Abstract
Objective:
Our objective was to determine whether altered naive CD4 T-cell biology contributes to development of disease progression in secondary progressive multiple sclerosis (SPMS).
Methods:
We compared the naive CD4 T-cell gene expression profiles of 19 patients with SPMS and 14 healthy controls (HCs) using a whole-genome microarray approach. We analyzed surface protein expression of critical genes by flow cytometry after T-cell receptor (TCR) stimulation of naive CD4 T cells isolated from HCs and patients with SPMS.
Results:
Hierarchical clustering segregated patients with SPMS into 2 subgroups: SP-1, which had a short duration of relapsing-remitting multiple sclerosis (MS), and SP-2, which had a long duration of relapsing-remitting MS. SP-1 patients upregulated numerous immune genes, including genes within TCR and toll-like receptor (TLR) signaling pathways. SP-2 patients showed immune gene downregulation in comparison with HCs. We identified an SP-1–specific transcriptional signature of 3 genes (TLR4, TLR2, and chemokine receptor 1), and these genes had higher surface protein expression in SP-1 than in SP-2. After TCR stimulation for 48 hours, only SP-1 showed a progressive linear increase in TLR2 and TLR4 protein expression.
Conclusions:
Differences in naive CD4 T-cell biology, notably of TCR and TLR signaling pathways, identified patients with MS with more rapid conversion to secondary progression, a critical determinant of long-term disability in MS.
Immune cells, including T cells, contribute to CNS injury in relapsing-remitting multiple sclerosis (RRMS), but their role in progressive multiple sclerosis (MS) is uncertain. Many patients with RRMS eventually develop a progressive accumulation of neurologic disability irrespective of relapses: secondary progressive MS (SPMS). Our exploratory study focused on T-cell activation in SPMS. We compared the gene expression profiles of naive CD4 T cells from 14 healthy controls (HCs) and 19 patients with SPMS. We studied naive CD4 T cells for the following reasons: 1) this T subset includes the autoreactive T cells that initiate MS1; 2) some patients with MS have altered naive CD4 T-cell homeostasis with increased homeostatic proliferation2—some propose a link between altered T-cell homeostasis, homeostatic proliferation, and autoimmune disease3,4; and 3) several investigators defined a naive CD4 T-cell transcriptional signature in patients with clinically isolated syndromes (CIS). Decreased expression of TOB1 (transducer of ERBB2, 1), a repressor of naive T-cell quiescence, identified patients with CIS having rapid progression to RRMS.5 We report that opposing naive CD4 T-cell gene expression patterns segregated SPMS patients into 2 subgroups: SP-1, which had a short RRMS duration, and SP-2, which had a long RRMS duration. SP-1 patients upregulated numerous immune genes, including genes within T-cell receptor (TCR) and toll-like receptor (TLR) signaling pathways, compared with controls and SP-2 patients. Surface marker analyses before and after TCR stimulation confirmed naive T-cell immune activation in SP-1 patients. Thus, differences in naive CD4 T-cell biology identify patients with MS having different rates of development of secondary progression, the critical determinant of long-term prognosis in MS.6
METHODS
Patients and controls.
We recruited 19 patients with SPMS, 5 with primary progressive MS (PPMS), and 13 with RRMS from the Montreal Neurological Hospital MS Clinics, diagnosed by standard criteria,7 and 14 HCs (table 1). Inclusion criteria were females, age younger than 65 years, and Expanded Disability Status Scale (EDSS) score <7 for patients with progressive MS. Exclusion criteria included autoimmune diseases other than MS, relapses or treatment with immunomodulatory agents, cytotoxic agents, or corticosteroids within 3 months, or any infection in the previous 4 weeks. Initially, we analyzed only female HCs and patients with SPMS by microarrays, because of male-female differences in gene expression.8 Subsequently, we included patients with RRMS and PPMS in microarray studies to determine the stability of microarray-defined SPMS subgroups. For protein expression studies, we recruited additional patients with SPMS, based on RRMS durations.
Table 1.
Cohorts for microarray studies

Standard protocol approvals, registrations, and patient consents.
The Institutional Review Board of McGill University, Montreal, provided ethical approval. All participants provided written informed consent.
Naive CD4 T-cell isolation, RNA preparation, and hybridization.
We isolated peripheral blood CD4+ CD45RA+ (naive CD4) T cells by negative selection using MACS naive CD4 T-cell isolation kits, human II (Miltenyi Biotec, Bergisch Gladbach, Germany). We isolated total RNA using RNeasy Plus Mini Kits (Qiagen, Mississauga, Canada), amplified the RNA using Low RNA Input Linear Amplification Kit PLUS (Agilent Technologies, Mississauga, Canada), dye-labeled (Cy5) and hybridized on 4 × 44 K Oligochip Slides (Agilent Technologies) containing 41,000 unique probes. We used the Agilent 2100 Bioanalyzer to verify RNA quality.
Array data quality and analysis.
We analyzed microarray data using GeneSpring GX (version 11, Agilent Technologies) and R programs.9–11 We used Feature Extraction QC reports (FE 9.3.1, Agilent Technologies) to control RNA quality and applied LOWESS (locally weighted scatterplot smoothing) normalization. Using analysis of variance (ANOVA), we identified the most variable genes across SPMS and HC samples followed by unsupervised hierarchical clustering12 to identify sample subgroups. To identify differentially expressed genes between subgroups, we used GeneSpring GX, including multiple testing correction, Benjamini-Hochberg false discovery rate (FDR)-corrected p value ≤0.05, together with an absolute difference ≥1.7-fold and 1.3-fold in mean intensity for each gene. We did not use more stringent FDR or fold changes because these may overlook subtle but important pathway changes in complex diseases such as MS.13 We performed in-depth pathway analysis with GeneSpring GX, Gene Ontology, Ingenuity Pathway Analysis, and Gene Set Enrichment Analysis (GSEA).13,14 For GSEA, we used an FDR <0.25, as recommended.14
Real-time PCR.
We used real-time PCR to confirm the expression of selected genes (see the results section) using the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) and the comparative CT method.15
Surface marker expression and functional analyses.
We analyzed naive CD4 T-cell surface markers before and after in vitro culture of isolated naive CD4 T cells in flat-bottom plates (250 × 105 T cells/well), with and without CD3/CD28 Dynabeads human T-activator CD3/CD28 (Invitrogen, Grand Island, NY), under optimal stimulation conditions (25 µL beads/106 naive CD4 T cells) for 24 and 48 hours in complete RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin/streptomycin in 5% CO2 at 37°C. We used a FACSAria (Becton Dickinson, San Diego, CA) to analyze T-cell aliquots stained with the following antibodies: CD45RA PE-Cy7, TLR2 PE, TLR4 APC, CD28 PerCP-Cy 5.5 (eBioscience, San Diego, CA), and CCR1 Alexa Fluor 488 (R&D Systems, Minneapolis, MN). We defined CD4+ CD45RA+ CD45RO− CD62L+ T cells as naive. Purity was >90% in all samples, with the majority >95%.
Statistics.
Using GraphPad Prism 5 and the D'Agostino-Pearson omnibus test, we determined whether data had a normal distribution. We used the Student t test to analyze ages and MS durations, and the Mann-Whitney test to analyze EDSS scores, because the EDSS does not have a normal distribution. We analyzed surface protein expression data using FlowJo 7.6.4 and compared percent expression between HCs and 2 SPMS subgroups using 1-way ANOVA. We compared median fluorescence intensity (MFI) between subgroups using Kruskal-Wallis and Dunn multiple comparison tests. To compare temporal changes within subgroups, we used Friedman and Dunn posttests for mean fluorescence intensity (MFI). We used repeated-measures ANOVA plus Tukey multiple comparison test and an ANOVA posttest for linear trends to compare percent expression. A p value ≤0.05 was considered statistically significant. We used Pvclust16 to confirm reproducibility of microarray-defined clusters.
RESULTS
Hierarchical clustering discriminated HCs from SPMS and identified 2 SPMS subgroups: Association of subgroups with RRMS duration.
Unsupervised hierarchical clustering (Ward clustering method and Euclidean distance metric) based on 1,439 genes having the greatest variance across all HCs and patients with SPMS segregated HCs and patients with SPMS from one another (figure 1). Notably, we identified 2 SPMS subgroups, which we termed SP-1 and SP-2. Pvclust, a multiscale bootstrap resampling method, gave an “approximately unbiased” p value of 0.95, indicating that all subgroups are significantly reproducible16 and not due to random partitioning of samples.
Figure 1. Hierarchical clustering segregated HCs from patients with SPMS and identified 2 SPMS subgroups.
Unsupervised hierarchical clustering (Ward clustering method and Euclidean distance metric) of 1,439 genes having the greatest variance across all samples segregated HCs from patients with SPMS and identified 2 SPMS subgroups (SP-1 and SP-2). Patients with SPMS clustering with HCs are indicated in gray. HC = healthy control; SPMS = secondary progressive multiple sclerosis.
In a limited study, we investigated whether the segregation of patients with SPMS into subgroups was temporally stable. We repeated microarray analyses for one SP-1 and one SP-2 patient on peripheral blood samples taken 11 and 6 months after the initial blood draws. Hierarchical clustering using the most variable genes across all samples segregated the 2 patients as found previously (data not shown). That is, the gene expression differences between patients with SPMS seem stable over time.
Patients in the SP-1 and SP-2 subgroups did not differ in mean ages, EDSS scores, age at onset, or mean SPMS durations. Interestingly, SP-1 patients had a shorter mean RRMS duration than SP-2 patients (12.17 vs 24.27 years; p < 0.001). These differences in RRMS duration explain the shorter total mean MS duration in SP-1 vs SP-2 (21.8 vs 31.4 years, p = 0.008) (table 1).
Transcriptional program associated with T-cell activation in SP-1.
We found significant differences in gene expression between HCs and total SPMS (data not shown), between HCs and the 2 SPMS subgroups, and between SP-1 and SP-2. SP-1 upregulated most differentially expressed genes between SP-1 and HCs (1,901 upregulated, 100 downregulated, FDR-corrected p ≤ 0.05, ≥1.7-fold change), and most genes were immune-related. SP-2 downregulated most differentially expressed genes between SP-2 and HCs, and few genes were immune-related (figure e-1 on the Neurology® Web site at www.neurology.org). SP-1 upregulated most differentially expressed genes between SP-1 and SP-2 (2,537 upregulated, 398 downregulated, p ≤ 0.05, ≥1.7-fold change).
Comparisons of SP-1 vs HCs and SP-1 vs SP-2 showed that approximately 40% and 70% of the differentially expressed genes in SP-1 overlapped with a T-cell activation signature obtained by TCR stimulation of CD4 T cells.17 That is, many genes upregulated during T-cell activation17,18 had increased expression in SP-1 vs both HCs and SP-2: upregulated genes included proapoptotic genes CASP1 (1.8× up) and FAS (1.8× up), chemokine receptors CCR1 (7.3× up) and CCR2 (4.8× up), and IL1R2 (4.8× up) (table e-1A). TOB1, a gene with reduced expression in patients with CIS having rapid conversion to RRMS,5 was downregulated in SP-1 vs SP-2 (FDR = 6.6E-4, 1.4× down).
We compared patients with stable RRMS with HCs and our now-defined SP-1 and SP-2 subgroups to see whether the patients with RRMS differed from our SPMS subgroups. SP-1 and SP-2 patients continued to represent separate clusters (data not shown). We questioned whether SP-2 patients upregulated genes involved in neurodegeneration. GSEA showed no altered expression of genes in neurodegeneration pathways, in keeping with recent findings from genome-wide association studies.19
Altered expression of genes involved in TCR and TLR signaling in SP-1.
Several tyrosine kinase genes involved in TCR-induced signaling (including LYN, 7.2× up; ZAP70, 1.3× down; and SYK, 4.3× up) and genes involved in calcium signaling (table e-1B) had altered expression in SP-1 vs HCs. The vitamin D receptor (VDR), normally expressed on naive CD4 T cells only after TCR signaling,20 showed increased gene expression in SP-1 vs HCs (FDR = 1.7E-7, 3.8× up).
Cells of the innate immune system, as well as activated and memory T cells, express TLRs.21,22 Normally, naive CD4 T cells express very low levels of TLR messenger RNA but TCR signaling upregulates TLR expression.23 TCR and TLR share T-cell signaling pathways.21 TLR-2 is a costimulatory receptor for TCR activation22 and TLR-4 induction regulates TCR signaling events through the mitogen-activated protein kinase (MAPK) cascade.21 TLR1, TLR2, TLR4, and TLR7 (FDR <1E-5; 2×, 4.2×, 8.1×, and 6× up, respectively) were among the genes showing the highest expression in SP-1 vs HCs (table e-1B). SP-1 patients upregulated many downstream targets of TLR signaling including MYD88, IRF5, IRF7, TANK (TRAF family member-associated NF-κB activator), TBK1, and MAPK. Thus, our pathway analyses suggest that enhanced TLR-TCR signaling results in increased activation of naive CD4 T cells in SP-1.
T-cell activation transcriptional signatures define SP-1 patients.
Using Gene Ontology, GSEA, and Ingenuity Pathway Analysis, we generated a gene list of 130 T-cell activation-associated genes and performed unsupervised hierarchical clustering of all HCs and patients with SPMS and RRMS. We included 5 patients with PPMS in this comparison to determine whether this gene list is SP-1–specific. SP-1 patients segregated from all other individuals (data not shown). Comparison of this 130-gene list with the gene set in reference 5 identified TOB1 as the only gene overlapping with our dysregulated pathways. From the 130-gene list, we selected 3 genes (TLR2, TLR4, and CCR1) because their expression levels (figure 2A) suggested that they could be used for further microarray studies and for protein expression analyses; unlike most genes in the list, these genes had corresponding T-cell surface membrane products recognized by commercially available monoclonal antibodies. Real-time PCR confirmed the relative upregulation of the 3 genes in SP-1 (data not shown). Unsupervised hierarchical clustering using this 3-gene list segregated SP-1 patients from all other individuals (figure 2B), indicating that this 3-gene list is SP-1–specific and suggests that HCs, SP-2 patients, patients with stable RRMS, and patients with PPMS have similar low levels of naive T-cell activation.
Figure 2. Transcriptional signature of 3 genes distinguishes SP-1 patients from other patients with MS and HCs.
(A) SP-1 patients have higher expression of TLR2, TLR4, and CCR1 genes than either HCs or SP-2 patients. Mean fold change values are shown for SP-1 vs SP-2. (B) Hierarchical clustering based on the 3-gene list distinguishes SP-1 patients. Hierarchical clustering segregated SP-1 from HCs, SP-2, patients with stable RRMS, and patients with PPMS. HC = healthy control; MS = multiple sclerosis; PPMS = primary progressive MS; RRMS = relapsing-remitting MS.
Naive CD4 T-cell surface markers before and after CD3/CD28 stimulation.
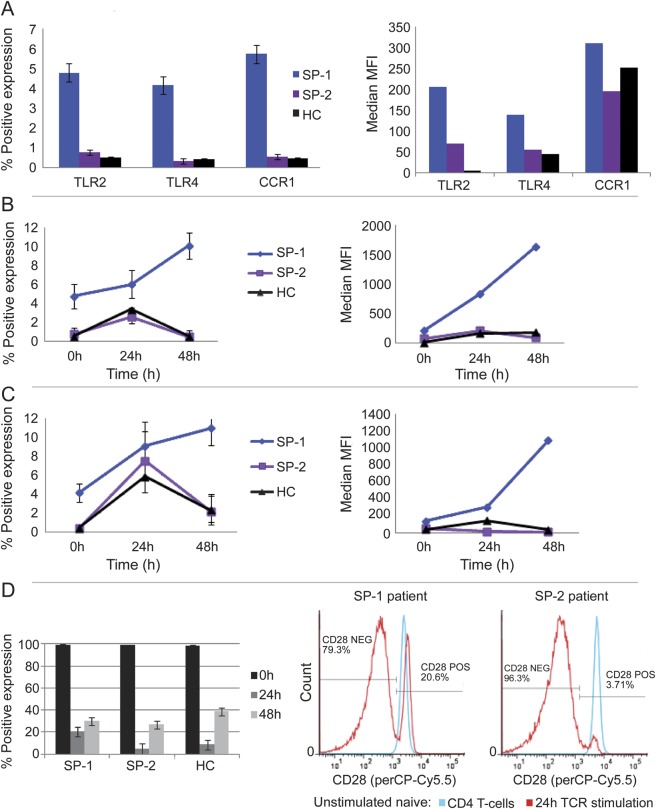
Initially, to confirm gene expression results at the protein expression level, we analyzed naive CD4 T-cell surface expression of the 3 SP-1 signature genes (TLR2, TLR4, and CCR1), before and after anti-CD3/CD28 stimulation. We also studied surface expression of the CD28 costimulatory molecule. We included 3 SP-1 and 2 SP-2 patients from our initial microarray studies (bled approximately 2 years later). We recruited 2 new SP-1 patients, 3 new SP-2 patients, and 5 HCs. The 5 SP-1 and 5 SP-2 patients had respective RRMS durations of <11 years and >20 years (table 2). In SP-1, freshly isolated naive CD4 T cells had significantly higher percentage of TLR2, TLR4, and CCR1 expression than SP-2 and HCs (figure 3A). Because monocytes have high TLR expression, we costained naive CD4 T cells for CD14, a monocyte/macrophage marker,24 and for CD4, CD45RA, TLR2, and TLR4 before and after 24 hours of stimulation with Dynabeads. The percentage of CD14 expression varied from 0% to 0.125%, with no CD14 expression on TLR2- or TLR4-positive cells, i.e., TLR positivity does not reflect monocyte contamination. Thus, these surface marker studies confirm our microarray findings at the protein expression level.
Table 2.
Cohorts for surface protein expression analysis

Figure 3. Surface expression of 3-gene signature and CD28.
(A) Increased protein expression on freshly isolated naive CD4 T cells from SP-1. SP-1 patients had significantly higher percentage of TLR2, TLR4, and CCR1 than HCs and SP-2 patients (95% confidence interval not shown) and higher TLR4 MFI than HCs and SP-2 patients (p < 0.05, both comparisons). (B) TCR stimulation increased TLR2 surface expression in SP-1. At 48 hours, SP-1 had increased percentage of TLR2 and MFI (p < 0.05, both comparisons) compared with unstimulated cells, with a positive linear increase in percentage of TLR2 from 0 to 48 hours (p = 0.02). At 48 hours, SP-1 had higher percentage of TLR2 expression than HCs and SP-2 (p < 0.001, both comparisons) and higher TLR2 MFI than HCs and SP-2 (p < 0.05, both comparisons). (C) TCR stimulation increased TLR4 surface expression in SP-1 patients. The percentage of TLR4 increased in HCs (p < 0.05) and SP-2 (p < 0.01) from 0 to 24 hours, then decreased from 24 to 48 hours. In SP-1, percentage TLR4 showed a positive linear increase from 0 to 48 hours (p = 0.0025). At 48 hours, SP-1 had higher percentage of TLR4 than HCs or SP-2 (p < 0.01, both comparisons) and higher TLR4 MFI than HCs or SP-2 (p < 0.05, both comparisons). (D) CD28 surface expression. TCR stimulation for 24 hours decreased percentage CD28 (p < 0.001) and CD28 MFI (p < 0.05, not shown) in all groups, but at 24 hours, SP-1 had higher percentage CD28 than HCs (p < 0.05) or SP-2 (p < 0.01) plus higher CD28 MFI than HCs (p < 0.05, not shown). HC = healthy control; MFI = mean or median fluorescence intensity; TCR = T-cell receptor.
After TCR stimulation, TLR2 and TLR4 expression (MFI and %) peaked at 24 hours in HCs and the SP-2 subgroup and then decreased to prestimulation levels by 48 hours, whereas TLR2 and TLR4 expression increased linearly in SP-1 from 0 to 24 to 48 hours; at 48 hours, these TLRs had higher expression in SP-1 than in HCs or SP-2 (Figure 3, B and C). The increasing TLR expression in SP-1 after TCR stimulation supports our microarray data and suggests that these cells are preprogrammed toward T-cell activation. In all 3 groups, CD28 expression (percentage and MFI) decreased at 24 hours and then increased at 48 hours, but at 24 hours, percentage of CD28 positivity was significantly higher in SP-1 (figure 3D). This relative retention of CD28 in SP-1 at 24 hours may have facilitated a sustained response to TCR signaling leading to an increase in TLR expression from 24 to 48 hours.
Validation of surface protein expression.
To validate our protein expression findings, we analyzed freshly isolated naive CD4 T cells from a second set of 5 HCs (4 newly recruited females and 1 male), 5 SP-1 patients (one from the microarray study, 2 newly enrolled females, and 2 males), and 5 SP-2 patients (3 from the microarray study and 2 newly enrolled females), based on their RRMS durations as defined above. Ages did not differ between subgroups. SP-1 patients had a significantly shorter RRMS duration but significantly higher percentage expression of TLR2, TLR4, and CCR1 than HCs or SP-2 (figure e-2). Thus, we confirm increased surface protein expression of the 3-gene signature in SP-1.
DISCUSSION
We report that differences in naive CD4 T-cell biology, identified by unsupervised hierarchical clustering, segregated patients with SPMS into 2 significantly reproducible subgroups. We did not preselect patients on the basis of their RRMS durations for these gene expression studies, but these durations differed significantly between subgroups (SP-1 vs SP-2; 12.17 vs 24.27 years). One question is whether patients with RRMS would also separate into 2 distinct subgroups, but to address that question, a larger prospective study focusing on RRMS is needed. Our data suggest that naive CD4 T-cell upregulation/T-cell activation identifies patients with MS having a short vs protracted RRMS course. The downregulation of TOB1 in SP-1 vs SP-2 supports this interpretation, as this repressor of naive CD4 T-cell activation is downregulated in patients with CIS having rapid progression to RRMS.5 Possibly, TOB1 downregulation persists from CIS to RRMS to SPMS and identifies patients with rapid MS progression. Some researchers suggest that a short RRMS phase predicts rapid disease progression in SPMS.25 Thus, SP-1 patients could potentially have a more active/aggressive SPMS disease course.
The upregulation of genes involved in TCR and TLR signaling in SP-1 is noteworthy as various TLRs function as costimulatory receptors that can enhance proliferation and cytokine production by TCR-activated T cells.21,22 Gandhi et al.26 reported T-cell dysregulation in all MS subtypes. Our findings modify this conclusion by showing, at least for naive CD4 T cells, that only a subgroup of patients with SPMS has immune gene dysregulation. Moreover, the findings suggest either global naive CD4 T-cell activation in SP-1 or activation of sufficient naive CD4 T cells to permit detection of differences between patient subgroups. Of interest is the basis of the immune gene dysregulation in SP-1. Some claim a link between homeostatic T-cell proliferation and autoimmunity.3,4 We previously reported reduced thymic output with increased homeostatic naive CD4 T-cell proliferation in RRMS.2,27,28 Possibly, SP-1 patients had increased homeostatic proliferation but we have no data on this issue.
A potential limitation of our study is that we did not repeat the microarray study to validate our gene expression findings. Instead, we opted to study the protein expression of the 3-gene signature on 2 separate cohorts of HCs and patients with SPMS. The significantly increased TLR2, TLR4, and CCR1 expression on freshly isolated naive CD4 T cells in SP-1 is consistent with the findings of an activated transcriptional program in this patient subgroup. The differences in TLR expression between subgroups seem to be small, but are entirely consistent with another report that demonstrated biologically significant differences in T-cell TLR expression between patients and controls.29 The progressive linear increase in TLR expression under optimal stimulation conditions provides further evidence of T-cell activation in SP-1. CD28 stimulation transiently downregulates CD28 messenger RNA and surface expression levels in CD4 T cells, with resulting temporary unresponsiveness to CD28 signaling.30 The decrease in TLR protein levels from 24 to 48 hours in HCs and SP-2 may reflect a normal feedback response to TCR signaling due to loss of CD28. Conversely, the relative CD28 retention in SP-1 at the 24-hour time point may have permitted a continued naive CD4 T-cell response to TCR signaling, leading to a sustained increase in TLR protein expression. Together, the microarray and surface phenotypic marker findings suggest in vivo naive CD4 T-cell activation in SP-1 patients and that this activation is responsible for the increased response to in vitro TCR signaling seen in these patients. Microarray-based classification has predictive value in CIS5,31 and MS.32 For example, T-cell transcriptional profiling can predict the response to interferon β (IFNβ) treatment in RRMS.32 Our microarray findings have potential clinical implications for SPMS. The global naive CD4 T-cell downregulation in SP-2, i.e., in the larger SPMS subgroup, suggests that patients with this phenotype will be unresponsive to anti-inflammatory treatments. A North American SPMS trial showed no reduction in disability progression with IFNβ treatment.33 In a European trial, which included younger patients with shorter disease duration, higher relapse rate, and more aggressive SPMS, some patients showed reduced disability progression.34,35 The patients who benefited from IFNβ treatment may correspond, in part, to our SP-1 subgroup with T-cell activation and rapid progression. Thus, our 3-gene T-cell activation signature may identify patients with SPMS who would benefit from anti-inflammatory therapies, but further validation of this signature is indicated.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Julie Livingstone (McGill University, Montreal, Canada) for her input on the microarray analysis and Dr. Maria Cortes (Montreal Neurological Hospital, Montreal, Canada) for her assistance with the clinical data analysis. The authors also thank the patients with MS and healthy controls who participated in this study.
GLOSSARY
- ANOVA
analysis of variance
- CIS
clinically isolated syndrome
- EDSS
Expanded Disability Status Scale
- FDR
false discovery rate
- GSEA
Gene Set Enrichment Analysis
- HC
healthy control
- IFNβ
interferon β
- MAPK
mitogen-activated protein kinase
- MFI
mean or median fluorescence intensity
- MS
multiple sclerosis
- PPMS
primary progressive multiple sclerosis
- RRMS
relapsing-remitting multiple sclerosis
- SPMS
secondary progressive multiple sclerosis
- TCR
T-cell receptor
- TLR
toll-like receptor
- TOB1
transducer of ERBB2, 1
Footnotes
Supplemental data at www.neurology.org
AUTHOR CONTRIBUTIONS
E. Zastepa, PhD graduate student: designed, optimized, and performed the cell isolation, microarray experiments, real-time PCR experiments, array quality control, and data analysis; performed the protein expression and T-cell activation experiments and data analysis; critically interpreted the data and wrote the majority of the manuscript. L. Fitz-Gerald, MSc student: participated in patient recruitment; critically evaluated, discussed, and corrected the manuscript; helped with protein expression experiments. M. Hallett, bioinformatist: actively participated in discussion of class discovery and class distinction methods, including appropriate usage of pvclust to evaluate unbiased hierarchical clustering, involved extensively in discussion and evaluation of data, and critically evaluated and discussed the manuscript. J. Antel, A. Bar-Or, and Y. Lapierre, neurologists with expertise in MS: reviewed and selected patients with MS who satisfied the currently accepted diagnostic criteria and the requirements for recruitment into the study, provided relevant clinical data, critically evaluated and discussed the data and data interpretation, and discussed the manuscript and review. S. Baranzini, human genomics expert: provided microarray data from previous study identifying TOB1 as a predictive marker of CIS disease progression, facilitated comparison of our data set with his with insights into the relevance of TOB1 in our progressive MS study, and provided a helpful and critical review of the manuscript. D.G. Haegert, principal investigator and supervisor of this research: intimately involved in experimental design, writing of the manuscript, discussion and interpretation of the data, communication and discussion with neurologists, and extensive involvement in manuscript revisions.
STUDY FUNDING
Supported by the Multiple Sclerosis Society of Canada. E.Z. was funded by a PhD fellowship from NSERC. The authors acknowledge infrastructure support and technical assistance from the Breast Cancer Functional Genomics Group, which is supported by funds from the Terry Fox Foundation and CIHR.
DISCLOSURE
E. Zastepa, L. Fitz-Gerald, and M. Hallett report no disclosures. J. Antel (* = compensation received) reports the following disclosures. Commercial: advisory boards or data safety monitoring committees: Biogen Idec*, TEVA*, emdSerono*, Genzyme*, Sanofi-Aventis*, Novartis*; nonprofit: advisory boards or data safety monitoring committees: Cleveland Clinic Foundation*, MS Society (Canada), MSIF, Glaucoma Research Foundation (USA), Hertie Foundation (Germany)*, Burroughs Wellcome Medical Scientist Programme. Editorial: editor for the Americas Multiple Sclerosis Journal (SAGE Publishing)*. Research support: MS Society of Canada, CIHR (Industry Partnership Program), Novartis. A. Bar-Or has participated as a speaker at meetings sponsored by, received consulting fees and/or received grant support from: Amplimmune, Bayhill Therapeutics, Berlex/Bayer, Biogen Idec, BioMS, DioGenix, Eli Lilly, Genentech, GlaxoSmithKline, Guthy-Jackson/GGF, Merck/EMD Serono, Medimmune, Mitsubishi Pharma, Novartis, Ono Pharma, Receptos, Roche, Sanofi-Genzyme, Teva Neuroscience, Wyeth. S. Baranzini reports no disclosures. Y. Lapierre has received consulting fees and serves on advisory boards from Biogen Idec, Bayer, EMD Serono, Genzyme, Novartis, Teva. D. Haegert reports no disclosures. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Muraro PA, Pette M, Bielekova B, McFarland HF, Martin R. Human autoreactive CD4+ T cells from naive CD45RA+ and memory CD45RO+ subsets differ with respect to epitope specificity and functional antigen avidity. J Immunol 2000;164:5474–5481 [DOI] [PubMed] [Google Scholar]
- 2.Haegert DG, Hackenbroch JD, Duszczyszyn D, et al. Reduced thymic output and peripheral naive CD4 T-cell alterations in primary progressive multiple sclerosis (PPMS). J Neuroimmunol 2011;233:233–239 [DOI] [PubMed] [Google Scholar]
- 3.Khoruts A, Fraser JM. A causal link between lymphopenia and autoimmunity. Immunol Lett 2005;98:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King C, Ilic A, Koelsch K, Sarvetnick N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell 2004;117:265–277 [DOI] [PubMed] [Google Scholar]
- 5.Corvol JC, Pelletier D, Henry RG, et al. Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc Natl Acad Sci USA 2008;105:11839–11844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Confavreux C, Vukusic S, Adeleine P. Early clinical predictors and progression of irreversible disability in multiple sclerosis: an amnesic process. Brain 2003;126:770–782 [DOI] [PubMed] [Google Scholar]
- 7.McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol 2001;50:121–127 [DOI] [PubMed] [Google Scholar]
- 8.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 2003;100:1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–315 [DOI] [PubMed] [Google Scholar]
- 10.R Development Core Team R: a language and environment for statistical computing [online]. Available at: http://www.r-project.org/. Accessed February 1, 2011
- 11.Yang Y, Paquet A, Dudoit S. Marray: exploratory analysis for two-colour spotted microarray data [online]. Available at: http://www.bioconductor.org/packages/2.12/bioc/html/marray.html. Accessed February 10, 2011
- 12.Jain A. Data clustering: 50 years beyond K-means. Pattern Recognit Lett 2010;31:651–666 [Google Scholar]
- 13.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–273 [DOI] [PubMed] [Google Scholar]
- 14.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 2007;23:3251–3253 [DOI] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 16.Suzuki R, Shimodaira H. Pvclust: an R package for hierarchical clustering with p-values [online]. Available at: http://www.is.titech.ac.jp/∼shimo/prog/pvclust. Accessed April 15, 2010 [DOI] [PubMed]
- 17.Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics 2008;9:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chtanova T, Newton R, Liu SM, et al. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol 2005;175:7837–7847 [DOI] [PubMed] [Google Scholar]
- 19.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Essen MR, Kongsbak M, Schjerling P, Olgaard K, Odum N, Geisler C. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat Immunol 2010;11:344–349 [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Navajas JM, Fine S, Law J, et al. TLR4 signaling in effector CD4+ T cells regulates TCR activation and experimental colitis in mice. J Clin Invest 2010;120:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc Natl Acad Sci USA 2004;101:3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLeod H, Wetzler LM. T cell activation by TLRs: a role for TLRs in the adaptive immune response. Sci STKE 2007;2007:pe48. [DOI] [PubMed] [Google Scholar]
- 24.Wahlstrom J, Berlin M, Skold CM, Wigzell H, Eklund A, Grunewald J. Phenotypic analysis of lymphocytes and monocytes/macrophages in peripheral blood and bronchoalveolar lavage fluid from patients with pulmonary sarcoidosis. Thorax 1999;54:339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremlett H, Yinshan Z, Devonshire V. Natural history of secondary-progressive multiple sclerosis. Mult Scler 2008;14:314–324 [DOI] [PubMed] [Google Scholar]
- 26.Gandhi KS, McKay FC, Cox M, et al. The multiple sclerosis whole blood mRNA transcriptome and genetic associations indicate dysregulation of specific T cell pathways in pathogenesis. Hum Mol Genet 2010;19:2134–2143 [DOI] [PubMed] [Google Scholar]
- 27.Duszczyszyn DA, Beck JD, Antel J, et al. Altered naive CD4 and CD8 T cell homeostasis in patients with relapsing-remitting multiple sclerosis: thymic versus peripheral (non-thymic) mechanisms. Clin Exp Immunol 2006;143:305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duszczyszyn DA, Williams JL, Mason H, Lapierre Y, Antel J, Haegert DG. Thymic involution and proliferative T-cell responses in multiple sclerosis. J Neuroimmunol 2010;221:73–80 [DOI] [PubMed] [Google Scholar]
- 29.Babu S, Blauvelt CP, Kumaraswami V, Nutman TB. Cutting edge: diminished T cell TLR expression and function modulates the immune response in human filarial infection. J Immunol 2006;176:3885–3889 [DOI] [PubMed] [Google Scholar]
- 30.Linsley PS, Bradshaw J, Urnes M, Grosmaire L, Ledbetter JA. CD28 engagement by B7/BB-1 induces transient down-regulation of CD28 synthesis and prolonged unresponsiveness to CD28 signaling. J Immunol 1993;150:3161–3169 [PubMed] [Google Scholar]
- 31.Achiron A, Grotto I, Balicer R, Magalashvili D, Feldman A, Gurevich M. Microarray analysis identifies altered regulation of nuclear receptor family members in the pre-disease state of multiple sclerosis. Neurobiol Dis 2010;38:201–209 [DOI] [PubMed] [Google Scholar]
- 32.Satoh J, Nakanishi M, Koike F, et al. T cell gene expression profiling identifies distinct subgroups of Japanese multiple sclerosis patients. J Neuroimmunol 2006;174:108–118 [DOI] [PubMed] [Google Scholar]
- 33.Panitch H, Miller A, Paty D, Weinshenker B. Interferon beta-1b in secondary progressive MS: results from a 3-year controlled study. Neurology 2004;63:1788–1795 [DOI] [PubMed] [Google Scholar]
- 34.European Study Group on Interferon Beta-1b in Secondary Progressive MS Placebo-controlled multicentre randomised trial of interferon beta-1b in treatment of secondary progressive multiple sclerosis. Lancet 1998;352:1491–1497 [PubMed] [Google Scholar]
- 35.Kappos L, Weinshenker B, Pozzilli C, et al. Interferon beta-1b in secondary progressive MS: a combined analysis of the two trials. Neurology 2004;63:1779–1787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.