009;21:509–22. doi: 10.1016/j.cellsig.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Bourdon JC, Coates PJ, Sjostrom B, Nylander K. Expression of p53 isoforms in squamous cell carcinoma of the head and neck. Eur J Cancer. 2007;43:617–23. doi: 10.1016/j.ejca.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005a;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005b;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budihardjo, Poirier GG, Kaufmann SH. Apparent cleavage of poly(ADP-ribose) polymerase in non-apoptotic mouse LTA cells: an artifact of cross-reactive secondary antibody. Mol Cell Biochem. 1998;178:245–9. doi: 10.1023/a:1006808001462. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr Protoc Pharmacol. 2011;Chapter 14(Unit 14):6. doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, et al. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–8. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- Crescenzi E, Palumbo G, de Boer J, Brady HJ. Ataxia telangiectasia mutated and p21CIP1 modulate cell survival of drug-induced senescent tumor cells: implications for chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:1877–87. doi: 10.1158/1078-0432.CCR-07-4298. [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–43. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–13. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012 doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Stewart D, Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24:7987–97. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Marcel V, Olivier M, Oren M, Rotter V, Hainaut P. Understanding wild-type and mutant p53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Ther. 2011;18:2–11. doi: 10.1038/cgt.2010.63. [DOI] [PubMed] [Google Scholar]
- Grover R, Candeias MM, Fahraeus R, Das S. p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene. 2009;28:2766–72. doi: 10.1038/onc.2009.138. [DOI] [PubMed] [Google Scholar]
- Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–98. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- Grover R, Sharathchandra A, Ponnuswamy A, Khan D, Das S. Effect of mutations on the p53 IRES RNA structure: implications for de-regulation of the synthesis of p53 isoforms. RNA Biol. 2011;8:132–42. doi: 10.4161/rna.8.1.14260. [DOI] [PubMed] [Google Scholar]
- Gwosdz C, Scheckenbach K, Lieven O, Reifenberger J, Knopf A, Bier H, et al. Comprehensive analysis of the p53 status in mucosal and cutaneous melanomas. Int J Cancer. 2006;118:577–82. doi: 10.1002/ijc.21366. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Hainaut P. Massively regulated genes: the example of TP53. The Journal of pathology. 2010;220:164–73. doi: 10.1002/path.2637. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Houben R, Hesbacher S, Schmid CP, Kauczok CS, Flohr U, Haferkamp S, et al. High-level expression of wild-type p53 in melanoma cells is frequently associated with inactivity in p53 reporter gene assays. PLoS One. 2011;6:e22096. doi: 10.1371/journal.pone.0022096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Kiyosawa N, Kumagai K, Manabe S, Matsunuma N, Yamoto T. Molecular mechanism investigation of cycloheximide-induced hepatocyte apoptosis in rat livers by morphological and microarray analysis. Toxicology. 2006;219:175–86. doi: 10.1016/j.tox.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol. 2012;132:356–64. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- Kaufmann WK, Nevis KR, Qu P, Ibrahim JG, Zhou T, Zhou Y, et al. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–87. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichina JV, Rauth S, Das Gupta TK, Gudkov AV. Melanoma cells can tolerate high levels of transcriptionally active endogenous p53 but are sensitive to retrovirus-transduced p53. Oncogene. 2003;22:4911–7. doi: 10.1038/sj.onc.1206741. [DOI] [PubMed] [Google Scholar]
- Knopf A, Plettenberg C, Pickhard A, Bas M, Reifenberger J, Bier H, et al. Analysis of the functional integrity of the p53 tumor-suppressor gene in malignant melanoma. Melanoma Res. 2011 doi: 10.1097/CMR.0b013e328347ee04. [DOI] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harbor perspectives in biology. 2010;2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Li W, Sanki A, Karim RZ, Thompson JF, Soon Lee C, Zhuang L, et al. The role of cell cycle regulatory proteins in the pathogenesis of melanoma. Pathology. 2006;38:287–301. doi: 10.1080/00313020600817951. [DOI] [PubMed] [Google Scholar]
- Machado-Silva A, Perrier S, Bourdon JC. p53 family members in cancer diagnosis and treatment. Semin Cancer Biol. 2010;20:57–62. doi: 10.1016/j.semcancer.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004a;18:306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004b;18:306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–24. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Perrier S, Aoubala M, Ageorges S, Groves MJ, Diot A, et al. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS letters. 2010;584:4463–8. doi: 10.1016/j.febslet.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging. 2009a;30:483–97. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging. 2009b;30:483–97. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. p53: link to the past, bridge to the future. Genes Dev. 2005;19:2091–9. doi: 10.1101/gad.1362905. [DOI] [PubMed] [Google Scholar]
- Montano X, Shamsher M, Whitehead P, Dawson K, Newton J. Analysis of p53 in human cutaneous melanoma cell lines. Oncogene. 1994;9:1455–9. [PubMed] [Google Scholar]
- Muthusamy V, Hobbs C, Nogueira C, Cordon-Cardo C, McKee PH, Chin L, et al. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer. 2006;45:447–54. doi: 10.1002/gcc.20310. [DOI] [PubMed] [Google Scholar]
- Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. The Journal of biological chemistry. 1998;273:33533–9. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harbor perspectives in biology. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Powell DJ, Hrstka R, Candeias M, Bourougaa K, Vojtesek B, Fahraeus R. Stress-dependent changes in the properties of p53 complexes by the alternative translation product p53/47. Cell Cycle. 2008;7:950–9. doi: 10.4161/cc.7.7.5626. [DOI] [PubMed] [Google Scholar]
- Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkman M, Tolchinsky S, Lederkremer GZ. ER stress induces alternative nonproteasomal degradation of ER proteins but not of cytosolic ones. Cell Stress Chaperones. 2007;12:373–83. doi: 10.1379/CSC-281.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–81. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto JL, Cabrera CM, Serrano S, Lopez-Nevot MA. Mutation analysis of genes that control the G1/S cell cycle in melanoma: TP53, CDKN1A, CDKN2A, and CDKN2B. BMC Cancer. 2005;5:36. doi: 10.1186/1471-2407-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Giannini C, Sarkaria JN, Schroeder M, Rogers J, Mastroeni D, et al. p53 isoform profiling in glioblastoma and injured brain. Oncogene. 2012 doi: 10.1038/onc.2012.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T, Torchia EC, Dai D, Robinson SE, Murao K, Stiegmann RA, et al. p53 prevents progression of nevi to melanoma predominantly through cell cycle regulation. Pigment Cell Melanoma Res. 2010;23:781–94. doi: 10.1111/j.1755-148X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010a;24:2408–19. doi: 10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010b;24:2408–19. doi: 10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Weiss J, Heine M, Arden KC, Korner B, Pilch H, Herbst RA, et al. Mutation and expression of TP53 in malignant melanomas. Recent Results Cancer Res. 1995;139:137–54. doi: 10.1007/978-3-642-78771-3_10. [DOI] [PubMed] [Google Scholar]
- Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–7. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- Zerp SF, van Elsas A, Peltenburg LT, Schrier PI. p53 mutations in human cutaneous melanoma correlate with sun exposure but are not always involved in melanomagenesis. Br J Cancer. 1999;79:921–6. doi: 10.1038/sj.bjc.6690147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.