Abstract
Wild birds are the primary source of genetic diversity for influenza A viruses that eventually emerge in poultry and humans. Much progress has been made in the descriptive ecology of avian influenza viruses (AIVs), but contributions are less evident from quantitative studies (e.g., those including disease dynamic models). Transmission between host species, individuals and flocks has not been measured with sufficient accuracy to allow robust quantitative evaluation of alternate control protocols. We focused on the United States of America (USA) as a case study for determining the state of our quantitative knowledge of potential AIV emergence processes from wild hosts to poultry. We identified priorities for quantitative research that would build on existing tools for responding to AIV in poultry and concluded that the following knowledge gaps can be addressed with current empirical data: (1) quantification of the spatio-temporal relationships between AIV prevalence in wild hosts and poultry populations, (2) understanding how the structure of different poultry sectors impacts within-flock transmission, (3) determining mechanisms and rates of between-farm spread, and (4) validating current policy-decision tools with data. The modeling studies we recommend will improve our mechanistic understanding of potential AIV transmission patterns in USA poultry, leading to improved measures of accuracy and reduced uncertainty when evaluating alternative control strategies.
Keywords: Avian influenza, USA, Between-farm spread, Disease-dynamic model, Quantitative data, Poultry
1. Introduction
Emergence of avian influenza viruses (AIVs) in poultry remains a global problem that can cost hundreds of millions of US dollars (Halvorson, 2009, Lupiani and Reddy, 2009). In the USA, even low-pathogenic avian influenza viruses (LPAIVs) can cost millions of dollars to control once detected in commercial poultry (Davison et al., 1999, Halvorson, 2009). Major goals of the USA national plan (Foreign Animal Diseases Preparedness and Response Plan) for minimizing losses due to AIVs are: (1) to prevent the introduction of AIVs into poultry, (2) to identify infected flocks as quickly as possible, and (3) to eliminate the virus as quickly as possible once it is detected (USDA, 2012). These goals are achieved through biosecurity (management procedures that minimize introduction or dissemination of infectious diseases), diagnostics and surveillance (detection of AIVs), depopulation and controlled slaughter, education of flock owners/workers and public outreach, all of which occur in a planned, coordinated manner (USDA, 2012).
In developing and implementing specific prevention and response activities, multiple biological, political and economic factors are considered, such as virus pathotype (either highly pathogenic avian influenza virus: HPAIV, or LPAIV), the poultry commodity or commodities affected, the type of operation (i.e., commercial, backyard or live-bird market), the density of poultry in a geographic area, the demands of export markets, federal versus state regulatory authority, availability of financial compensation, public perception and potential for zoonotic transmission of the virus. Thus, the numerous response activities that occur depend on scenario-specific circumstances. The success of any strategy is dependent on trust, co-operation and interaction between industry and government (Swayne and Akey, 2005). Consequently, it can be challenging to assimilate all of the necessary information during an emergency. Sound quantitative tools are essential for preparedness and response planning.
Preparedness and response modeling are two complementary quantitative approaches for informing policy-based decisions made during an AIV event. During preparedness modeling, there is more time for model formulation, evaluation and “situational analysis”, but appropriate data from previous outbreaks may be unavailable or irrelevant. In response modeling, appropriate quantitative data are likely being collected and analyzed as the outbreak unfolds, but time for detailed evaluation of quantitative methods is very limited. Because preparedness and response modeling involve similar methods and data, preparedness modeling can and does facilitate response modeling. The development, detailed investigation, and validation of several sound quantitative approaches prior to an event are important for performing response analyses with high confidence in a short period of time.
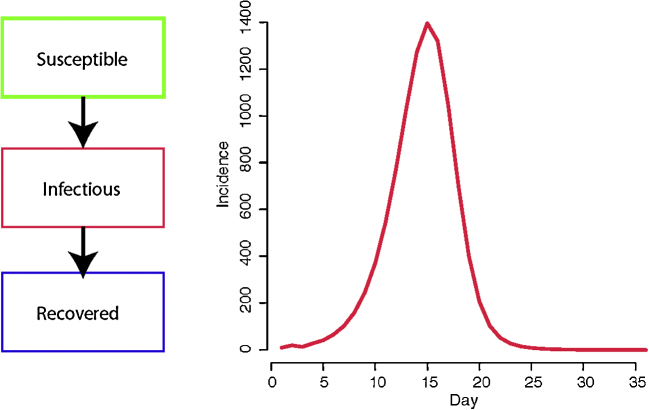
Disease-dynamic models are useful for informing control policies (Anderson and May, 1992) because they incorporate a quantitative description of how transmission changes during the course of an epidemic (Fig. 1). Adding additional components, such as age-structure or life-history stage, to simple disease-stage models (i.e., models with different disease states such as susceptible, infectious or recovered; Fig. 1) allows determination of how alternative control strategies, implemented at different stages of the transmission process, will impact epidemic dynamics. Disease-dynamic models are characterized by the presence of a force of infection (rate at which a susceptible individual acquires disease) term that defines precisely how the infection hazard experienced by a susceptible individual (or farm) depends on the current number of infectious individuals (or farms), their proximity and their type. A key parameter that can usually be derived using the force if infection term is the basic reproductive number, R0, defined as the expected number of secondary infections generated by one infectious individual (or farm) in an otherwise susceptible population. R0 is used to assess the required proportion of a population that must be rendered non-transmissible for an outbreak to be controlled (Heesterbeek and Roberts, 2007) and is predictive of the impact of interventions in reducing the attack rate, even when full control is not achieved (Wu et al., 2006). Sometimes, models are too complex in their assumptions about the population or the pathogen for the derivation of R0 to be tractable (e.g., individual-based spatial simulations).
Fig. 1.
Diagram illustrating a simple dynamic-disease model. The host population is divided into “compartments” that differ by disease status (left); in this case susceptible, infectious or recovered (and presumed to be immune). Disease-dynamic models are a mathematical description of pathogen transmission. Solving or conducting simulations with such a model gives an estimate of how the risk of infection within a population (i.e., flock) changes over time. The example shown here is for a pathogen with a basic reproductive number of 5 and a generation time of 5 days spreading through a population of 10,000 individuals. Here the force of infection is proportional to the number of currently infectious birds expressed as a ratio of the total population size.
Below, we outline some of the key policy decisions related to minimizing AIV emergence in poultry, for which quantitative disease-dynamic models could be of service. The article is organized in three sections to reflect the stages of emergence: (1) wildlife reservoir dynamics and spillover to poultry, (2) transmission within poultry flocks and (3) transmission between poultry flocks (Fig. 2). For each stage of emergence, we highlight key quantitative data that are currently available and could be incorporated into models focused on reducing uncertainty in policy decisions. The quantitative data we present are not comprehensive, rather we focus on presenting the range of values that have been observed. We also mention important data gaps, especially with regards to spillover processes because this is the least understood stage of emergence. Although transmission and evolution are both important in outbreaks of AIV in poultry, we focus primarily on transmission because key epidemiological processes such as the depletion of susceptible hosts are far more predictable than evolutionary processes, such as mutation and reassortment.
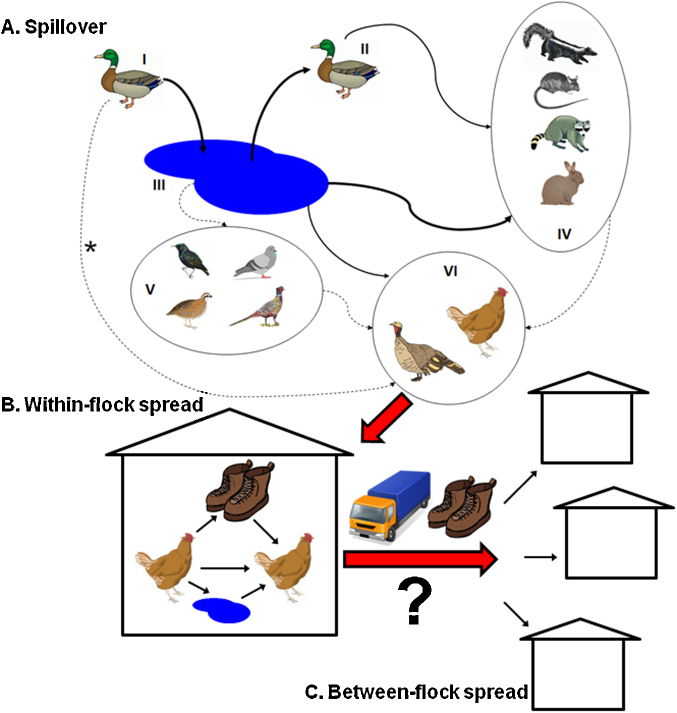
Fig. 2.
Pathways of emergence of AIVs in commercial poultry operations. Red arrows indicate transitions between the different processes in emergence: AIV spillover from wildlife to AIV spread within poultry flocks on a single operation to AIV spread between poultry operations. (A) Spillover mechanisms from wildlife (adapted from Franklin, 2008). Arrows represent movement of AIVs. Bold arrows indicate transmission links that are strongly supported by empirical studies, thin arrows indicate connections that are supported by limited studies and dotted arrows indicate pathways that remain unexplored. Indirect AIV transmission pathways from wild waterfowl (I and II) to poultry (VI) include: drinking contaminated water (III) or contacting non-waterfowl bird species (V) or wild mammals (IV) that were infected through III. IV may also be infected by scavenging infected waterfowl carcasses by wild mammals. *Note that the importance of direct transmission routes from waterfowl to poultry is well-supported in mixed-species backyard flocks but the importance of this connection in transmission to commercial poultry remains to be determined. It is possible that any of these links involve an intermediate link such as human shoes, etc. (B) AIV spread within poultry operations. Once AIV infects poultry in a farm, it can be transmitted directly to other individuals or indirectly through contaminated water, fomites or air. (C) AIV spread between poultry operations. The mechanisms are variable and currently uncertain, particularly for airborne and local spread. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2. Reservoir dynamics and spillover to poultry
2.1. Control policies
Control measures for preventing either maintenance within wildlife populations or spillover to poultry are not currently conducted in wildlife populations (Cardona, 2005, Clark and Hall, 2006). In fact, some wildlife species are not even considered when evaluating biosecurity measures in commercial poultry operations (McQuiston et al., 2005), and operations with low biosecurity (i.e., backyard flocks or gamebird facilities) allow open access for wildlife species (Slota et al., 2011). Typical biosecurity activities, which are common in commercial operations, aim to prevent direct contact between high-risk wildlife species and poultry (e.g., rodent control and wild bird exclusion; USDA, 2012). Many facilities also implement measures to prevent indirect contact with wildlife (e.g., contamination of food and water supplies by wildlife), which may also be important (Fig. 2A). Developing general policies to prevent spillover from wildlife to domestic poultry will require additional quantitative information on wildlife contact rates with poultry and risk assessment to determine which wildlife species have the highest potential to transmit virus to poultry.
2.2. Quantitative host population data
The key quantities required to understand transmission of AIVs include host demographic dynamics, population sizes, contact structure among host individuals, susceptibility and host species composition. The picture for wild hosts is complicated relative to poultry populations because it involves multiple species, each with ecologically determined demographics, spatial interactions and movement patterns. Furthermore, numerous species are known to be susceptible to AIVs but their role in maintenance within the wild-host reservoir system and/or transmission to poultry is unclear (Boyce et al., 2009; Fig. 2A). Population movement coupled with environmental transmission is the main reason for this lack of understanding: multiple species that move freely about their environment become contaminated or get infected by environmental sources (Henaux and Samuel, 2011, Brown et al., 2013a, Brown et al., 2013b). Another reason is that maintenance and spillover are distinct processes that are both important for AIV outbreaks in poultry: spillover force of infection is driven by the product of contact rate between reservoir and target-host populations, probability of infection of the target-host given contact, and pathogen prevalence in the reservoir at the time of contact (Lloyd-Smith et al., 2009).
Field surveys suggest that dabbling ducks, especially mallards (Anas platyrhynchos), are an important reservoir for most AIV subtypes, including subtypes that can lead to HPAI in poultry (Hinshaw et al., 1980, Stallknecht and Shane, 1988b, Stallknecht et al., 1990, Olsen et al., 2006, Stallknecht and Brown, 2008, DeLiberto et al., 2009, Wilcox et al., 2011, Bahl et al., 2013). In addition to being a primary reservoir for AIVs, mallards are also the most abundant duck species in North America (Drilling et al., 2002). Furthermore, most experimental studies of AIVs in wild birds have been in mallards, and thus there are more quantitative data on AIVs in mallards than in any other wildlife host. Dose–response and contact experiments support the idea that mallards are highly susceptible to most wild-bird origin LPAIVs based on: (1) mallards can be infected after exposure to low inoculation doses with multiple subtypes of LPAIVs (Swayne and Slemons, 2008, Brown et al., 2013a, Brown et al., 2013b), (2) high rates of infection occur after exposure to high doses, and (3) transmission to contact birds occurs readily (Webster et al., 1978, Hinshaw et al., 1980, Kida et al., 1980, Alexander et al., 1986, Swayne, 2007, Mundt et al., 2009). Thus, further progress in understanding LPAIV persistence mechanisms and spillover risk could be made by parameterizing dynamic models with population ecology data from mallards. We focus on mallards as the “wild hosts” for the remainder of this article.
Population size over space and time can be extremely variable and difficult to measure for mallard populations, in part because this species is very abundant and widely distributed across North America (USFWS, 2012b). Breeding population size from aerial surveys conducted over parts of Canada and the north-central USA (∼1.3 million sq. mi.) was estimated at 10.6 ± 0.3 million birds in 2012, an estimate about 15% above the 2011 estimate, 40% above long-term averages, but slightly lower than the maximum of ∼11.2 million breeding birds reported in 1958 (USFWS, 2012b). Reproductive success and annual survival are also similarly variable. For example, estimates of nest success range from 1 to 17% nests hatching at least one egg and vary with habitat type and breeding chronology (Emery et al., 2005). Survival probabilities for females after fledging vary by age and breeding status (i.e., breeding survival tends to be higher than nonbreeding survival; Coluccy et al., 2008) and population growth rate is sensitive to variability in nonbreeding survival and other vital rates (Coluccy et al., 2008). Also, mallard populations may increase or decrease depending on environmental conditions (e.g., availability of prairie-pond habitat) or variability in phenology (e.g., timing of nest initiation), which can vary by precipitation or habitat availability (Greenwood et al., 1995). Thus a constant population size over space and time cannot be assumed, as it can for commercial poultry populations.
Challenge studies in mallards have shown that age-structure does not seem to be a major determinant of infection dynamics, except on a very broad scale: mallards < 1 month old show higher viral loads (Costa et al., 2010b) and mallards < 1 year are at least twice as likely to be found infected (Munster et al., 2007, Farnsworth et al., 2012) than those in other age-classes, suggesting that they may be more susceptible or infectious. Similarly, field data demonstrate a difference between hatch year and after-hatch year birds. The difference in the field is likely due to immunity in adults that have been previously exposed to multiple LPAIV strains (Wilcox et al., 2011). With regards to differences between the sexes, results are mixed. For example, two studies found that males can shed up to 2 orders of magnitude more virus by the cloacal route in challenge experiments (Pepin et al., 2012) and are 15% more likely to be found infected in natural populations as compared to females (Farnsworth et al., 2012), but other studies have found no differences between the sexes (Krauss et al., 2004, Munster et al., 2007, Wilcox et al., 2011). Because mallard populations tend to be male-biased, averaging 1.33 males for every female in over-wintering populations (Drilling et al., 2002), determining whether there exists significant transmission biases due to sex may be important.
As with most host populations, host mixing is likely not homogenous but the quantitative, or even qualitative nature of host contact patterns is not yet documented sufficiently to justify a different assumption. Bird banding data are available for quantifying mallard movement patterns (Miller et al., 2013b) and an ongoing network analysis will provide a much needed quantitative understanding of annual mallard movement patterns throughout continental USA (CT Webb, unpublished data). An additional factor is the ability for wild migratory birds to introduce a complete genome (unreassorted) or segments of AIV intercontinentally. The observed frequency of AIV reassortment events between continents varies greatly between studies ranging from 0.25 to 45.0 percent (Miller et al., 2013a). Such translocation via migratory birds contributed in part to the spread of highly pathogenic Asian strain H5N1 out of Southern China and Southeast Asia across Central Asia, into Europe and Africa (Kilpatrick et al., 2006, Salzberg et al., 2007, Prosser et al., 2009, Gaidet et al., 2010, Gilbert et al., 2010, Takekawa et al., 2010). Also, recent phylogenetic analyses of A(H7N9) indicates that migratory birds from at least two distinct and distant flyways (Mediterranean-Black Sea and East Asian) may have contributed to its emergence (Gao et al., 2013, Kageyama et al., 2013, Liu et al., 2013). The potential dissemination of newly emerging AIVs within Eurasia to North America via migratory movements of birds remains a concern (Feare, 2007, Gauthier-Clerc et al., 2007) because phylogenetic analyses suggest that reassortment between North American and Eurasian AIVs occurs in several groups of migratory birds including sea ducks (Fries et al., 2013), dabbling ducks (Koehler et al., 2008, Wahlgren et al., 2008), shorebirds and gulls (Makarova et al., 1999). Thus, if these introductions are frequent, they could be important in AIV persistence and should be considered in mechanistic descriptions of AIV dynamics.
A quantitative understanding of interactions between wild hosts and poultry (i.e., frequency or rate of contact) is fundamental for parameterizing the spillover process in a transmission model. Also, for AIV, contact must be quantified in two ways: directly through bird-to-bird interactions and indirectly through sharing the same environment during a time period that virus can remain stable outside the host. For example, an experiment employing camera traps that record the number of direct contacts between wild hosts and poultry over a set time period in a setting with low biosecurity (i.e., backyard flock) would be one way to quantify direct contact rates. Data on indirect contact rates could also be extracted from the same experiment by summarizing the amount of time spent by wild hosts in the same environment or water sources as poultry. Other indirect contact data could be derived from longitudinal sampling of environmental DNA typed to species. In the USA, areas where mallard populations overlap poultry populations at a coarse resolution are being described qualitatively (CT Webb, unpublished data), but direct measurement of wild host-poultry interaction remains to be conducted.
2.3. Quantitative AIV surveillance data
The most common LPAIV subtypes found in mallards are: H3N8, H4N6 and H6N4 (Krauss et al., 2004, Olsen et al., 2006, Munster et al., 2007, Wilcox et al., 2011), although one study has found H5N2 to be common in the USA (40% of all virus isolations; Pedersen et al., 2010). Although HPAIV strains of H5N1 occur in dabbling ducks in some parts of Asia, northern Africa and Eastern Europe; in the USA only LPAIV strains of H5N1 viruses (from the North-American lineage) have been detected – despite numerous large-scale sampling studies (Hanson et al., 2003, Krauss et al., 2004, Pedersen et al., 2010, Wilcox et al., 2011). In fact, considering all subtypes, only LPAIV strains have been found in wild birds to date in the USA.
Prevalence of LPAIVs in mallards varies spatially along north-south portions of migratory flyways from >10 to 15% in the northern staging grounds to <1–2% on wintering grounds (Stallknecht and Shane, 1988a, Goekjian et al., 2011). LPAIVs are most prevalent in late summer/early fall, after the breeding season and during pre-migrational staging (Wilcox et al., 2011). Annual prevalence ranges from 1 to 5% during early winter and the spring migration to as high as 30–40% in early fall and during the fall migration (Stallknecht and Shane, 1988a, Webster et al., 1992, Hanson et al., 2003, Krauss et al., 2004, Wilcox et al., 2011). More long-term longitudinal sampling in areas where prevalence is highest at the annual peak is needed to examine whether these patterns are shared by all subtypes equally and to understand the environmental, population or immunity factors that cause them. These data also would provide crucial information for verifying whether or not current disease-dynamic models accurately capture transmission processes in important wild hosts and poultry.
2.4. Quantitative experimental data
Numerous LPAIV challenge studies have been conducted in mallards. These quantitative data are the main empirical data source for parameterizing disease-dynamic models (Rohani et al., 2009). While there is inherent variation in data among studies due to differences in virus, host individual and experimental design, there are emergent patterns in mallards (Table 1) that are different from those in gallinaceous poultry (e.g., chickens and turkeys; Mundt et al., 2009, Brown et al., 2011). Most challenge experiments to date have characterized readily measurable parameters of infection, such as incubation period, duration of infection, clinical outcome, viral shedding rates, viral persistence in water and antibody response (Slemons and Easterday, 1978, Webster et al., 1978, Kida et al., 1980, Mundt et al., 2009, Spackman et al., 2009, Davidson et al., 2010, Nazir et al., 2010, VanDalen et al., 2010, Lebarbenchon et al., 2012). More recent studies have even begun to quantify factors that explain infection variability in nature such as host-individual health status (e.g., stress, nutritional condition; Arsnoe et al., 2011, Reperant et al., 2011) and infectious doses (Swayne and Slemons, 2008, Brown et al., 2013a, Brown et al., 2013b). However, few studies quantify other immunity and transmission-related quantities, such as the duration and strength of immunity to the same (homosubtypic) or different (heterosubtypic) subtypes (Park et al., 2004, Fereidouni et al., 2009, Costa et al., 2010a, Jourdain et al., 2010, Pepin et al., 2012) and transmission rates and probabilities (Achenbach and Bowen, 2011), which are fundamental components of LPAIV epidemiology. This is mainly due to difficulties with maintaining wild hosts in captivity for longer than 2–3 months, and simulating contact rates between wild hosts and poultry under natural conditions. While it is challenging to interpret absolute measures of laboratory-based transmission rates, quantifying the relative importance of different mechanisms of transmission on individual-level infection parameters is both interpretable and important for describing AIV dynamics quantitatively.
Table 1.
Quantitative values of the infection and transmission processes in mallards as determined from experimental data. We focused on results from cloacal and fecal samples since these are known to have the highest LPAIV titers in mallards. Note that not all of these values would be used in a single model. Also, the table is not meant to be comprehensive in terms of capturing every sound experimental study, rather we aimed to capture the range of values that have been observed using several examples.
| Parameter | Rangea-VI | Rangeb-PCR | Units | Experimental measure | LPAIV strainsc | Dose | Host age | References | Caveatsd |
|---|---|---|---|---|---|---|---|---|---|
| Infection | |||||||||
| Minimum infectious dose or BID50 | ≤10, BID50: 103.1, 101.9 | Virions | Minimum number of viral particles that lead to an infectious individual | H6N1, H6N3, H8N4, H4N8, H5N1 | Variable | 3 weeks to 1 month | Swayne and Slemons (2008), Brown et al., 2013a, Brown et al., 2013b | Brown: Other strains were tested and did not cause infection at doses as high as 101.9; Swayne: BID50 (rather than minimum infectious dose) for mallard-origin viruses. A chicken-origin virus (H4N8) had a BID50 of 103.3. Swayne's study was performed in Pekin ducks (mallard-derived domestic duck). | |
| Incubation period | 0.5–2 days | 1–2 | Days | Number of days between inoculation and detection of virus | H5N2, H3N6, H7N2; H3N8 | 106–108 | 1–8-months | Webster et al. (1978), Kida et al. (1980), Spackman et al. (2009), Costa et al. (2010b) | The first sample was collected at 1 day, thus earlier incubation times could not be determined; Webster and Kida manuscripts detected viral shedding at <1 DPI |
| Detection period (IP) | 7 | 7 | Days | Number of days that virus is detected in >75% of infected ducks | H5N2, H3N8, H4N6, H7N7 | 106 | 3–4 weeks, 5–6 months | Spackman et al. (2009), Costa et al. (2010b), Jourdain et al. (2010), Brown et al. (2012), Pepin et al. (2012) | Mean across replicates and experiments |
| Peak shedding period | 1–7 | 1–7 | Days | Time points during which shedding rates are >103 | H3N8, H5N2; H7N2; H1N2; H4N1; | 106–108 | 3 weeks to 8 months | Webster et al. (1978), Kida et al. (1980), Spackman et al. (2009), Brown et al. (2012) | Means across replicates and experiments. |
| Peak shedding titer | 103.4–107 | 104–107 | Virions | Number of viral particles shed at the peak of infection | H3N8, H5N2; H7N2; H1N2; H4N1; | 106–108 | 3 weeks to 8 months | Webster et al. (1978), Kida et al. (1980), Spackman et al. (2009), Brown et al. (2012) | Means across replicates and experiments. Some birds excrete up to 109 EID50/g feces. |
| Length of homosubtypic immunity | ? | ? | Days | Number of days that infection with a strain of the same H/N subtype is suppressed | No studies to our knowledge | ||||
| Length of heterosubtypic immunity | ? | ? | Days | Number of days that infection with a strain from a different H/N subtype is suppressed | No studies to our knowledge | ||||
| Strength of homosubtypic immunity on shedding | Not quantified (but not 100%) | % | Percent reduction in total viral load that occurs during secondary infection with a strain of the same H/N subtype compared with primary infection | H7N7 | 108.7 | 3 months | Jourdain et al. (2010) | Data are not presented quantitatively | |
| Strength of heterosubtypic immunity on shedding | 99.2 | % | Percent reduction in total viral load that occurs during secondary infection with a strain from a different H/N subtype compared with primary infection | H3N8 → H4N6; H5N2 & H3N8 | 106–108.7 | 1–6 months | Costa et al. (2010a), Jourdain et al. (2010), Pepin et al. (2012) | Second infections were conducted 21 days after first. Arrow represents one-way cross-immunity assay; & represents two-way. | |
| Strength of homosubtypic immunity on IP | 75 | % | Percent reduction in IP that occurs during secondary infection with a strain of the same H/N subtype compared with primary infection | H7N7 | 108.7 | 3 months | Jourdain et al. (2010) | ||
| Strength of heterosubtypic immunity on IP | 0–60, 45 | % | Percent reduction in IP that occurs during secondary infection with a strain from a different H/N subtype compared with primary infection | H5N2 & H3N8, H3N8 → H4N6 | 106 | 4 months | Costa et al. (2010a), Pepin et al. (2012) | Second infections were conducted 21 days after first. Arrow represents one-way cross-immunity assay; & represents two-way. | |
| Direct transmission | |||||||||
| Transmission probability | 100 | % | % of hosts infected given contact with an infected host (assumes that probability is constant throughout the infectious period) | H5N2, H7N3 | NR | 2–4 months | Achenbach and Bowen (2011) | There were only 4 contact ducks in each experiment. Results are likely strain-dependent (Sturm-Ramirez et al., 2005). This experiment tested contact transmission in 23 different strains of H5N1 isolated from a variety of domestic and wild bird host species. 22/23 strains transmitted by contact and one of these only transmitted to 1/2 contacts. | |
| Transmission rate | 4+ | Infections/day | Number of hosts infected per day by one infected bird (assumes a constant daily rate over the infectious period) | H5N2, H7N3 | NR | 2–4 months | Achenbach and Bowen (2011) | There were only 4 contact ducks in each experiment and all 4 were infected in 1–2 days following exposure. | |
| Transmission probability function | f(x) = ? | f(x) = ? | % | % of hosts infected given contact with an infected host and viral load in the infected host over the entire infectious period | This relationship remains unknown and thus most models do not include effects of within-host viral dynamics on epidemiological outcome. | ||||
| Environmental transmission | |||||||||
| Water uptake rate | 0.3 | L/kg body mass/day | Amount of water drunk per day per host | Isanhart et al. (2011) | |||||
| Virus decay rate in water | 0.05 ± 0.04 at 5 °C; 2.8 ± 3.1 40 °Ce | Virions/day | Number of infectious viral particles that decay per day | H1N1; H2N4; H3N2; H4N6; H5N2; H6N4; H7N6; H8N4; H9N2; H10N7; H11N6; H12N5 | NR | NR | Handel et al. (2013) | Decay rates vary dramatically depending on strain and water temperature. Other factors such as salinity, pH and organic content affect decay rates too but these were not tested in Handel et al. (2013). | |
| Transmission probability | 75% at 102.8–103.1 | 100% at 102.8–103.1 | % Infected at virions/ml water | Quantitative relationship between the likelihood of infection given exposure to a given viral concentration | H4N6 | NR | 3 or 6 months | VanDalen et al. (2010) | There were 8 contact ducks. Ideally, we need to understand this relationship over range of viral concentrations. We also need to understand how transmission from a given viral concentration impacts subsequent infection dynamics over range of viral concentrations. For example, Van Dalen et al. determined that exposure to viral concentrations in water between 102.8 and 103.1 leads to infectious periods of 2.3–4.3 days and peak viral loads of 102.1 and 104.2 PCR EID50 depending on whether birds were 3 or 6 months. Thus, subsequent transmission may be possible following infection at these levels of water exposure. |
Range-VI: gives the measured range of values in the referenced studies that are from virus isolation results.
Range-VI: gives the measured range of values in the referenced studies that are from RT-PCR results.
Strains – host species: lists the strain-host species combinations that were used in the referenced studies.
Caveats: lists the experimental conditions that may have led to the highest and lowest values.
Mean ± standard deviation.
In Table 1, we have focused on LPAIV in mallards because HPAIV strains are not thought to contribute to the epidemiology of AIV in North American wild bird populations (Hanson et al., 2003, Krauss et al., 2004, Pedersen et al., 2010, Wilcox et al., 2011). However, experiments have investigated the effects of HPAIVs nonetheless. Most HPAIV trials in mallards have been conducted with H5N1 HPAIV and these data indicate infection differs significantly from LPAIV, both in regards to clinical disease and patterns of shedding. As opposed to LPAIV, the predominate route of H5N1 HPAIV viral shedding in mallards and other waterfowl is via the respiratory tract (Sturm-Ramirez et al., 2004, Brown et al., 2006, Keawcharoen et al., 2008). Viral shedding also occurs in the feces, but is at a lower titer and shorter duration. In addition to differences in shedding, virulence in mallards and other ducks can range from no clinical signs to 100% mortality depending on the clade of H5N1 HPAIV, host species and age (Perkins and Swayne, 2002, Sturm-Ramirez et al., 2005, Brown et al., 2006, Pantin-Jackwood and Swayne, 2007).
2.5. Models
Transmission models have been used to synthesize subsets of the data described above in order to generate additional quantitative insight into different aspects of LPAIV transmission among wild birds in the USA. Example applications are quantification of the role of environmental transmission in maintenance of LPAIVs (Breban et al., 2009) and the relative importance of trade routes and migration as sources of introduction of HPAIV strains from Asia (Winker and Gibson, 2010). However, risk models of spillover of LPAIVs to poultry have yet to include the spillover mechanism explicitly, largely due to the paucity of appropriate quantitative data describing interactions or transmission at the wildlife-poultry interface. With suitable empirical information at hand, such models would take into account known geographic distributions of wild bird migration routes, staging areas and breeding sites in decomposing the transmission risk of LPAIVs to poultry operations.
2.6. Priorities for quantitative research
Although there are a number of data gaps, there is an appropriate literature base and databases for which large-scale models could be developed. For example, continental determinants of LPAIV prevalence in wild birds have been identified by analytical models (Farnsworth et al., 2012). Further quantification with available data includes: (1) linking the density of poultry to that of mallard abundance indices (USFWS, 2012a) at a continental scale in space and time using hunter-harvest and National Agricultural Statistics Service data (e.g., Fig. 3), and (2) examining the effects of annual changes in the number of newly susceptible birds on annual continental distributions of LPAIV in waterfowl using national data on waterfowl recruitment (e.g., indices used to set harvest regulations such as female age ratios; Johnson et al., 1997, USFWS, 2012a) and LPAIV distributions from the U.S. Wild Bird AIV Surveillance program (DeLiberto et al., 2009, Farnsworth et al., 2012). The first analysis would help to identify regions where commercial poultry may have a high risk of spillover and thus where surveillance should be increased and where contact experiments would help to quantify actual risk. It should be noted that because there is currently no database summarizing the spatial locations of backyard poultry, the results would exclude risk from spillover to this sector. Thus, the overall risk to commercial poultry may be underestimated because backyard holdings, which could be a source of virus in commercial operations, have a higher risk of spillover from wild birds due to their lower biosecurity practices. The second analysis will help with understanding how mallard population dynamics affect LPAIV risk across space and time. A dynamical model parameterized with the recruitment data as well as mallard movement data would allow for a quantitative understanding of the spatio-temporal distribution of LPAIV prevalence, which is important for risk assessment.
Fig. 3.
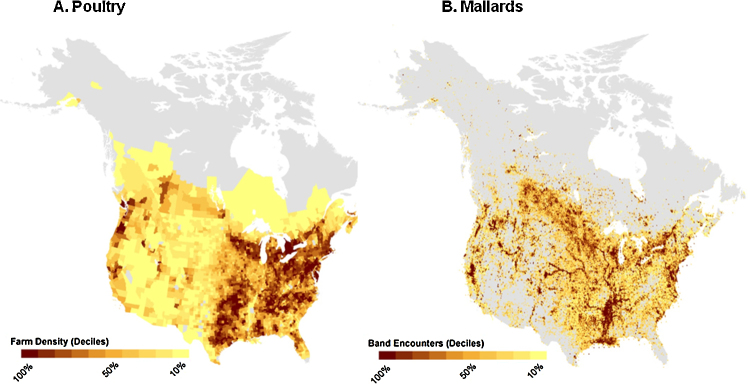
Distribution of poultry and wild mallards in the USA. (A) Density of poultry farms. (B) Thirty year annual average mallard band encounters between 1980 and 2010 based on hunter harvest data.
The next step would be to design empirical surveillance studies that temporally monitor mallard-poultry contact rates, coupled with estimating prevalence of infectious LPAIVs in mallard populations in select locations where there is the greatest overlap of poultry and high LPAIV prevalence in mallards. These studies should include both backyard and commercial operations with appropriate sampling designs that account for the differences in the density of holdings and biosecurity. For example, surveillance in backyard operations should quantify both direct and indirect contact rates between mallards and poultry and monitor LPAIV prevalence in both populations. In contrast, surveillance strategies in more secure commercial poultry operations would benefit from monitoring all wildlife and domestic species on the premises as well as environmental surfaces because LPAIVs are rarely detected and the direct role of mallards in spillover is less clear. This would provide crucial data for quantifying the risk of transmission given prevalence on the premises as well as identifying sources (biological or environmental) where LPAIVs are detected most frequently in close proximity to poultry.
3. Transmission within poultry flocks
We now turn from the potential spillover of LPAIVs from wild birds into poultry to flock-level control methods, demographics and LPAIV prevalence.
3.1. Control policies
Detection of AIV in poultry occurs at the flock-level: once confirmed in a single surveillance sample, the whole flock is determined to be infected. When HPAIV (see note below for Office International des Epizooties (OIE) definition)2 is detected on a farm, depopulation and other control activities outlined in the HPAI Response Plan are conducted in an effort to halt within-farm spread and prevent transmission to other farms (USDA, 2012).
Similar control strategies are applied to LPAIV strains that are considered to be high-risk for mutation to HPAIV (i.e., H5 or H7), although LPAIV outbreaks are usually managed without depopulation (Halvorson, 2009). The key decisions at the flock level involve determining depopulation or controlled-marketing strategies that will minimize transmission, contamination and time to virus elimination. Detailed response actions for H5 or H7 LPAIV in USA poultry are carried out in accordance with each state's H5/H7 LPAIV Initial State Response and Containment Plan.
In the case of detection of non-notifiable LPAIV strains (i.e., non-H5 and non-H7 strains LPAIV), options for on-farm control methods are more numerous and do not require federal action according to national or state response plans. The state in which the outbreak is located is in charge of the response. In these cases, cost-effectiveness of different control options within an operation and effects on continuity of business are primary considerations. The main on-farm control options considered are controlled marketing (marketing birds after clinical signs are apparent, when shedding is very low), trade/movement bans, disinfection and, rarely, vaccination (Halvorson, 2002, Halvorson et al., 2003). For example, because indemnity was not available to producers for depopulation of non-H5 or -H7 LPAIV outbreaks in California in 2000–2002, controlled marketing was used successfully to control an H6N2 LPAI outbreak (Cardona, 2005). This involved on-farm, active surveillance testing of flocks prior to movement to slaughter, thus reducing the risk of moving actively shedding flocks. If the flock was determined to be clinically infected, movement was prohibited until after the flock had seroconverted. Live-haul operators were required to follow designated routes that were chosen to avoid exposure of poultry flocks on facilities adjacent to roadways. Most of the influenza virus that is shed from a positive flock occurs during the first two weeks of infection, and orderly marketing 3–4 weeks post-onset of clinical signs is currently used to reduce economic loss to turkey producers (Halvorson, 2003). Similar, voluntary controlled-marketing programs have been used to control seasonal LPAI outbreaks in turkey flocks in Minnesota (Cardona, 2005).
3.2. Quantitative host population data
In contrast to mallard populations, the population structure and demographic dynamics are well defined for commercial poultry (Fig. 4A). Nearly all poultry operations with greater than $1000 USD of agricultural sales are surveyed by the Census of Agriculture (www.agcensus.usda.gov), excluding public, industrial and grazing association land (USDA, 2008a). The industry is 84.6% production farms (65.6% broiler chickens, 2.3% table-egg chickens and 16.7% meat turkeys) and 15.4% breeder farms (USDA, 2011). Flock sizes in production farms vary from <50,000 to >100,000 birds (Table 2, USDA, 2011). The lifespans and ages of each poultry type within a flock are homogeneous in production farms. Meat poultry are very short-lived; on average marketing or slaughter occurs at 7.2 weeks for broiler chickens, 14.2 weeks for turkey hens and 19.7 weeks for turkey toms (USDA, 2011). Table-egg layers live for an average of 88.7 weeks (molted) and 64.2 weeks (non-molted) from placement in the house until removal (roughly 50% of commercial flocks are molted; USDA, 2011). In about 67% of table-egg layer flocks, birds are kept in cages such that contact between birds in the same cage occurs readily while between-cage bird contacts are possible but more limited. However, more widespread contact is possible indirectly through shared water systems, feed systems and ventilation. In the remaining table-egg layer flocks and all broiler chickens and turkeys, birds are reared on the floor and could potentially contact a larger number of other birds.
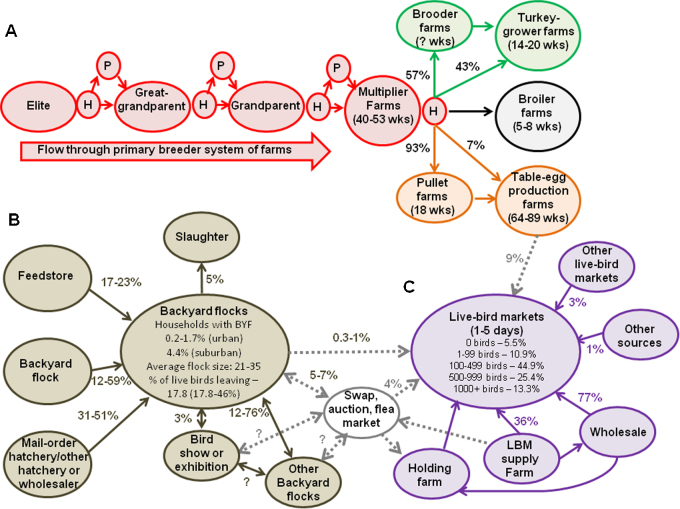
Fig. 4.
Structure of the poultry industry in the USA. The industry consists of 3 main sectors: commercial (A; multiple colors), backyard (B; brown) and live-bird market system (C; purple). Dotted gray arrows indicate connections between different sectors. Numbers on arrows indicate the percentage of poultry that are typically moved between the indicated locations. (A) Production begins with several rounds of breeding to control host genetics. Elite breeder flocks send eggs to the hatchery (H) where chicks hatch, which takes about 3–5 weeks. Chicks leave hatcheries at 1 day-old. Most chicks will go to a pullet farm (P) for 18 weeks before the next breeder farm for variable amounts of time (determined by the breeder), although some may go directly to the next breeder farm. Some table-egg layers will skip the great-grandparent breeding phase. At the final stage of breeding (i.e., multiplier farms), broiler chickens and 43% of turkeys will go directly from a hatchery to a broiler farm or turkey grower, while table-egg layers typically go to a pullet farm before being transferred to a table-egg production farm. The three different types of commercial poultry (turkey, green; broiler chickens, black; and table-egg layers, orange) are bred and reared on separate farms, often by separate companies. (B) and (C) The supply chain for backyard flocks (BYF) and live-bird markets (LBM) involves multiple sources and poultry within each of these holdings incoming and outgoing poultry within each of these sources may contact each other (i.e., none of these are all-in, all-out operations). Note that “bird, swap, auction, flea market” events are the main mechanisms for interaction between BYF and LBMS and thus do not belong exclusively to either BYF or LBMS. Data in this figure were derived from USDA censuses (USDA, 2005, USDA, 2011, Garber, 2006) and unpublished data from surveys of small-scale poultry traders (K. Pabilonia).
Table 2.
Population size distribution of production farms in the USA (USDA, 2011). Turkey farms are meat production. Values are means with standard errors shown in brackets.
| Number of birds | Broiler % (SE) | Table egg % (SE) | Turkey % (SE) |
|---|---|---|---|
| <50,000 | 11.7 (0.4) | 34.3 (2.5) | 73.4 (0.5) |
| 50,000–99,999 | 56.3 (0.6) | 12.0 (1.9) | 23.9 (0.4) |
| 100,000+ | 32.0 (0.5) | 53.7 (2.7) | 2.7 (0.3) |
All production poultry are bred through the “primary breeding system” which makes up the smallest proportion of all breeders. For example, 10% of breeder farms for chickens (broilers and layers) are primary breeders compared with 90% that are multiplier farms (USDA, 2011). Thus, well under 3% of all farms are primary breeders. Elite primary breeders make up the smallest number of farms and the number of farms per breeder category increases with flow through the system (Fig. 4A). It is estimated that genetic traits from the elite flocks swamp the production flocks in about 5 years, meaning that there is roughly a five year lag between initial breeding and the time that a potentially different genotype reaches production farms. Breeding farms can also have more complex age-structure than production flocks but the details depend on the particular farm. In general, eggs are collected daily from broiler breeders and brought to the hatchery. Once hatched, chicks will be transferred to a pullet farm or directly to the next breeder farm at one day of age to be reared to reproductive maturity. Thus, there is no direct transfer of birds between breeders or between breeders and production farms. The only direct movement of birds is from pullet farms to production farms.
Although the majority of commercial poultry are included in the agricultural census, only a small proportion of the total poultry operations are included because many small non-commercial operations, or “backyard flocks” (21–35 birds on average) exist in the USA (Fig. 4B). A recent study found that commercial poultry operations in the USA had an average of 1.9 backyard flocks within 1 mile of their perimeters (USDA, 2005, Garber et al., 2007), while in Colorado alone, 69% of poultry operations contained 1–50 birds and only 6% had >200 birds (Smith et al., 2012). In Denver and the surrounding metropolitan cities, 2.2% of residences had backyard flocks with a higher prevalence in suburban areas (1.55% of residences in metropolitan Denver, as compared with suburbs (4.4%)). Most flocks were layers (80.6%) and 37.3% contained multiple species, including waterfowl, pigeons, peafowl, turkey or exhibition chickens (KL Pabilonia, unpublished data). The multi-host and other structural differences of the backyard flocks require a different model description because these differences could impact subtype-specific transmission dynamics (Duan et al., 2007).
Live-bird markets (LBMs) are another important small holder poultry system in the USA (Fig. 4C). The majority of the markets are located in a few states (California, Florida, New Jersey, New York, Pennsylvania; Mullaney, 2003). However, the origin of birds that enter the markets could be almost anywhere in the USA because of a complex supply system for the numerous species that may be stocked (Bulaga et al., 2003) (Fig. 4C). Chickens (spent layers, spent breeder hens, and broilers) are the most commonly stocked species, with ducks and guinea fowl among the next most common species (Garber, 2006, Yee et al., 2008). Other avian species may include turkeys, quail, chukkars/partridge, pheasant geese and peafowl. Mammals, most often rabbits, are also common in the live-animal markets (Trock et al., 2003). The structure of the LBM system is different from backyard poultry although there may be informal interactions between them (Fig. 4). Uniform standards for surveillance, cleaning and disinfection (including rest days where the markets are completely depopulated and cleaned) were implemented in 2008 and have decreased the incidence of AIV in these markets (Trock and Huntley, 2010).
3.3. Quantitative AIV surveillance data
There is a well-defined process for AIV surveillance in poultry that includes both passive and active methods. The National Poultry Improvement Plan (NPIP) documents 2.0–2.7 million tests per year for AIV in commercial poultry. Results are reported in the NPIP annual report. Samples taken from monitored flocks are sent to the National Animal Health Laboratory Network (NAHLN) where they are assayed for type A influenza. Type A influenza-positive samples are then submitted to the National Veterinary Services Laboratories (NVSL; Ames, Iowa) to screen for the presence of H5 or H7 subtypes, and the findings are reported to the National Animal Health Policy and Programs (NAHPP) staff in Riverdale, Maryland. Each year, two or three H5/H7 LPAIV cases are reported in commercial poultry across the USA. Since 2004, all cases have been LPAIV. Many detections are antibody positive cases, where live virus is not recovered. There have been four documented HPAIV outbreaks in the US, one in the upper Midwest and northeastern LBMs in 1924–1925, a limited outbreak in New Jersey in 1929, one in commercial farms in Pennsylvania in 1983–84 (where the initial outbreak strain was a LPAIV), and one in a LBM supplier in Gonzales (Pelzel et al., 2006), Texas, and two associated LBMs in Houston, Texas in 2004 (Swayne, 2008).
Assessing incidence of AIVs in the backyard and LBM sectors was more challenging historically due to the low level of testing being conducted as compared with commercial poultry. However, since 1986, states have routinely conducted surveillance in LBMs because the markets were shown to be a source of AIV infection for commerial poultry. In 2004, the US Department of Agriculture's Animal and Plant Health Inspection Service (USDA-APHIS) published uniform program standards (USDA, 2008b) to prevent and control H5 and H7 LPAIV subtypes in the LBMs. The standards cover (1) licensing, (2) AIV testing, (3) recordkeeping, (4) sanitation, (5) biosecurity, (6) surveillance, (7) inspection, (8) trace backs, (9) premises registration, (10) trace outs when positives occur, and (11) response to positive facilities. These standards apply to LBMs, auctions, and small sales, as well as to producers and distributors who supply the markets. Overall, since the initiation of the H5/H7 LPAIV LBM program, the total number of LBM-positive premises has decreased steadily from 63 in 2005 to between 2 and 6 since 2009. Only LPAIVs have been detected and the predominant subtype varies from year to year (NVSL annual report, Proceedings from the annual meeting of the USAHA, 2009, 2010, 2011). In 2007, LPAIV H7N2, which had been circulating in LBMs in the Northeast United States since 1994, was declared eradicated.
3.4. Quantitative experimental data
In contrast to mallards where the dominant route of shedding is through the cloaca or in feces, AIVs are mainly shed oropharyngeally in gallinaceous poultry (primarily chickens and turkeys). Oropharyngeal shedding rates of LPAIV in experimental studies range from undetectable to 100% of the birds shedding. Typical titers from oral samples range from 101 to 105 50% egg infectious doses per ml (EID50/ml) depending on the virus isolate and dose (Morales et al., 2009, Pillai et al., 2010, Spackman et al., 2010). Cloacal shedding rates of LPAIV also occurs with the following differences relative to oral shedding: (1) more birds have shedding rates below detection limits, (2) detectable shedding is about 1–3 orders of magnitude lower and (3) peak loads are usually 1–2 days later (Morales et al., 2009, Jackwood et al., 2010, Ladman et al., 2010, Spackman et al., 2010). HPAIVs are mostly shed orally with titers as high as 105 to 108 EID50/ml (Lee et al., 2005, Swayne and Pantin-Jackwood, 2006, Grund et al., 2011). The incubation period for both LPAIV and HPAIV is 24–48 h. The infectious period for LPAIV ranges from 1 to 7 days post-infection (DPI), with a peak of oral shedding at about 2 DPI and a peak of cloacal shedding at approximately 4 DPI (Morales et al., 2009, Spackman et al., 2010, Spackman et al., 2013). Rarely, virus can be recovered from birds 10 DPI (Morales et al., 2009, Pillai et al., 2010, Spackman et al., 2010). The infectious profile for HPAIV is the same except that most birds (i.e., 75–100% of individual birds that are infected with HPAIV, including chickens, turkeys and domestic ducks) will not survive beyond the infectious period, dying between 1 and 5 DPI. As with LPAIV infections, HPAIV survivors will usually not be infectious past 7–10 DPI, but the absolute value depends on infectious dose (Swayne et al., 1997, Lee et al., 2005).
The duration of immunity from vaccines or natural infections in poultry is not well understood, although maternal immunity is known to last approximately two weeks after hatch. However, the role of pre-existing immunity to AIVs is probably of limited relevance to commercial poultry in the USA because vaccination is not practiced (except for vaccination against H1 and H3 in turkey breeders), life-span is short and flocks that become infected with HPAIV or H5 or H7 LPAIV are culled. Also, based on a routine surveillance, it is very rare for a commercial flock to have antibody to AIV of any subtype. Thus, an appropriate model structure includes disease states: susceptible-infectious-notified-depopulated, rather than including a compartment for immunity. However, in backyard operations and LBMs where there is opportunity for exposure to multiple strains, pre-existing immunity is likely very important.
AIVs can be transmitted via several mechanisms in gallinaceous poultry: direct contact (bird-to-bird contact), indirect contact (e.g., contact with feces, drinking water or other fomites) or aerosols (van der Goot et al., 2003, Leung et al., 2007, Yee et al., 2009, Spekreijse et al., 2011, Spekreijse et al., 2013). Few studies have approached the transmission process quantitatively. For example, quantification of transmission rates absolutely (e.g., Gonzales et al., 2012, Saenz et al., 2012), determining the likelihood of transmission from environmental sources or determining relative quantitative differences among transmission mechanisms have rarely been done for either LPAIV or HPAIV in poultry. One study using the LPAIV and HPAIV H5N2 outbreak isolates (chicken Pennsylvania 1983) found that 30% of the susceptible chickens in the LPAIV group were infected, whereas 100% were infected in the HPAIV group when cages were placed 1.5 m apart (van der Goot et al., 2003). Another study showed that transmission between cages occurred earlier when fecal material was allowed to drop into lower cages as compared with aerosol-only conditions (Yee et al., 2009). Finally, transmission to susceptible chickens in cages placed 0.2, 0.4 or 1.1 m from infected birds was rare (6/72 chickens; only in one replicate trial; Spekreijse et al., 2011). Collectively these data suggest that aerosol transmission is less important than contact, but more experiments are needed before the relative difference between different transmission mechanisms can be quantified.
Although this article focuses on transmission processes, it is important to mention that evolutionary changes should also be considered in quantitative analyses of the transmission process. However, this remains an extremely challenging aspect to understand because neither the conditions facilitating the genesis of reassortants nor those that impose selection for novel strains are understood well enough to predict risk of novel HPAIVs, although some progress has been made using phylogenetic analyses (e.g., Duan et al., 2007). These gaps highlight a key priority for empirical quantitative research. For example, quantification of rates of co-infection and the propensity for different subtypes to co-infect poultry (e.g., Pepin et al., 2013) and wild hosts are important for estimating rates of reassortment and understanding ecological conditions that favor them. Likewise, in order to identify conditions that facilitate adaptation, more experimental evolution studies (e.g., Giannecchini et al., 2010) and fitness measures in poultry with isolates from wild hosts are needed.
3.5. Models
Within-flock outbreaks in poultry have been extensively studied using mechanistic models (Savill et al., 2006, Savill et al., 2008, Bos et al., 2007, Bos et al., 2009, Tiensin et al., 2007, Bouma et al., 2009, Malladi et al., 2011, Malladi et al., 2012, Weaver et al., 2012, Reeves et al., 2013). Increased insight from within-flock models compared to wild-bird models or system-wide models is largely because of the precision with which the population dynamics of single flock can be described. As with the between-flock models described in the next section, within-flock models typically employ either of two approaches: (1) developing a simulation model that used condition-specific parameters as inputs and produces outbreak dynamics or transmission parameters as outputs, or (2) fitting a disease dynamic model to outbreak data and estimating transmission parameters (or other epidemiological quantities) from the outbreak data. The first approach allows for a scenario-based understanding of within-flock transmission behavior, whereas the second allows for direct estimation of quantities such as transmission rate.
The versatility of within-flock dynamic-model approaches has allowed for evaluation of both efficacy and consequences of alternative control methods. For an example of the former, a within-flock model was used to generate plausible outbreak data against which the efficacy of various outbreak detection methods were compared (Malladi et al., 2011). For an example of the latter, a within-flock model showed that vaccines conferring incomplete protection could counterintuitively increase the infectiousness of a flock, by delaying the time to detection and depopulation (Savill et al., 2006). Within-flock models have also been used to: (1) infer important events – such as the day of introduction – from mortality data (Bos et al., 2007), (2) estimate the risk of HPAIV transmission due to the movement of poultry-industry products from monitored flocks (i.e., flocks not known to be infected with HPAIV) into commerce (Malladi et al., 2012), (3) estimate important epidemiological parameters (Bouma et al., 2009) and (4) estimate mean time to detection for HPAIV strains under different surveillance-detection triggers (Weaver et al., 2012).
3.6. Priorities for quantitative research
In the USA, within-flock transmission data are largely limited to a few small-scale transmission studies. Thus, the accuracy of within-flock transmission models cannot be quantified, and our data-based understanding of transmission dynamics (i.e., estimates of transmission derived from outbreak data) in different poultry sectors remains weak. However, because the details of USA poultry-population demography and structure are well-defined, simulation models can be used to understand how the structure of different poultry sectors impacts transmission rates of AIV throughout different poultry sectors using situation-specific details as inputs. Indeed, such dynamic models (not validated by outbreak data) specifically tailored to the USA situation have been used to improve our mechanistic understanding of within-flock transmission from a theoretical standpoint (Malladi et al., 2012, Weaver et al., 2012). However, the tools used in these studies have been underused relative to the number of pressing questions that could be addressed. For example: (1) how does the type of poultry sector impact within-flock transmission rates?; and (2) how does the type of surveillance data – active versus passive – impact estimates of infection of the index case?. In addition, as more within-flock data become available results from these simulation studies should be validated.
4. Transmission between poultry flocks
Now we summarize AIV-response decisions and quantitative data pertaining to transmission between different poultry flocks.
4.1. Control policies
AIV management goals at the between-flock level involve conducting epidemiological investigations, surveillance, depopulation, movement restrictions, biosecurity and public awareness campaigns in a manner that will identify at-risk premises and minimize between-farm spread. Extensive trace-back and trace-forward epidemiological investigations are conducted in which the movement of people, animals, equipment and other materials between flocks in the period leading up to the detection of infection on premises are investigated. Thus, a key immediate decision is to define a maximum period prior to detection that should be considered for trace-back and trace-forward investigations. This decision is often informed through estimation of the date of the initial case using daily mortality data, production data, current diagnostic results (i.e., continued shed of virus versus antibody only persistence) and date of last negative flock test. The spatial area surrounding infected premises where surveillance should be intensified during and following the event is usually unclear. Recent movements into the flock (e.g., spiking roosters), bird placement dates, and recent handling of birds on the farm are the main considerations for determining the spatial boundaries of surveillance. However, these issues could be well-served by quantitative models, especially in the first few days when data are limited.
An equally important consideration is to identify the temporal and spatial intensity at which surveillance should be conducted in epidemiologically linked premises and how the surveillance protocol should change on premises that are closed versus those that continue with business. These surveillance protocols, which are implemented to maintain continuity of business during an outbreak, can be labor- and resource-intensive. Advance planning is used to determine how laboratories will leverage their critical diagnostic resources and laboratory support (e.g., Secure Egg Supply Plan www.secureeggsupply.com) to manage the significant increase in incoming samples and urgent need for results. Parsimonious quantitative models that rapidly determine the cost-effectiveness for alternative surveillance protocols would be valuable.
4.2. Quantitative host population data
The vast majority of commercial poultry farms operate as all-in, all-out with no access to the outside environment (Fig. 4A). The turnover rates for broilers and meat turkeys are about 5.5 and 2.8 times per year respectively (8.9 billion sold to 1.6 billion inventory; 296 million sold to 107 million inventory; respectively) based on a census conducted in 2007 (USDA, 2007). However, for caged-layer operations, all-in all-out only applies to individual barns (which are considered to be separate flocks with multiple barns on one layer complex), while for broilers, it refers to all birds on an operation (which comprise one whole flock divided amongst several barns). For table-egg layers, all birds are the same age in 38.4% of farms, while 60.7% have birds of different ages in different barns and 0.9% have birds of different ages in the same barn. Also, 7.2% of farms raise their own pullets, which is another potential means for poultry of different ages to be present on layer farms (USDA, 2011). Meat-type turkey operations may be either single-age premises (i.e., all-in, all-out) or multi-age complexes in which younger brooder birds are raised on the same premises as grow-out birds, ensuring that these premises are almost never empty (USDA, 2011). For table-egg layers, the barns remain empty for an average of 17 days after poultry are removed (USDA, 2000). For transportation of birds off farms, nearly all broiler farms and about 80% of turkey farms use vehicles dedicated to the company (USDA, 2011). However, roughly 90% of table-egg farms use vehicles that are also used by other companies (USDA, 2011).
Sources of new birds also differ between types of operations. Most (76.9%) table-egg farms received pullets that were produced by multiplier flocks owned by a primary breeder company. In contrast, almost all broiler farms and ∼50% of turkey farms receive chicks/poults from company-owned multiplier farms.
The biosecurity of backyard flocks contrasts sharply with that of commercial operations. A recent study in Colorado found that 97% of backyard flocks have access to the outside environment and birds in 35% of the flocks are able to leave the property (Smith et al., 2012). The vast majority of backyard flocks do not practice all-in, all-out management, which allows for significant bird movement to and from these flocks. Up to 46% of owners move birds off their properties annually, primarily to attend bird shows, sell birds at swap meets, auctions or flea markets, take birds to slaughter or LBMs or move birds to/from other backyard flocks (Smith et al., 2012), but there are little data on the precise rates and location of these movements.
4.3. Quantitative AIV surveillance data
Data on the between-farm spread of AIV in the USA is challenging to obtain because: (1) these data are commercially sensitive, (2) data collection during emergency response efforts is usually not prioritized, and (3) surveillance and response are so effective that disease rarely spreads after detection. Nevertheless, 10 multi-flock outbreaks of LPAIV have occurred since the 1920s, one of which included evolution from a LPAIV strain to a HPAIV strain (Pennsylvania 1983; Halvorson, 2009). In total, these outbreaks have cost the industry $368 million, which does not include costs to consumers for increases in egg and poultry prices (Halvorson, 2009). One of the more recent and widespread multi-flock LPAIV outbreaks, which occurred in Virginia in 2002, was especially well-documented and included data from a detailed epidemiological investigation (McQuiston et al., 2005). Initially, the H7N2 strain was detected in a single commercial turkey-breeder flock and the operation immediately depopulated. However, 4 additional farms were confirmed positive within a week of initial detection and 60 farms within a month. By the end of the outbreak, 197 total farms had been affected, including facilities with both turkeys and chickens, and an estimated cost of $149 million (McQuiston et al., 2005).
An epidemiological investigation of the Virginia outbreak collected data from 151 infected farms and 199 control farms in an effort to identify risk factors of infection. Some of the data recorded, which could be important for development of mechanistic models, were: farm location, date of reported infection, date of depopulation or recovery, age of infected birds, number of birds, number of poultry barns, type of barns, worker activities, biosecurity measures and presence of wild, livestock and peridomestic host species. Although information on the origin and destination of traffic coming onto and leaving farms was not reported in the McQuiston et al. (2005) study, it was documented by the USDA. There were also genetic analyses of the isolates collected during the outbreak (Spackman et al., 2003). Together, these data can be used to: (1) develop a parsimonious model that quantifies rates and patterns of between-farm spread, (2) test predictions, quantify uncertainty and validate crucial flock-level simulation-modeling tools such as the North American Animal Disease Spread Model (NAADSM) (Harvey et al., 2007), InterSpread Plus (Stevenson et al., 2013) or AusSpread (Garner and Beckett, 2005), and (3) evaluate outbreak response strategies (Fig. 5).
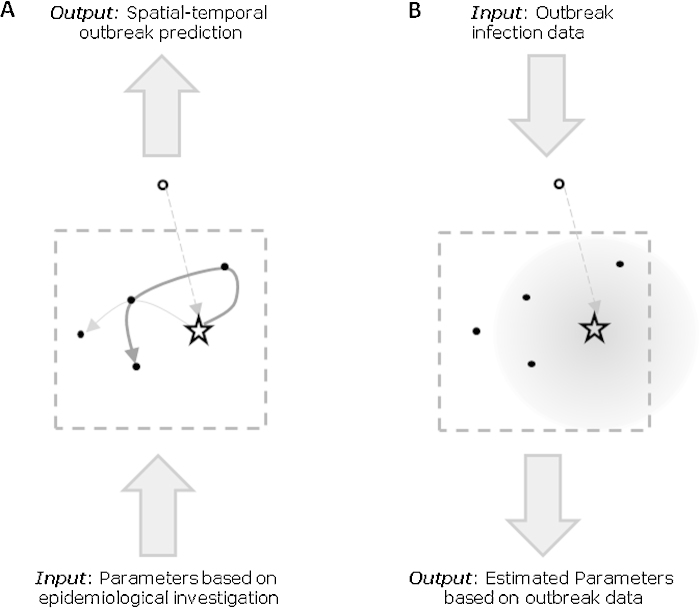
Fig. 5.
Conceptual representations of two different approaches to modeling poultry disease outbreaks. (A) Bottom-up approach. Parameters from epidemiological investigations and other sources are input into a detailed simulation of routes of transmission. Within the simulation, a local area is represented by the dotted box. In this example, contact of an infectious premises (open circle) with a focal premises that becomes infected (open star) is via long distance transmission represented by the dotted line. Other types of contact and potential routes of transmission from the focal infected premises to susceptible premises (filled circles) may be by different routes represented by the black and gray solid lines. The detailed simulation output is often a spatial-temporal prediction of outbreak dynamics. (B) Top-down approach. Spatial-temporal outbreak infection data are the input for model fitting of a local spread model. Some features of the detailed simulation model, such as long distance transmission represented by the dotted line may be maintained. Other features such as specific, local transmission routes can be subsumed into a general, local transmission kernel represented by the shaded circle with highest transmission risk near the infectious premises and decreasing transmission risk with distance. This approach may result in a simpler model that can more easily be fit to outbreak data. The output of a local spread model is often estimated outbreak parameters, such as between-premises transmission rates or the time between initial infection and notification.
4.4. Quantitative experimental data
To our knowledge, there are no experimental data quantifying rates and mechanisms of between-farm spread using a model system, mainly because such an experiment would be too costly and risky to conduct. The lack of such data underscores the importance of quantifying these processes using naturally produced outbreak data and verifying our understanding of the between-farm processes that are currently described mathematically in highly-parameterized simulation models.
4.5. Models
Between-farm transmission of agricultural animal diseases is an archetypical process for development of quantitative methods in spatial infectious-disease dynamics (Schoenbaum and Terry Disney, 2003, Garner and Beckett, 2005, Karsten et al., 2005a, Karsten et al., 2005b, Harvey et al., 2007, Riley, 2007, Sharkey et al., 2008, Tildesley et al., 2008, Tildesley et al., 2010, Stevenson et al., 2013). Models of between-farm spread of HPAIV H7N7 in The Netherlands (Boender et al., 2007) and HPAIV H7N1 in Italy (Dorigatti et al., 2010) have quantified spread rates and patterns using maximum likelihood estimation, and have determined control strategies that would minimize spatial spread in future outbreaks by simulation. Recently, progress has also been made in developing a theoretical framework for estimating between-farm spread of AIV and identifying effective control strategies using contact-tracing data, outbreak data and Bayesian inference (Jewell and Roberts, 2012). Bayesian inference of an underlying spatial transmission model has also been used to show that although backyard farms in Thailand were a key element of the successful propagation of the 2004–2005 wave of H5N1 infections, it is likely that improved biosecurity within the commercial farming sector was responsible for the subsequent overall reduction in transmissibility in the system.
For AIVs in the USA, disease-dynamic simulation models have been used to: (1) understand which epidemiologic parameters have large impacts on potential spread of HPAIV (Patyk et al., 2013), (2) estimate the consequences of a potential incursion of HPAIV into the USA (Patyk et al., 2013), and (3) estimate the time to detection on farms (Dorea et al., 2010). Flock-level simulation models have been used to estimate the economic consequences of HPAIV introductions in specific areas as well as the cost-effectiveness of situation-specific alternative control strategies (Patyk et al., 2013). This model includes numerous empirically determined parameters that describe details of poultry population demographics, structure and movement between facilities, and has undergone many tests to validate the model structure and to measure output variation (due to stochasticity).
4.6. Priorities for quantitative research
Much of the previous and current quantitative research on the between-farm spread of AIV in the USA involves flock-based simulation models with numerous parameters (Harvey et al., 2007, Patyk et al., 2013; Fig. 5A). This approach is valuable because it permits the examination of outcomes under particular circumstances which may be important when preparing for unknown events that could occur anywhere in the country. That is, geographic variation in the distribution of poultry facilities and how these facilities may be connected varies widely across the country which means that transmission patterns occurring from any particular introduction of AIV could depend on where the introduction occurs. Although, the detailed models mentioned above are constructed with field-measured parameters, their output is only as accurate as our a priori assumptions about how the parameters are connected. Thus, an important priority is to assess the accuracy of these models using data (i.e., how well these models predict real-world outcomes). Complimentary decision-support tools such as the development of simpler quantitative models of between-farm spread are also needed (e.g., Fig. 5B): models with large numbers of parameters inherently produce output with higher uncertainty. Also, complex models can be computationally intense limiting their utility during an emergency when a reliable answer may be needed within hours (although the complex models listed above are intended for use in advance of an outbreak). In our opinion, the data similar to those collected during the 2002 Virginia outbreak provide an opportunity for developing simpler models that: (1) may produce results with reasonable uncertainties, (2) estimate important quantities such as the time between infection and notification or the radius for surveillance around infected premises, and (3) validate output of simulation models. This research is critical for gaining a data-based understanding of the processes that determine AIV spread between poultry operations.
5. Conclusion
We discussed how dynamical models can be used to inform AIV prevention and response policies in a large commercial poultry industry, using the USA as a case study. We identified the following important goals for modeling research that can be accomplished with current data: (1) quantify the spatio-temporal relationship between wild hosts and poultry in terms of population sizes and AIV prevalence, (2) understand how the structure of different poultry sectors impacts AIV transmission within poultry flocks, (3) quantify processes responsible for AIV spread between poultry operations, and (4) validate current policy-decision tools with data.
While the focus of this article has been on using currently available data for the development of quantitative tools that serve animal and public health policy decisions, data resources for such tools are clearly limited and could be improved substantially with relatively limited additional investment during key periods. Usually, when an outbreak occurs, the situation is an emergency and data collection for the purpose of informing future (or even current) quantitative analyses takes low priority. Although some data are always collected, the types (and quality) of data vary by outbreak due to situation-specific circumstances. Most outbreak data sets exclude simple pieces of information that could greatly improve our ability to estimate important control parameters. One way to enhance data resources would be through standardization of data collection during outbreaks. With the appropriate collaboration of expertise, including quantitative scientists, response personnel and policymakers; the development of such an outbreak-data collection policy could be achieved in a way that minimizes risk of contamination during response, maximizes cost-effectiveness and operational efficiency, and includes data that are essential for improving quantitative tools. Such a policy could generate high-value data resources that would facilitate the development of accurate response-oriented quantitative preparedness tools.
Acknowledgements
Thanks to the RAPIDD program of the Science and Technology Directorate, U.S. Department of Homeland Security, and the Fogarty International Center, NIH for funding the workshop and this output. SR thanks The Wellcome Trust (093488/Z/10/Z) and The Medical Research Council (UK, MR/J008761/1) for support. Also, thanks to Kim Forde-Folle for providing expert opinion and helpful comments on the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
“For the purpose of disease control programs and international trade in domestic poultry products, HPAI is defined in the Code of Federal Regulations, Title 9, Section 53.1 as: (a) Any influenza virus that kills at least 75 percent of eight, 4- to 6-week-old susceptible chickens, within ten days following intravenous inoculation with 0.2 ml of a 1:10 dilution of a bacteria-free, infectious allantoic fluid. (b) Any H5 or H7 virus that does not meet the criteria in paragraph (a) of this definition, but has an amino acid sequence at the hemagglutinin cleavage site that is compatible with HPAI viruses. (c) Any influenza virus that is not a H5 or H7 subtype and that kills one to five chickens and grows in cell culture in the absence of trypsin.”
Contributor Information
K.M. Pepin, Email: kimpepin@gmail.com, Kim.M.Pepin@aphis.usda.gov.
E. Spackman, Email: Erica.Spackman@ars.usda.gov.
J.D. Brown, Email: jubrown1@uga.edu.
K.L. Pabilonia, Email: Kristy.Pabilonia@ColoState.edu.
L.P. Garber, Email: Lindsey.p.Garber@aphis.usda.gov.
J.T. Weaver, Email: Todd.Weaver@aphis.usda.gov.
D.A. Kennedy, Email: dak30@psu.edu.
K.A. Patyk, Email: Kelly.A.Patyk@aphis.usda.gov.
K.P. Huyvaert, Email: kate.huyvaert@colostate.edu.
R.S. Miller, Email: Ryan.S.Miller@aphis.usda.gov.
A.B. Franklin, Email: alan.b.franklin@aphis.usda.gov.
K. Pedersen, Email: Kerri.Pedersen@aphis.usda.gov.
T.L. Bogich, Email: tiff.bogich@gmail.com.
P. Rohani, Email: rohani@uga.edu.
S.A. Shriner, Email: susan.a.shriner@aphis.usda.gov.
C.T. Webb, Email: Colleen.Webb@colostate.edu.
S. Riley, Email: s.riley@imperial.ac.uk.
References
- Achenbach J.E., Bowen R.A. Transmission of avian influenza A viruses among species in an artificial barnyard. PLoS One. 2011;6:e17643. doi: 10.1371/journal.pone.0017643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D.J., Parsons G., Manvell R.J. Experimental assessment of the pathogenicity of eight avian influenza A viruses of H5 subtype for chickens, turkeys, ducks and quail. Avian Pathol. 1986;15:647–662. doi: 10.1080/03079458608436328. [DOI] [PubMed] [Google Scholar]
- Anderson R.M., May R.M., editors. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press Inc.; New York: 1992. [Google Scholar]
- Arsnoe D.M., Ip H.S., Owen J.C. Influence of body condition on influenza A virus infection in mallard ducks: experimental infection data. PLoS One. 2011;6:e22633. doi: 10.1371/journal.pone.0022633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl J., Krauss S., Kuhnert D., Fourment M., Raven G., Pryor S.P., Niles L.J., Danner A., Walker D., Mendenhall I.H., Su Y.C.F., Dugan V.G., Halpin R.A., Stockwell T.B., Webby R.J., Wentworth D.E., Drummond A.J., Smith G.J.D., Webster R.G. Influenza A virus migration and persistence in North American wild birds. PLoS Pathog. 2013;9:e1003570. doi: 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boender G.J., Hagenaars T.J., Bouma A., Nodelijk G., Elbers A.R., de Jong M.C., van Boven M. Risk maps for the spread of highly pathogenic avian influenza in poultry. PLoS Comput. Biol. 2007;3:e71. doi: 10.1371/journal.pcbi.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos M.E.H., Van Boven M., Nielen M., Bouma A., Elbers A.R.W., Nodelijk G., Koch G., Stegeman A., De Jong M.C.M. Estimating the day of highly pathogenic avian influenza (H7N7) virus introduction into a poultry flock based on mortality data. Vet. Res. 2007;38:493–504. doi: 10.1051/vetres:2007008. [DOI] [PubMed] [Google Scholar]
- Bos M.E.H., Nielen M., Koch G., Bouma A., De Jong M.C.M., Stegeman A. Back-calculation method shows that within-flock transmission of highly pathogenic avian influenza (H7N7) virus in the Netherlands is not influenced by housing risk factors. Prev. Vet. Med. 2009;88:278–285. doi: 10.1016/j.prevetmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Bouma A., Claassen I., Natih K., Klinkenberg D., Donnelly C.A., Koch G., van Boven M. Estimation of transmission parameters of H5N1 avian influenza virus in chickens. PLoS Pathog. 2009;5:e1000281. doi: 10.1371/journal.ppat.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce W.M., Sandrock C., Kreuder-Johnson C., Kelly T., Cardona C. Avian influenza viruses in wild birds: a moving target. Comp. Immunol. Microbiol. Infect. Dis. 2009;32:275–286. doi: 10.1016/j.cimid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Breban R., Drake J.M., Stallknecht D.E., Rohani P. The role of environmental transmission in recurrent avian influenza epidemics. PLoS Comput. Biol. 2009;5:e1000346. doi: 10.1371/journal.pcbi.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., Stallknecht D.E., Beck J.R., Suarez D.L., Swayne D.E. Susceptibility of north american ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Swayne D.E., Costa T., Stallknecht D.E. Experimental infection studies of avian influenza in wild birds as a complement to surveillance. In: Majumdar S.K., Brenner F.J., Huffman J.E., McLean R.G., Panah A.I., Pietrobon P.J., Keeler S.P., Shive S., editors. Pandemic Influenza Viruses: Science, Surveillance, and Public Health. The Pennsylvania Academy of Sciences; Easton, PA: 2011. pp. 176–200. [Google Scholar]
- Brown J.D., Berghaus R.D., Costa T.P., Poulson R., Carter D.L., Lebarbenchon C., Stallknecht D.E. Intestinal excretion of a wild bird-origin H3N8 low pathogenic avian influenza virus in mallards (Anas Platyrhynchos) J. Wildl. Dis. 2012;48:991–998. doi: 10.7589/2011-09-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., Poulson R., Carter D.L., Lebarbenchon C., Stallknecht D.E. Infectivity of avian influenza virus-positive field samples for mallards: what do our diagnostic results mean? J. Wildl. Dis. 2013;49:180–185. doi: 10.7589/2011-11-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V.L., Drake J.M., Stallknecht D.E., Brown J.D., Pedersen K., Rohani P. Dissecting a wildlife disease hotspot: the impact of multiple host species, environmental transmission and seasonality in migration, breeding and mortality. J. Roy. Soc. Interfaces. 2013;10(79):20120804. doi: 10.1098/rsif.2012.0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulaga L.L., Garber L., Senne D., Myers T.J., Good R., Wainwright S., Suarez D.L. Descriptive and surveillance studies of suppliers to New York and New Jersey retail live-bird markets. Avian Dis. 2003;47:1169–1176. doi: 10.1637/0005-2086-47.s3.1169. [DOI] [PubMed] [Google Scholar]
- Cardona C.J. Low-pathogenicity avian influenza A outbreaks in commercial poultry in California. In: Knobler S.L., Mack A., Mahmoud A., Lemon S.M., editors. The Threat of Pandemic Influenza: Are We Ready? National Academies Press; Washington, DC: 2005. pp. 243–253. [Google Scholar]
- Clark L., Hall J. Avian influenza in wild birds: status as reservoirs, and risks to humans and agriculture. Ornithol. Monogr. 2006;60:3–29. [Google Scholar]
- Coluccy J.M., Yerkes T., Simpson R., Simpson J.W., Armstrong L., Davis J. Population dynamics of breeding mallards in the Great Lakes States. J. Wildl. Manage. 2008;72:1181–1187. [Google Scholar]
- Costa T.P., Brown J.D., Howerth E.W., Stallknecht D.E. Effect of a prior exposure to a low pathogenic avian influenza virus in the outcome of a heterosubtypic low pathogenic avian influenza infection in mallards (Anas platyrhynchos) Avian Dis. 2010;54:1286–1291. doi: 10.1637/9480-072210-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T.P., Brown J.D., Howerth E.W., Stallknecht D.E. The effect of age on avian influenza viral shedding in mallards (Anas platyrhynchos) Avian Dis. 2010;54:581–585. doi: 10.1637/8692-031309-ResNote.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I., Nagar S., Haddas R., Ben-Shabat M., Golender N., Lapin E., Altory A., Simanov L., Ribshtein I., Panshin A., Perk S. Avian influenza virus H9N2 survival at different temperatures and pHs. Avian Dis. 2010;54:725–728. doi: 10.1637/8736-032509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Davison S., Galligan D., Eckert T.E., Ziegler A.F., Eckroade R.J. Economic analysis of an outbreak of avian influenza, 1997–1998. J. Am. Vet. Med. Assoc. 1999;214:1164–1167. [PubMed] [Google Scholar]
- DeLiberto T.J., Swafford S.R., Nolte D.L., Pedersen K., Lutman M.W., Schmit B.B., Baroch J.A., Kohler D.J., Franklin A. Surveillance for highly pathogenic avian influenza in wild birds in the USA. Integr. Zool. 2009;4:426–439. doi: 10.1111/j.1749-4877.2009.00180.x. [DOI] [PubMed] [Google Scholar]
- Dorea F.C., Vieira A.R., Hofacre C., Waldrip D., Cole D.J. Stochastic model of the potential spread of highly pathogenic avian influenza from an infected commercial broiler operation in Georgia. Avian Dis. 2010;54:713–719. doi: 10.1637/8706-031609-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Dorigatti I., Mulatti P., Rosa R., Pugliese A., Busani L. Modelling the spatial spread of H7N1 avian influenza virus among poultry farms in Italy. Epidemics. 2010;2:29–35. doi: 10.1016/j.epidem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Drilling N., Titman R., McKinney F. Mallard (Anas platyrhynchos). In: Poole A., editor. The Birds of North America Cornell Lab of Ornithology; Ithaca; 2002. [Google Scholar]
- Duan L., Campitelli L., Fan X.H., Leung Y.H., Vijaykrishna D., Zhang J.X., Donatelli I., Delogu M., Li K.S., Foni E., Chiapponi C., Wu W.L., Kai H., Webster R.G., Shortridge K.F., Peiris J.S., Smith G.J., Chen H., Guan Y. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 2007;81:7529–7539. doi: 10.1128/JVI.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery R.B., Howerter D.W., Armstrong L.M., Anderson M.G., Devries J.H., Joynt B.L. Seasonal variation in waterfowl nesting success and its relation to cover management in the Canadian prairies. J. Wildl. Manage. 2005;69:1181–1193. [Google Scholar]
- Farnsworth M.L., Miller R.S., Pedersen K., Lutman M.W., Swafford S.R., Riggs P.D., Webb C.T. Environmental and demographic determinants of avian influenza viruses in waterfowl across the contiguous United States. PLoS One. 2012;7:e32729. doi: 10.1371/journal.pone.0032729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feare C.J. The role of wild birds in the spread of HPAI H5N1. Avian Dis. 2007;51:440–447. doi: 10.1637/7575-040106R1.1. [DOI] [PubMed] [Google Scholar]
- Fereidouni S.R., Starick E., Beer M., Wilking H., Kalthoff D., Grund C., Hauslaigner R., Breithaupt A., Lange E., Harder T.C. Highly pathogenic avian influenza virus infection of mallards with homo- and heterosubtypic immunity induced by low pathogenic avian influenza viruses. PLoS One. 2009;4:e6706. doi: 10.1371/journal.pone.0006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.B. In: Center N.W.R., editor. Ecology of Emerging Viral and Bacterial Diseases in Wildlife; Fort Collins, CO; 2008. [Google Scholar]
- Fries A.C., Nolting J.M., Danner A., Webster R.G., Bowman A.S., Krauss S., Slemons R.D. Evidence for the circulation and inter-hemispheric movement of the H14 subtype influenza A virus. PLoS One. 2013;8:e59216. doi: 10.1371/journal.pone.0059216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidet N., Cappelle J., Takekawa J.Y., Prosser D.J., Iverson S.A., Douglas D.C., Perry W.M., Mundkur T., Newman S.H. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: dispersal ranges and rates determined from large-scale satellite telemetry. J. Appl. Ecol. 2010;47:1147–1157. [Google Scholar]
- Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G.F., Wu G., Wang Y., Yuan Z., Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Garber L. In: Poultry’04 Part III: Reference of management practices in live-poultry markets in the United States, 2004. Nahms A., editor. US Department of Agriculture; 2006. [Google Scholar]
- Garber L., Hill G., Rodriguez J., Gregory G., Voelker L. Non-commercial poultry industries: surveys of backyard and gamefowl breeder flocks in the United States. Prev. Vet. Med. 2007;80:120–128. doi: 10.1016/j.prevetmed.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Garner M.G., Beckett S.D. Modelling the spread of foot-and-mouth disease in Australia. Aust. Vet. J. 2005;83:758–766. doi: 10.1111/j.1751-0813.2005.tb11589.x. [DOI] [PubMed] [Google Scholar]
- Gauthier-Clerc M., Lebarbenchon C., Thomas F. Recent expansion of highly pathogenic avian influenza H5N1: a critical review. IBIS. 2007;149:202–214. [Google Scholar]
- Giannecchini S., Clausi V., Di Trani L., Falcone E., Terregino C., Toffan A., Cilloni F., Matrosovich M., Gambaryan A.S., Bovin N.V., Delogu M., Capua I., Donatelli I., Azzi A. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology. 2010;408:167–173. doi: 10.1016/j.virol.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Gilbert M., Newman S.H., Takekawa J.Y., Loth L., Biradar C., Prosser D.J., Balachandran S., Rao M.V.S., Mundkur T., Yan B. Flying over an infected landscape: distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. EcoHealth. 2010;7:448–458. doi: 10.1007/s10393-010-0672-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goekjian V.H., Smith J.T., Howell D.L., Senne D.A., Swayne D.E., Stallknecht D.E. Avian influenza viruses and avian paramyxoviruses in wintering and breeding waterfowl populations in North Carolina, USA. J. Wildl. Dis. 2011;47:240–245. doi: 10.7589/0090-3558-47.1.240. [DOI] [PubMed] [Google Scholar]
- Gonzales J.L., Elbers A.R.W., Bouma A., Koch G., de Wit J.J., Stegeman J.A. Transmission characteristics of low pathogenic avian influenza virus of H7N7 and H5N7 subtypes in layer chickens. Vet. Microbiol. 2012;155:207–213. doi: 10.1016/j.vetmic.2011.09.016. [DOI] [PubMed] [Google Scholar]
- Greenwood R.J., Sargeant A.B., Johnson D.H., Cowardin L.M., Shaffer T.L. Factors associated with duck nest success in the prairie pothole region of Canada. Wildl. Monogr. 1995:1–57. [Google Scholar]
- Grund C., Abdelwhab el S.M., Arafa A.S., Ziller M., Hassan M.K., Aly M.M., Hafez H.M., Harder T.C., Beer M. Highly pathogenic avian influenza virus H5N1 from Egypt escapes vaccine-induced immunity but confers clinical protection against a heterologous clade 2.2.1 Egyptian isolate. Vaccine. 2011;29:5567–5573. doi: 10.1016/j.vaccine.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Halvorson D.A. Twenty-five years of avian influenza in Minnesota. Proceedings of the 53rd North Central Avian Disease Conference; Minneapolis, USA; 2002. pp. 65–69. [Google Scholar]
- Halvorson D.A. Avian Diseases, Fourth International Symposium on Avian Influenza, 1997, Proceedings, vol. 47. 2003. Strengths and weakness of vaccines as a control tool; pp. 223–227. (special issue) [Google Scholar]
- Halvorson D.A. Prevention and management of avian influenza outbreaks: experiences from the United States of America. Rev. Sci. Tech. OIE. 2009;28:359–369. doi: 10.20506/rst.28.1.1866. [DOI] [PubMed] [Google Scholar]
- Halvorson D.A., Frame D.D., Friendshuh K.A.J., Shaw D.P. Outbreaks of low pathogenicity avian influenza in U.S.A. Avian Dis. 2003;47:36–46. [Google Scholar]
- Handel A., Brown J., Stallknecht D., Rohani P. A Multi-scale analysis of influenza a virus fitness trade-offs due to temperature-dependent virus persistence. PLoS Comput. Biol. 2013;9:e1002989. doi: 10.1371/journal.pcbi.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B.A., Stallknecht D.E., Swayne D.E., Lewis L.A., Senne D.A. Avian influenza viruses in Minnesota ducks during 1998–2000. Avian Dis. 2003;47:867–871. doi: 10.1637/0005-2086-47.s3.867. [DOI] [PubMed] [Google Scholar]
- Harvey N., Reeves A., Schoenbaum M.A., Zagmutt-Vergara F.J., Dub C., Hill A.E., Corso B.A., McNab W.B., Cartwright C.I., Salman M.D. The North American Animal Disease Spread Model: a simulation model to assist decision making in evaluating animal disease incursions. Prev. Vet. Med. 2007;82:176–197. doi: 10.1016/j.prevetmed.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Heesterbeek J.A., Roberts M.G. The type-reproduction number T in models for infectious disease control. Math. Biosci. 2007;206:3–10. doi: 10.1016/j.mbs.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Henaux V., Samuel M.D. Avian influenza shedding patterns in waterfowl: implications for surveillance, environmental transmission, and disease spread. J. Wildl. Dis. 2011;47:566–578. doi: 10.7589/0090-3558-47.3.566. [DOI] [PubMed] [Google Scholar]
- Hinshaw V.S., Webster R.G., Turner B. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 1980;26:622–629. doi: 10.1139/m80-108. [DOI] [PubMed] [Google Scholar]
- Isanhart J.P., Wu H.M., Pandher K., MacRae R.K., Cox S.B., Hooper M.J. Behavioral, clinical, and pathological characterization of acid metalliferous water toxicity in mallards. Arch. Environ. Contam. Toxicol. 2011;61:653–667. doi: 10.1007/s00244-011-9657-z. [DOI] [PubMed] [Google Scholar]
- Jackwood M.W., Suarez D.L., Hilt D., Pantin-Jackwood M.J., Spackman E., Woolcock P., Cardona C. Biologic characterization of chicken-derived H6N2 low pathogenic avian influenza viruses in chickens and ducks. Avian Dis. 2010;54:120–125. doi: 10.1637/8987-070909-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Jewell C.P., Roberts G.O. Enhancing Bayesian risk prediction for epidemics using contact tracing. Biostatistics. 2012;13:567–579. doi: 10.1093/biostatistics/kxs012. [DOI] [PubMed] [Google Scholar]
- Johnson F.A., Moore C.T., Kendall W.L., Dubovsky J.A., Caithamer D.F., Kelley J.R., Williams B.K. Uncertainty and the management of mallard harvests. J. Wildl. Manage. 1997;61:202–216. [Google Scholar]
- Jourdain E., Gunnarsson G., Wahlgren J., Latorre-Margalef N., Brojer C., Sahlin S., Svensson L., Waldenstrom J., Lundkvist A., Olsen B. Influenza virus in a natural host, the mallard: experimental infection data. PLoS One. 2010;5:e8935. doi: 10.1371/journal.pone.0008935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T., Fujisaki S., Takashita E., Xu H., Yamada S., Uchida Y., Neumann G., Saito T., Kawaoka Y., Tashiro M. Genetic analysis of novel avian A (H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18:15. [PMC free article] [PubMed] [Google Scholar]
- Karsten S., Rave G., Krieter J. Monte Carlo simulation of classical swine fever epidemics and control. I. General concepts and description of the model. Vet. Microbiol. 2005;108:187–198. doi: 10.1016/j.vetmic.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Karsten S., Rave G., Krieter J. Monte Carlo simulation of classical swine fever epidemics and control. II. Validation of the model. Vet. Microbiol. 2005;108:199–205. doi: 10.1016/j.vetmic.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Keawcharoen J., van Riel D., van Amerongen G., Bestebroer T., Beyer W.E., van Lavieren R., Osterhaus A.D.M.E., Foucheir R.A.M., Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg. Infect. Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Yanagawa R., Matsuoka Y. Duck influenza lacking evidence of disease signs and immune-response. Infect. Immun. 1980;30:547–553. doi: 10.1128/iai.30.2.547-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick A.M., Chmura A.A., Gibbons D.W., Fleischer R.C., Marra P.P., Daszak P. Predicting the global spread of H5N1 avian influenza. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A.V., Pearce J.M., Flint P.L., Franson J.C., Ip H.S. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta) Mol. Ecol. 2008;17:4754–4762. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- Krauss S., Walker D., Pryor S.P., Niles L., Chenghong L., Hinshaw V.S., Webster R.G. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 2004;4:177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- Ladman B.S., Driscoll C.P., Pope C.R., Slemons R.D., Gelb J., Jr. Potential of low pathogenicity avian influenza viruses of wild bird origin to establish experimental infections in turkeys and chickens. Avian Dis. 2010;54:1091–1094. doi: 10.1637/9228-010410-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Lebarbenchon C., Sreevatsan S., Lefevre T., Yang M., Ramakrishnan M.A., Brown J.D., Stallknecht D.E. Reassortant influenza A viruses in wild duck populations: effects on viral shedding and persistence in water. Proc. R. Soc. B: Biol. Sci. 2012;279:3967–3975. doi: 10.1098/rspb.2012.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Suarez D.L., Tumpey T.M., Sung H.W., Kwon Y.K., Lee Y.J., Choi J.G., Joh S.J., Kim M.C., Lee E.K., Park J.M., Lu X., Katz J.M., Spackman E., Swayne D.E., Kim J.H. Characterization of highly pathogenic H5N1 avian influenza A viruses isolated from South Korea. J. Virol. 2005;79:3692–3702. doi: 10.1128/JVI.79.6.3692-3702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y.H.C., Zhang L.J., Chow C.K., Tsang C.L., Ng C.F., Wong C.K., Guan Y., Peiris J.S.M. Poultry drinking water used for avian influenza surveillance. Emerg. Infect. Dis. 2007;13:1380–1382. doi: 10.3201/eid1309.070517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W., Bi Y., Wu Y., Li X., Yan J. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith J.O., George D., Pepin K.M., Pitzer V.E., Pulliam J.R., Dobson A.P., Hudson P.J., Grenfell B.T. Epidemic dynamics at the human–animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiani B., Reddy S.M. The history of avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009;32:311–323. doi: 10.1016/j.cimid.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Makarova N., Kaverin N., Krauss S., Senne D., Webster R. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J. Gen. Virol. 1999;80:3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- Malladi S., Weaver J.T., Clouse T.L., Bjork K.E., Trampel D.W. Moving-average trigger for early detection of rapidly increasing mortality in caged table-egg layers. Avian Dis. 2011;55:603–610. doi: 10.1637/9636-122910-Reg.1. [DOI] [PubMed] [Google Scholar]
- Malladi S., Weaver J.T., Goldsmith T., Hueston W., Voss S., Funk J., Der C., Bjork K.E., Clouse T.L., Hennessey M., Sampedro F., Lee B., Halvorson D.A. The impact of holding time on the likelihood of moving internally contaminated eggs from a highly pathogenic avian influenza infected but undetected commercial table-egg layer flock. Avian Dis. 2012;56:897–904. doi: 10.1637/10191-041012-Reg.1. [DOI] [PubMed] [Google Scholar]
- McQuiston J.H., Garber L.P., Porter-Spalding B.A., Hahn J.W., Pierson F.W., Wainwright S.H., Senne D.A., Brignole T.J., Akey B.L., Holt T.J. Evaluation of risk factors for the spread of low pathogenicity H7N2 avian influenza virus among commercial poultry farms. J. Am. Vet. Med. 2005;226:767–772. doi: 10.2460/javma.2005.226.767. [DOI] [PubMed] [Google Scholar]
- Miller R.S., Sweeney S.J., Akkina J.E., Saito E.K. U.S. Department of Agriculture; Fort Collins, CO: 2013. Assessment of Introduction Pathway for Novel Avian Influenza Virus into North America by Wild Birds From Eurasia; p. 46. [Google Scholar]
- Miller R.S., Sweeney S.J., Akkina J.E., Saito E.K. In: Assessment of Introduction Pathway for Novel Avian Influenza Virus into North America by Wild Birds. USDA:APHIS:VS:CEAH, editor. USDA; Fort Collins, CO: 2013. [Google Scholar]
- Morales A.C., Jr., Hilt D.A., Williams S.M., Pantin-Jackwood M.J., Suarez D.L., Spackman E., Stallknecht D.E., Jackwood M.W. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis. 2009;53:552–562. doi: 10.1637/8877-041509-Reg.1. [DOI] [PubMed] [Google Scholar]
- Mullaney R. Live-bird market closure activities in the northeastern United States. Avian Dis. 2003;47:1096–1098. doi: 10.1637/0005-2086-47.s3.1096. [DOI] [PubMed] [Google Scholar]
- Mundt E., Gay L., Jones L., Saavedra G., Tompkins S.M., Tripp R.A. Replication and pathogenesis associated with H5N1, H5N2, and H5N3 low-pathogenic avian influenza virus infection in chickens and ducks. Arch. Virol. 2009;154:1241–1248. doi: 10.1007/s00705-009-0437-2. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Baas C., Lexmond P., Waldenstrom J., Wallensten A., Fransson T., Rimmelzwaan G.F., Beyer W.E., Schutten M., Olsen B., Osterhaus A.D., Fouchier R.A. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAADSM technical papers (http://www.naadsm.org/documentation/techpapers).
- Nazir J., Haumacher R., Ike A., Stumpf P., Bohm R., Marschang R.E. Long-term study on tenacity of avian influenza viruses in water (distilled water, normal saline, and surface water) at different temperatures. Avian Dis. 2010;54:720–724. doi: 10.1637/8754-033109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Olsen B., Munster V.J., Wallensten A., Waldenstrom J., Osterhaus A.D., Fouchier R.A. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Swayne D.E. Pathobiology of Asian highly pathogenic avian influenza H5N1 virus infections in ducks. Avian Dis. 2007;50:250–259. doi: 10.1637/7710-090606R.1. [DOI] [PubMed] [Google Scholar]
- Park A.W., Wood J.L., Daly J.M., Newton J.R., Glass K., Henley W., Mumford J.A., Grenfell B.T. The effects of strain heterology on the epidemiology of equine influenza in a vaccinated population. Proc. R. Soc. B: Biol. Sci. 2004;271:1547–1555. doi: 10.1098/rspb.2004.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patyk K.A., Helm J., Martin M.K., Forde-Folle K.N., Olea-Popelka F.J., Hokanson J.E., Fingerlin T., Reeves A. An epidemiologic simulation model of the spread and control of highly pathogenic avian influenza (H5N1) among commercial and backyard poultry flocks in South Carolina, United States. Prev. Vet. Med. 2013;110:510–524. doi: 10.1016/j.prevetmed.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Pedersen K., Swafford S.R., DeLiberto T.J. Low pathogenicity avian influenza subtypes isolated from wild birds in the United States, 2006–2008. Avian Dis. 2010;54:405–410. doi: 10.1637/8693-031309-Reg.1. [DOI] [PubMed] [Google Scholar]
- Pelzel A.M., McCluskey B.J., Scott A.E. Review of the highly pathogenic avian influenza outbreak in Texas, 2004. J. Am. Vet. Med. Assoc. 2006;228:1869–1875. doi: 10.2460/javma.228.12.1869. [DOI] [PubMed] [Google Scholar]
- Pepin K.M., VanDalen K.K., Mooers N.L., Ellis J.W., Sullivan H.J., Root J.J., Webb C.T., Franklin A.B., Shriner S.A. Quantification of heterosubtypic immunity between avian influenza subtypes H3N8 and H4N6 in multiple avian host species. J. Gen. Virol. 2012;93:2575–2583. doi: 10.1099/vir.0.045427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K.M., Wang J., Webb C.T., Smith G.J., Poss M., Hudson P.J., Hong W., Zhu H., Riley S., Guan Y. Multiannual patterns of influenza A transmission in Chinese live bird market systems. Influenza Other Respir. Viruses. 2013;7:97–107. doi: 10.1111/j.1750-2659.2012.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.E., Swayne D.E. Pathogenicity of a Hong Kong-origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Dis. 2002;46:53–63. doi: 10.1637/0005-2086(2002)046[0053:POAHKO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Pillai S.P., Pantin-Jackwood M., Suarez D.L., Saif Y.M., Lee C.W. Pathobiological characterization of low-pathogenicity H5 avian influenza viruses of diverse origins in chickens, ducks and turkeys. Arch. Virol. 2010;155:1439–1451. doi: 10.1007/s00705-010-0727-8. [DOI] [PubMed] [Google Scholar]
- Prosser D.J., Takekawa J.Y., Newman S.H., Yan B., Douglas D.C., Hou Y., Xing Z., Zhang D., Li T., Li Y. Satellite-marked waterfowl reveal migratory connection between H5N1 outbreak areas in China and Mongolia. IBIS. 2009;151:568–576. [Google Scholar]
- Reeves, A., Talbert, M., Salman, M.D., Hill, A.E. Technical document available on-line. Development of a stochastic, individual-based modeling framework for within-unit transmission of highly infectious animal diseases. http://www.naadsm.org/naadsm/files/wh/whDescription-draft121001.pdf.
- Reperant L.A., van de Bildt M.W., van Amerongen G., Buehler D.M., Osterhaus A.D., Jenni-Eiermann S., Piersma T., Kuiken T. Highly pathogenic avian influenza virus H5N1 infection in a long-distance migrant shorebird under migratory and non-migratory states. PLoS One. 2011;6:e27814. doi: 10.1371/journal.pone.0027814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S. Large-scale spatial-transmission models of infectious disease. Science. 2007;316:1298–1301. doi: 10.1126/science.1134695. [DOI] [PubMed] [Google Scholar]
- Rohani P., Breban R., Stallknecht D.E., Drake J.M. Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10365–10369. doi: 10.1073/pnas.0809026106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz R.A., Essen S.C., Brookes S.M., Iqbal M., Wood J.L., Grenfell B.T., McCauley J.W., Brown I.H., Gog J.R. Quantifying transmission of highly pathogenic and low pathogenicity H7N1 avian influenza in Turkeys. PLoS One. 2012;7:e45059. doi: 10.1371/journal.pone.0045059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S.L., Kingsford C., Cattoli G., Spiro D.J., Janies D.A., Aly M.M., Brown I.H., Couacy-Hymann E., De Mia G.M., Dung D.H. Genome analysis linking recent European and African influenza (H5N1) viruses. Emerg. Infect. Dis. 2007;13:713. doi: 10.3201/eid1305.070013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill N.J., St Rose S.G., Keeling M.J., Woolhouse M.E.J. Silent spread of H5N1 in vaccinated poultry. Nature. 2006;442:757. doi: 10.1038/442757a. [DOI] [PubMed] [Google Scholar]
- Savill N.J., St Rose S.G., Woolhouse M.E. Detection of mortality clusters associated with highly pathogenic avian influenza in poultry: a theoretical analysis. J. R. Soc. Interface. 2008;5:1409–1419. doi: 10.1098/rsif.2008.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbaum M.A., Terry Disney W. Modeling alternative mitigation strategies for a hypothetical outbreak of foot-and-mouth disease in the United States. Prev. Vet. Med. 2003;58:25–52. doi: 10.1016/s0167-5877(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Sharkey K.J., Bowers R.G., Morgan K.L., Robinson S.E., Christley R.M. Epidemiological consequences of an incursion of highly pathogenic H5N1 avian influenza into the British poultry flock. Proc. Biol. Sci. 2008;275:19–28. doi: 10.1098/rspb.2007.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemons R.D., Easterday B.C. Virus replication in the digestive tract of ducks exposed by aerosol to type-A influenza. Avian Dis. 1978;22:367–377. [PubMed] [Google Scholar]
- Slota K.E., Hill A.E., Keefe T.J., Bowen R.A., Miller R.S., Pabilonia K.L. Human-bird interactions in the United States upland gamebird industry and the potential for zoonotic disease transmission. Vector Borne Zoonotic Dis. 2011;11:1115–1123. doi: 10.1089/vbz.2010.0114. [DOI] [PubMed] [Google Scholar]
- Smith E.I., Reif J.S., Hill A.E., Slota K.E., Miller R.S., Bjork K.E., Pabilonia K.L. Epidemiologic characterization of Colorado backyard bird flocks. Avian Dis. 2012;56:263–271. doi: 10.1637/9865-072811-Reg.1. [DOI] [PubMed] [Google Scholar]
- Spackman E., Senne D.A., Davison S., Suarez D.L. Sequence analysis of recent H7 avian influenza viruses associated with three different outbreaks in commercial poultry in the United States. J. Virol. 2003;77:13399–13402. doi: 10.1128/JVI.77.24.13399-13402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E., Pantin-Jackwood M.J., Swayne D.E., Suarez D.L. An evaluation of avian influenza diagnostic methods with domestic duck specimens. Avian Dis. 2009;53:276–280. doi: 10.1637/8520-111708-Reg.1. [DOI] [PubMed] [Google Scholar]
- Spackman E., Gelb J., Jr., Preskenis L.A., Ladman B.S., Pope C.R., Pantin-Jackwood M.J., McKinley E.T. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol. J. 2010;7:331. doi: 10.1186/1743-422X-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E., Pedersen J.C., McKinley E.T., Gelb J., Jr. Optimal specimen collection and transport methods for the detection of avian influenza virus and Newcastle disease virus. BMC Vet. Res. 2013;9:35. doi: 10.1186/1746-6148-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spekreijse D., Bouma A., Koch G., Stegeman J.A. Airborne transmission of a highly pathogenic avian influenza virus strain H5N1 between groups of chickens quantified in an experimental setting. Vet. Microbiol. 2011;152:88–95. doi: 10.1016/j.vetmic.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Spekreijse D., Bouma A., Koch G., Stegeman A. Quantification of dust-borne transmission of highly pathogenic avian influenza virus between chickens. Influenza Other Respir. Viruses. 2013;7:132–138. doi: 10.1111/j.1750-2659.2012.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht D.E., Shane S.M. Host range of avian influenza-virus in free-living birds. Vet. Res. Commun. 1988;12:125–141. doi: 10.1007/BF00362792. [DOI] [PubMed] [Google Scholar]
- Stallknecht D.E., Shane S.M. Host range of avian influenza virus in free-living birds. Vet. Res. Commun. 1988;12:125–141. doi: 10.1007/BF00362792. [DOI] [PubMed] [Google Scholar]
- Stallknecht D.E., Brown J.D. Ecology of avian influenza in wild birds. In: Swayne D.E., editor. Avian Influenza. Blackwell Publishing; Ames, IA: 2008. pp. 43–58. [Google Scholar]
- Stallknecht D.E., Shane S.M., Zwank P.J., Senne D.A., Kearney M.T. Avian influenza viruses from migratory and resident ducks of coastal Louisiana. Avian Dis. 1990;34:398–405. [PubMed] [Google Scholar]
- Stevenson M.A., Sanson R.L., Stern M.W., O’Leary B.D., Sujau M., Moles-Benfell N., Morris R.S. InterSpread Plus: a spatial and stochastic simulation model of disease in animal populations. Prev. Vet. Med. 2013;109:10–24. doi: 10.1016/j.prevetmed.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Sturm-Ramirez K.M., Ellis T., Bousfield B., Bissett L., Dyrting K., Rehg J.E., Poon L., Guan Y., Peiris M., Webster R.G. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J. Virol. 2004;78:4892–4901. doi: 10.1128/JVI.78.9.4892-4901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm-Ramirez K.M., Hulse-Post D.J., Govorkova E.A., Humberd J., Seiler P., Puthavathana P., Buranathai C., Nguyen T.D., Chaisingh A., Long H.T., Naipospos T.S., Chen H., Ellis T.M., Guan Y., Peiris J.S., Webster R.G. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 2007;51:242–249. doi: 10.1637/7763-110706-REGR.1. [DOI] [PubMed] [Google Scholar]
- Swayne D.E. High pathogenicity avian influenza in the Americas. In: Swayne D.E., editor. Avian Influenza. Blackwell Publishing; Ames, IA: 2008. [Google Scholar]
- Swayne D.E., Akey B.L. Avian influenza control strategies in the United States of America. In: Koch G., editor. Wageningen Frontis International Workshop on Avian Influenza Prevention and Control; Dordrecht; 2005. pp. 113–130. [Google Scholar]
- Swayne D.E., Pantin-Jackwood M. Pathogenicity of avian influenza viruses in poultry. Dev. Biol. (Basel) 2006;124:61–67. [PubMed] [Google Scholar]
- Swayne D.E., Slemons R.D. Using mean infectious dose of high- and low-pathogenicity avian influenza viruses originating from wild duck and poultry as one measure of infectivity and adaptation to poultry. Avian Dis. 2008;52:455–460. doi: 10.1637/8229-012508-Reg.1. [DOI] [PubMed] [Google Scholar]
- Swayne D.E., Perdue M.L., Garcia M., Rivera-Cruz E., Brugh M. Pathogenicity and diagnosis of H5N2 Mexican avian influenza viruses in chickens. Avian Dis. 1997;41:335–346. [PubMed] [Google Scholar]
- Takekawa J.Y., Newman S.H., Xiao X., Prosser D.J., Spragens K.A., Palm E.C., Yan B., Li T., Lei F., Zhao D. Migration of waterfowl in the East Asian Flyway and spatial relationship to HPAI H5N1 outbreaks. Avian Dis. 2010;54:466–476. doi: 10.1637/8914-043009-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiensin T., Nielen M., Vernooij H., Songserm T., Kalpravidh W., Chotiprasatintara S., Chaisingh A., Wongkasemjit S., Chanachai K., Thanapongtham W., Srisuvan T., Stegeman A. Transmission of the highly pathogenic avian influenza virus H5N1 within flocks during the 2004 epidemic in Thailand. J. Infect. Dis. 2007;196:1679–1684. doi: 10.1086/522007. [DOI] [PubMed] [Google Scholar]
- Tildesley M.J., Deardon R., Savill N.J., Bessell P.R., Brooks S.P., Woolhouse M.E.J., Grenfell B.T., Keeling M.J. Accuracy of models for the 2001 foot-and-mouth epidemic. Proc. R. Soc. B: Biol. Sci. 2008;275:1459–1468. doi: 10.1098/rspb.2008.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tildesley M.J., House T.A., Bruhn M.C., Curry R.J., O’Neil M., Allpress J.L.E., Smith G., Keeling M.J. Impact of spatial clustering on disease transmission and optimal control. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1041–1046. doi: 10.1073/pnas.0909047107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trock S.C., Huntley J.P. Surveillance and control of avian Influenza in the New York live bird markets. Avian Dis. 2010;54:340–344. doi: 10.1637/8728-032409-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Trock S.C., Senne D.A., Gaeta M., Gonzalez A., Lucio B. Low-pathogenicity avian influenza virus in live bird markets – what about the livestock area? Avian Dis. 2003;47:1111–1113. doi: 10.1637/0005-2086-47.s3.1111. [DOI] [PubMed] [Google Scholar]
- USDA In: APHIS:VS:CEAH, N.A.M.S., editor. Part II: Reference of 1999 table egg layer management in the US; Fort Collins, CO; 2000. [Google Scholar]
- USDA In: USDA:APHIS:VS C., National Animal Health Monitoring System, editors. Part I: Reference of Health and Management of Backyard/Small Production Flocks in the United States, 2004; Fort Collins, CO; 2005. [Google Scholar]
- USDA . Census of Agriculture; 2007. National Agricultural Statistics Service, 2007.http://www.agcensus.usda.gov [Google Scholar]
- USDA . 2008. National Agricultural Statistics Service. http://www.nass.usda.gov/About_NASS/History_of_Ag_Statistics/index.asp. [Google Scholar]
- USDA, 2008b. Prevention and control of H5 and H7 low pathogenicity avian influenza in the live bird marketing system. In: Uniform Standards for the Live Bird Marketing System, APHIS:VS D. 91-55-076.
- USDA In: APHIS:VS:CEAH:NAHMS, editor. Structure of the U.S. Poultry Industry, 2010; Fort Collins, CO; 2011. [Google Scholar]
- USDA Foreign animal disease preparedness & response plan: highly pathogenic avian influenza response plan – the red book. National Center for Animal Health Emergence Management is the group within APHIS:VS that writes this guide; Riverdale, MD; 2012. [Google Scholar]
- USFWS In: Service F.A.W., editor. Adaptive Harvest Management, 2012 Hunting Season; Laurel, MD; 2012. p. 58. [Google Scholar]
- USFWS In: Service F.A.W., editor. Waterfowl Population Status; Laurel, MD; 2012. p. 79. [Google Scholar]
- VanDalen K.K., Franklin A.B., Mooers N.L., Sullivan H.J., Shriner S.A. Shedding light on avian influenza H4N6 infection in mallards: modes of transmission and implications for surveillance. PLoS One. 2010;5:e12851. doi: 10.1371/journal.pone.0012851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot J.A., Koch G., de Jong M.C.M., van Boven M. Transmission dynamics of low- and high-pathogenicity A/Chicken/Pennsylvania/83 avian influenza viruses. Avian Dis. 2003;47:939–941. doi: 10.1637/0005-2086-47.s3.939. [DOI] [PubMed] [Google Scholar]
- Wahlgren J., Waldenström J., Sahlin S., Haemig P., Fouchier R., Osterhaus A., Pinhassi J., Bonnedahl J., Pisareva M., Grudinin M. Gene segment reassortment between American and Asian lineages of avian influenza virus from waterfowl in the Beringia area. Vector Borne Zoonotic Dis. 2008;8:783–790. doi: 10.1089/vbz.2007.0274. [DOI] [PubMed] [Google Scholar]
- Weaver J.T., Malladi S., Goldsmith T.J., Hueston W., Hennessey M., Lee B., Voss S., Funk J., Der C., Bjork K.E., Clouse T.L., Halvorson D.A. Impact of virus strain characteristics on early detection of highly pathogenic avian influenza infection in commercial table-egg layer flocks and implications for outbreak control. Avian Dis. 2012;56:905–912. doi: 10.1637/10189-041012-Reg.1. [DOI] [PubMed] [Google Scholar]
- Webster R.G., Yakhno M., Hinshaw V.S., Bean W.J., Murti K.G. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox B.R., Knutsen G.A., Berdeen J., Goekjian V., Poulson R., Goyal S., Sreevatsan S., Cardona C., Berghaus R.D., Swayne D.E., Yabsley M.J., Stallknecht D.E. Influenza-A viruses in ducks in northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One. 2011;6:e24010. doi: 10.1371/journal.pone.0024010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K., Gibson D.D. The Asia-to-America influx of avian influenza wild bird hosts is large. Avian Dis. 2010;54:477–482. doi: 10.1637/8741-032509-Reg.1. [DOI] [PubMed] [Google Scholar]
- Wu J.T., Riley S., Fraser C., Leung G.M. Reducing the impact of the next influenza pandemic using household-based public health interventions. PLoS Med. 2006;3:1532–1540. doi: 10.1371/journal.pmed.0030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee K.S., Carpenter T.E., Mize S., Cardona C.J. The live bird market system and low-pathogenic avian influenza prevention in Southern California. Avian Dis. 2008;52:348–352. doi: 10.1637/8138-101207-Reg.1. [DOI] [PubMed] [Google Scholar]
- Yee K.S., Carpenter T.E., Farver T.B., Cardona C.J. An evaluation of transmission routes for low pathogenicity avian influenza virus among chickens sold in live bird markets. Virology. 2009;394:19–27. doi: 10.1016/j.virol.2009.08.017. [DOI] [PubMed] [Google Scholar]