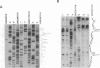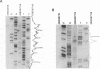Abstract
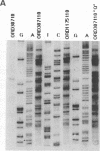
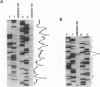
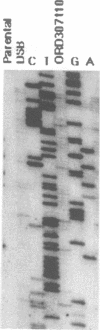
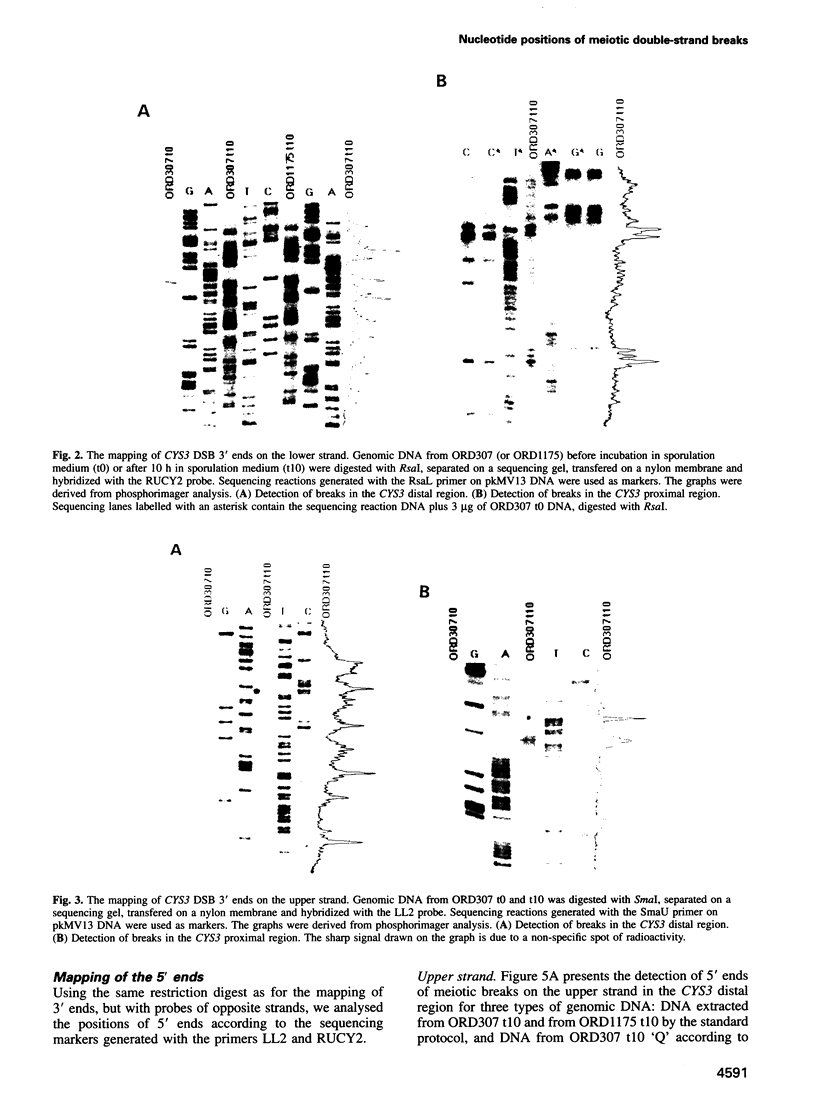
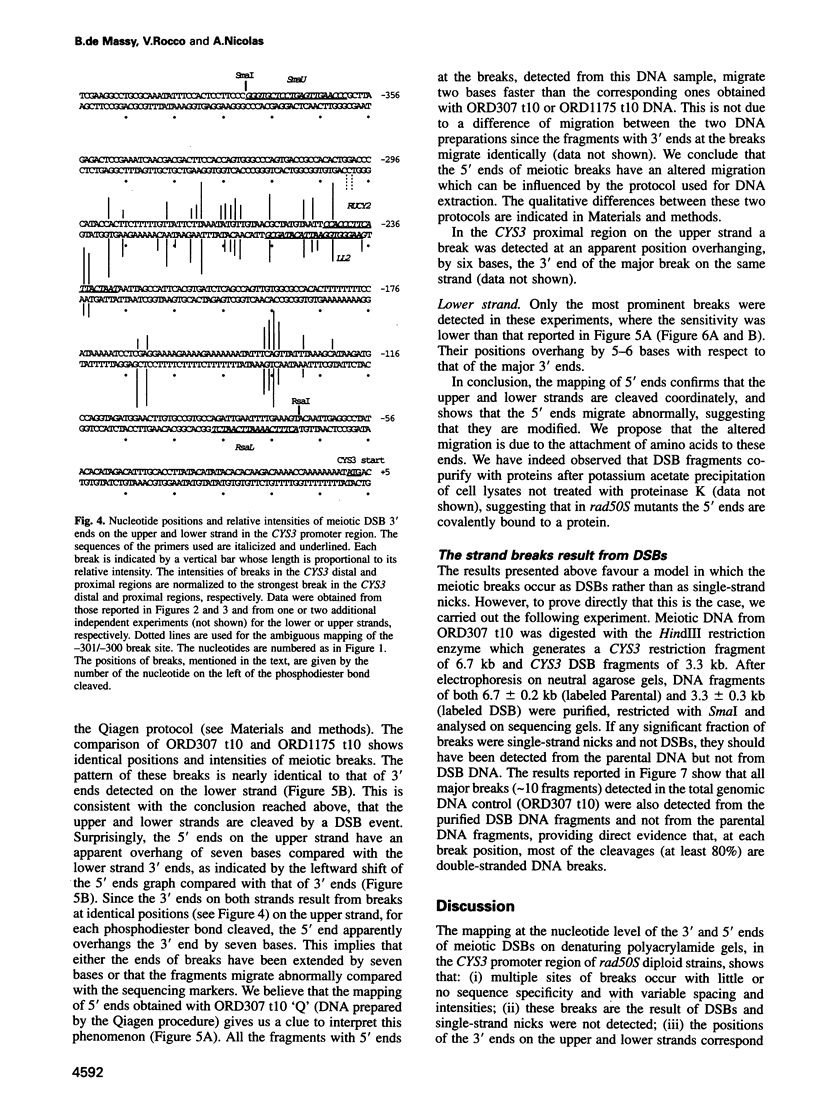
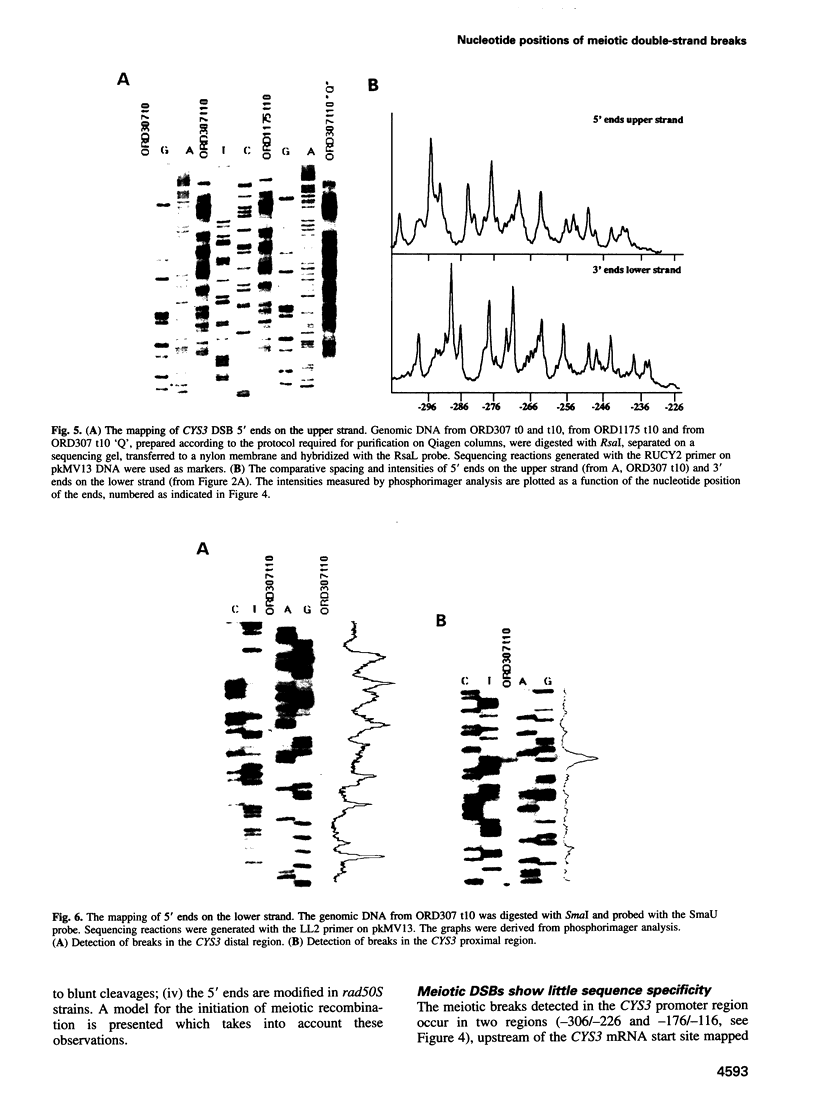
Initiation of meiotic recombination in the yeast Saccharomyces cerevisiae occurs by localized DNA double-strand breaks (DSBs) at several locations in the genome, corresponding to hot spots for meiotic gene conversion and crossing over. The meiotic DSBs occur in regions of chromatin that are hypersensitive to nucleases. To gain insight into the molecular mechanism involved in the formation of these DSBs, we have determined their positions at the nucleotide level at the CYS3 hot spot of gene conversion on chromosome I. We found four major new features of these DSBs: (i) sites of DSBs are multiple with varying intensities and spacing within the promoter region of the CYS3 gene; (ii) no consensus sequence can be found at these sites, indicating that the activity involved in DSB formation has little or no sequence specificity; (iii) the breaks are generated by blunt cleavages; and (iv) the 5' ends are modified in rad50S mutant strains, where the processing of these ends is known to be prevented. We present a model for the initiation of meiotic recombination taking into account the implications of these results.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Baker H. V. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9443–9447. doi: 10.1073/pnas.88.21.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Fitzgerald-Hayes M., O'Brien T. C. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989 Jun 25;264(18):10843–10850. [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992 May 1;69(3):439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994 Apr 8;77(1):1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Cai M. J., Davis R. W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989 Jun;9(6):2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Cherest H., Surdin-Kerjan Y. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics. 1992 Jan;130(1):51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Massy B., Baudat F., Nicolas A. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11929–11933. doi: 10.1073/pnas.91.25.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale S. S., Lane A. C. Compilation of sequence-specific DNA-binding proteins implicated in transcriptional control in fungi. Nucleic Acids Res. 1993 Dec 11;21(24):5537–5546. doi: 10.1093/nar/21.24.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Xu F., Petes T. D. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol Cell Biol. 1995 Mar;15(3):1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldway M., Sherman A., Zenvirth D., Arbel T., Simchen G. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double strand breaks. Genetics. 1993 Feb;133(2):159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993 Jan;13(1):373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R. S., Arbel T. Yeast genetics and the fall of the classical view of meiosis. Cell. 1993 Feb 12;72(3):301–303. doi: 10.1016/0092-8674(93)90108-3. [DOI] [PubMed] [Google Scholar]
- Ivanov E. L., Korolev V. G., Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992 Nov;132(3):651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. D., Philippsen P. Purification of a protein binding to the CDEI subregion of Saccharomyces cerevisiae centromere DNA. Mol Cell Biol. 1989 Dec;9(12):5585–5593. doi: 10.1128/mcb.9.12.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo K., Topal M. D. DNA topoisomerase and recombinase activities in Nae I restriction endonuclease. Science. 1995 Mar 24;267(5205):1817–1820. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995 Apr;139(4):1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Tsang J. S., Crowther D. J., Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol Cell Biol. 1994 Aug;14(8):5229–5241. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Drak J., Rice J. A., Crothers D. M. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990 May 1;29(17):4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- Käs E., Laemmli U. K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992 Feb;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wu T. C., Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995 Sep 15;14(18):4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Rowe T. C., Yang L., Tewey K. M., Chen G. L. Cleavage of DNA by mammalian DNA topoisomerase II. J Biol Chem. 1983 Dec 25;258(24):15365–15370. [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Morishima N., Shibata T. An endonuclease with multiple cutting sites, Endo.SceI, initiates genetic recombination at its cutting site in yeast mitochondria. EMBO J. 1992 Jul;11(7):2707–2715. doi: 10.1002/j.1460-2075.1992.tb05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell K. F., Surdin-Kerjan Y., Baker R. E. Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol Cell Biol. 1995 Apr;15(4):1879–1888. doi: 10.1128/mcb.15.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Shibata T., Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994 Dec 1;13(23):5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono B., Tanaka K., Naito K., Heike C., Shinoda S., Yamamoto S., Ohmori S., Oshima T., Toh-e A. Cloning and characterization of the CYS3 (CYI1) gene of Saccharomyces cerevisiae. J Bacteriol. 1992 May;174(10):3339–3347. doi: 10.1128/jb.174.10.3339-3347.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheroff N., Zechiedrich E. L., Gale K. C. Catalytic function of DNA topoisomerase II. Bioessays. 1991 Jun;13(6):269–273. doi: 10.1002/bies.950130603. [DOI] [PubMed] [Google Scholar]
- Ouellette B. F., Clark M. W., Keng T., Storms R. K., Zhong W., Zeng B., Fortin N., Delaney S., Barton A., Kaback D. B. Sequencing of chromosome I from Saccharomyces cerevisiae: analysis of a 32 kb region between the LTE1 and SPO7 genes. Genome. 1993 Feb;36(1):32–42. doi: 10.1139/g93-005. [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991 Sep 20;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Reitman M., Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol. 1990 Jun;10(6):2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V., de Massy B., Nicolas A. The Saccharomyces cerevisiae ARG4 initiator of meiotic gene conversion and its associated double-strand DNA breaks can be inhibited by transcriptional interference. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol Cell Biol. 1993 Jun;13(6):3445–3455. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T. C., Wang J. C., Liu L. F. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986 Apr;6(4):985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983 Jul 10;258(13):8421–8428. [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994 Jan 14;76(1):51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Shibata T., Watabe H., Kaneko T., Iino T., Ando T. On the nucleotide sequence recognized by a eukaryotic site-specific endonuclease, Endo.SceI from yeast. J Biol Chem. 1984 Aug 25;259(16):10499–10506. [PubMed] [Google Scholar]
- Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994 Nov;10(11):408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P. Chromatin structure, not DNA sequence specificity, is the primary determinant of topoisomerase II sites of action in vivo. Mol Cell Biol. 1991 Oct;11(10):4973–4984. doi: 10.1128/mcb.11.10.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe H., Iino T., Kaneko T., Shibata T., Ando T. A new class of site-specific endodeoxyribonucleases. Endo.Sce I isolated from a eukaryote, Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):4663–4665. [PubMed] [Google Scholar]
- West S. C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994 Jan 28;263(5146):515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- Yang L., Rowe T. C., Nelson E. M., Liu L. F. In vivo mapping of DNA topoisomerase II-specific cleavage sites on SV40 chromatin. Cell. 1985 May;41(1):127–132. doi: 10.1016/0092-8674(85)90067-4. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Denniger G. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr Genet. 1994 Feb;25(2):142–149. doi: 10.1007/BF00309540. [DOI] [PubMed] [Google Scholar]
- Zenvirth D., Arbel T., Sherman A., Goldway M., Klein S., Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992 Sep;11(9):3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Nicolas A. The control in cis of the position and the amount of the ARG4 meiotic double-strand break of Saccharomyces cerevisiae. EMBO J. 1993 Apr;12(4):1459–1466. doi: 10.1002/j.1460-2075.1993.tb05789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]