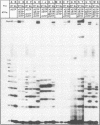
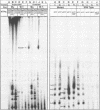
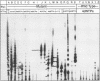
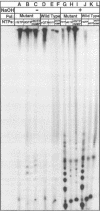
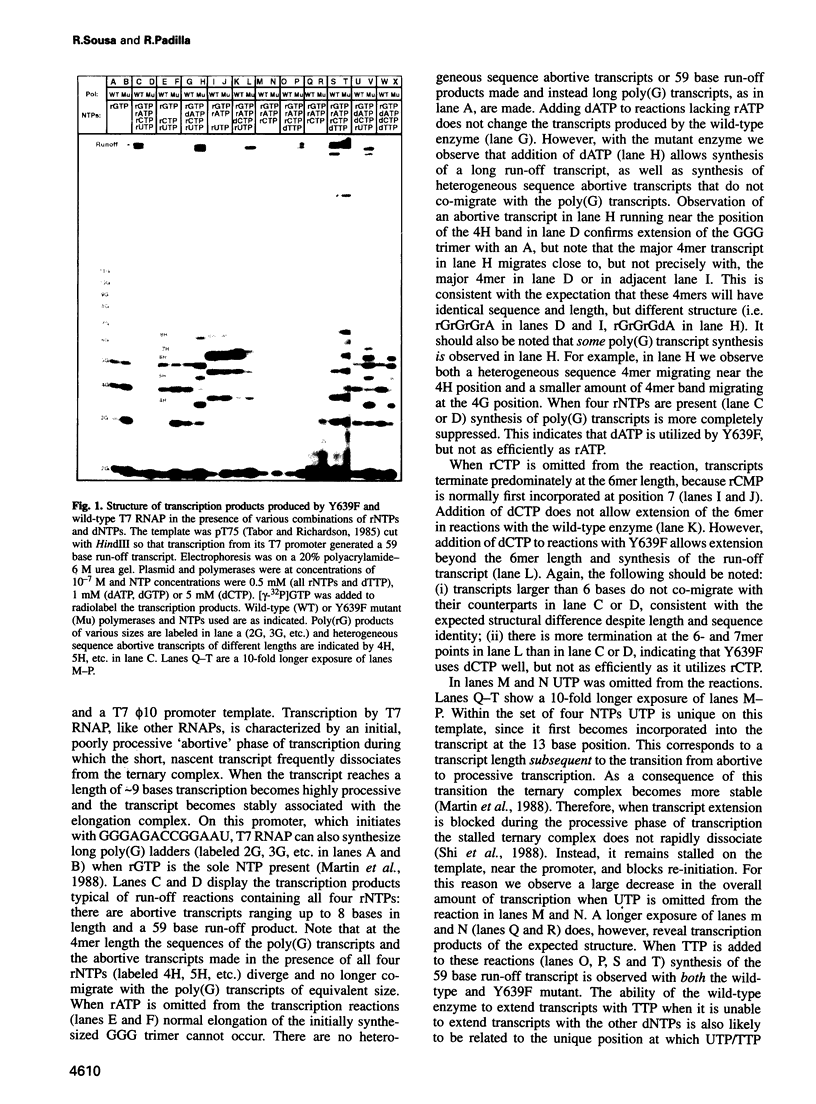
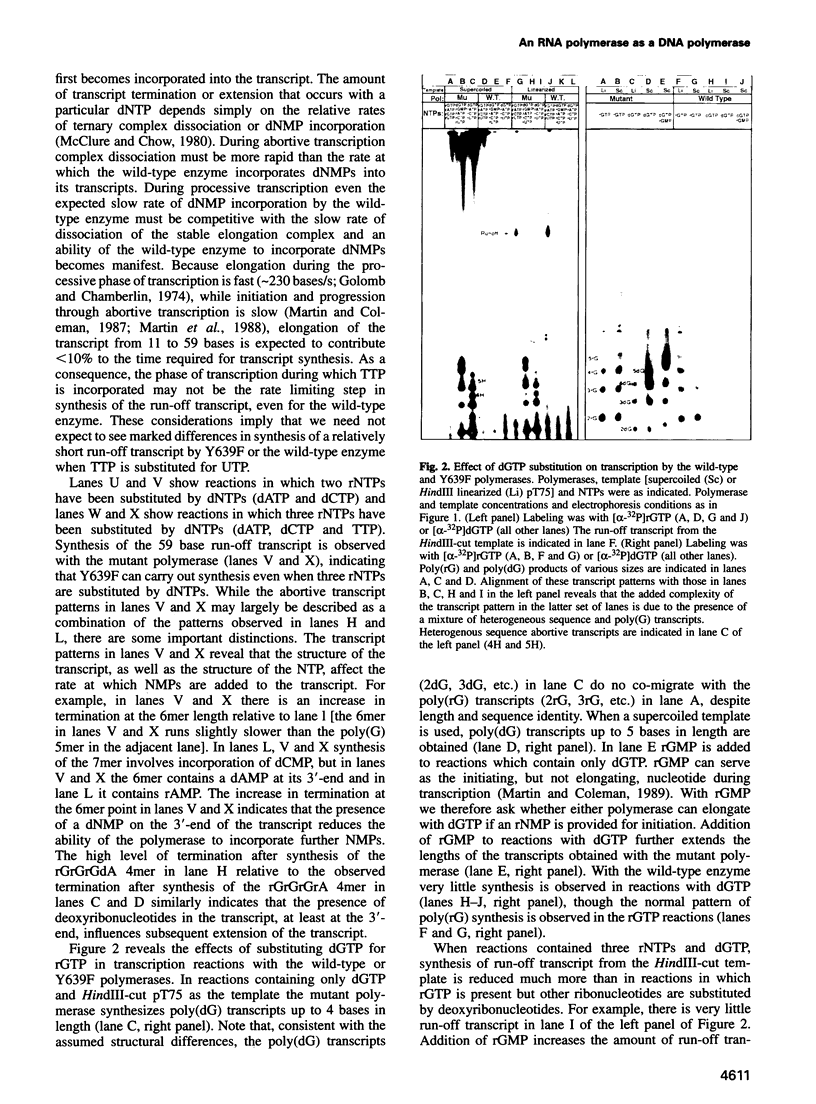
Abstract
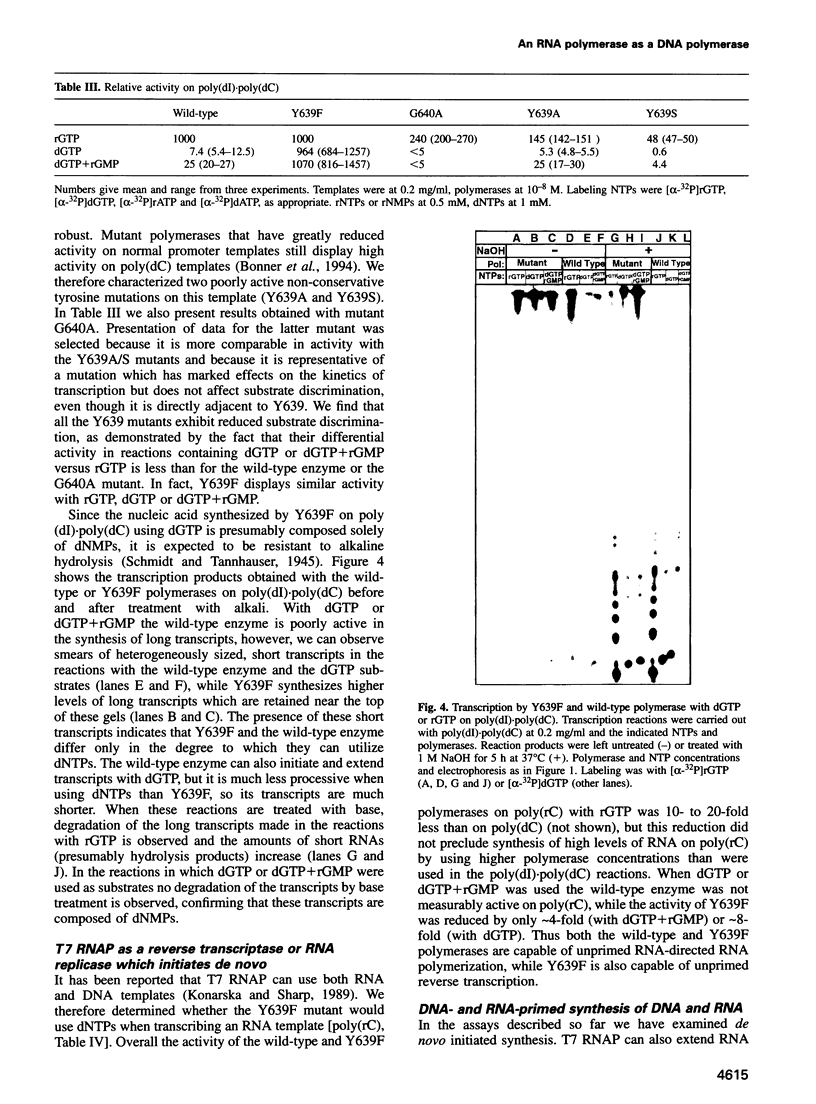
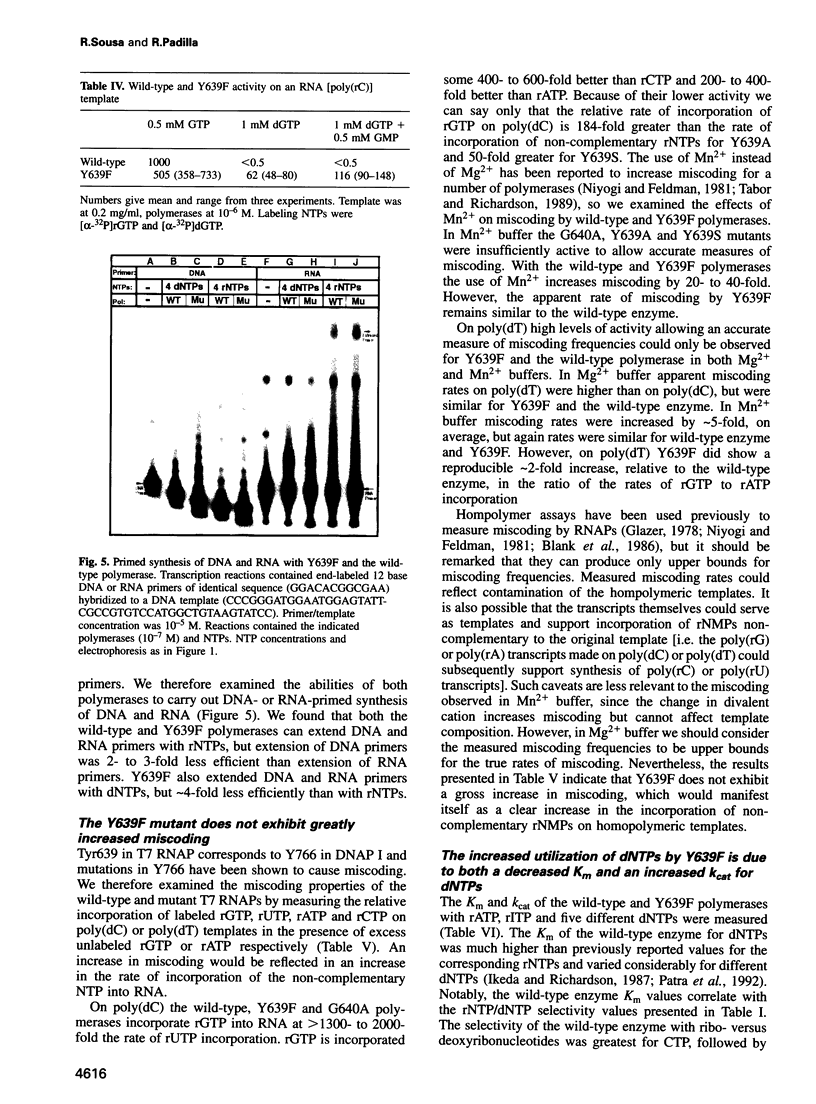
We have identified a T7 RNA polymerase (RNAP) mutant that efficiently utilizes deoxyribonucleoside triphosphates. In vitro this mutant will synthesize RNA, DNA or 'transcripts' of mixed dNMP/rNMP composition depending on the mix of NTPs present in the synthesis reaction. The mutation is conservative, changes Tyr639 within the active site to phenylalanine and does not affect promoter specificity or overall activity. Non-conservative mutations of this tyrosine also reduce discrimination between deoxyribo- and ribonucleoside triphosphates, but these mutations also cause large activity reductions. Of 26 mutations of other residues in and around the active site examined none showed marked effects on rNTP/dNTP discrimination. Mutations of the corresponding tyrosine in DNA polymerase (DNAP) I increase miscoding, though effects on dNTP/rNTP discrimination for the DNAP I mutations have not been reported. This conserved tyrosine may therefore play a similar role in many polymerases by sensing incorrect geometry in the structure of the substrate/template/product due to inappropriate substrate structure or mismatches. T7 RNAP can use RNA templates as well as DNA templates and is capable of both primer extension and de novo initiation. The Y639F mutant retains the ability to use RNA or DNA templates. Thus this mutant can display de novo initiated or primed DNA-directed DNA polymerase, reverse transcriptase, RNA-directed RNA polymerase or DNA-directed RNA polymerase activities depending simply on the templates and substrates presented to it in the synthesis reaction.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astatke M., Grindley N. D., Joyce C. M. Deoxynucleoside triphosphate and pyrophosphate binding sites in the catalytically competent ternary complex for the polymerase reaction catalyzed by DNA polymerase I (Klenow fragment). J Biol Chem. 1995 Jan 27;270(4):1945–1954. doi: 10.1074/jbc.270.4.1945. [DOI] [PubMed] [Google Scholar]
- Blank A., Gallant J. A., Burgess R. R., Loeb L. A. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986 Oct 7;25(20):5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- Bonner G., Lafer E. M., Sousa R. Characterization of a set of T7 RNA polymerase active site mutants. J Biol Chem. 1994 Oct 7;269(40):25120–25128. [PubMed] [Google Scholar]
- Bonner G., Patra D., Lafer E. M., Sousa R. Mutations in T7 RNA polymerase that support the proposal for a common polymerase active site structure. EMBO J. 1992 Oct;11(10):3767–3775. doi: 10.1002/j.1460-2075.1992.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. S., Cowart M., Benkovic S. J. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991 Jan 22;30(3):804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Glazer R. I. Comparisons of the fidelity of transcription of RNA polymerase I and II following N-hydroxy-2-acetylaminofluorene treatment. Nucleic Acids Res. 1978 Jul;5(7):2607–2616. doi: 10.1093/nar/5.7.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem. 1974 May 10;249(9):2858–2863. [PubMed] [Google Scholar]
- Ikeda R. A., Richardson C. C. Interactions of a proteolytically nicked RNA polymerase of bacteriophage T7 with its promoter. J Biol Chem. 1987 Mar 15;262(8):3800–3808. [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989 May 5;57(3):423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., Benkovic S. J. Kinetic mechanism of DNA polymerase I (Klenow). Biochemistry. 1987 Dec 15;26(25):8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Coleman J. E. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987 May 19;26(10):2690–2696. doi: 10.1021/bi00384a006. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Coleman J. E. T7 RNA polymerase does not interact with the 5'-phosphate of the initiating nucleotide. Biochemistry. 1989 Apr 4;28(7):2760–2762. doi: 10.1021/bi00433a002. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Muller D. K., Coleman J. E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988 May 31;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- McClure W. R., Chow Y. The kinetics and processivity of nucleic acid polymerases. Methods Enzymol. 1980;64:277–297. doi: 10.1016/s0076-6879(80)64013-0. [DOI] [PubMed] [Google Scholar]
- Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985 Jul 16;24(15):4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Moroney S. E., Piccirilli J. A. Abortive products as initiating nucleotides during transcription by T7 RNA polymerase. Biochemistry. 1991 Oct 22;30(42):10343–10349. doi: 10.1021/bi00106a036. [DOI] [PubMed] [Google Scholar]
- Niyogi S. K., Feldman R. P. Effect of several metal ions on misincorporation during transcription. Nucleic Acids Res. 1981 Jun 11;9(11):2615–2627. doi: 10.1093/nar/9.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Davis P. A., Sreerama N., Volkin D. B., Middaugh C. R., Woody R. W., Woody A. Y. Bacteriophage T7 RNA polymerase and its active-site mutants. Kinetic, spectroscopic and calorimetric characterization. J Mol Biol. 1994 Mar 18;237(1):5–19. doi: 10.1006/jmbi.1994.1205. [DOI] [PubMed] [Google Scholar]
- Patra D., Lafer E. M., Sousa R. Isolation and characterization of mutant bacteriophage T7 RNA polymerases. J Mol Biol. 1992 Mar 20;224(2):307–318. doi: 10.1016/0022-2836(92)90996-w. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Polesky A. H., Steitz T. A., Grindley N. D., Joyce C. M. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990 Aug 25;265(24):14579–14591. [PubMed] [Google Scholar]
- Ricchetti M., Buc H. E. coli DNA polymerase I as a reverse transcriptase. EMBO J. 1993 Feb;12(2):387–396. doi: 10.1002/j.1460-2075.1993.tb05670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya M. R., Pelletier H., Kumar A., Wilson S. H., Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994 Jun 24;264(5167):1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Hearst J. E. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J Biol Chem. 1988 Jan 5;263(1):527–534. [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Smerdon S. J., Jäger J., Joyce C. M. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994 Dec 23;266(5193):2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo C., Ding J., Jacobo-Molina A., Nanni R. G., Boyer P. L., Hughes S. H., Pauwels R., Andries K., Janssen P. A., Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994 Oct 28;243(3):369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]