Abstract
We have recently reported the three dimensional structure of the McpS chemoreceptor sensor domain in complex with its cognate ligands. The domain was characterized by a bimodular architecture, where ligand binding to each module caused a chemotactic response. This is a novel small molecule binding domain, which, however, is un-annotated in relevant databases. We report here the domain signature of the family of McpS-like sensor domains, which was termed helical bimodular (HBM) domain. The HBM domain was identified in Bacteria and Archaea and forms part of chemoreceptors and histidine kinases. The conservation of amino acids in the ligand binding sites of both modules suggests that HBM family members recognize similar ligands.
Keywords: domain profile, chemotaxis receptor, histidine kinase, sequence analysis
Introduction
Bacteria respond to changing environmental conditions through various signal transduction mechanisms. Genome analyses suggest that responses are primarily mediated by one-component systems, two-component systems, and chemoreceptor-based signaling pathways.1 These different systems share sensor domains for signal recognition. Chemoreceptors are typically transmembrane proteins and the molecular stimulus caused by ligand binding to the sensor domain is transmitted across the membrane causing a modulation of the Chemotaxis protein A (CheA) autophosphorylation activity and consequently transphosphorylation activity to the Chemotaxis protein Y (CheY).2 Chemosensory systems mediate flagellum-mediated chemotaxis, but are also involved in Type IV pili mediated taxis or in the regulation of alternative cellular processes.3,4
The study of chemosensory pathways from different organisms has revealed a high degree of diversity.4,5 This diversity is also reflected in the architecture of chemoreceptors, which were found to differ in their topologies and in the type of sensor domains.6 Chemoreceptor sensor domains can be classified according to their size: cluster I sensor domains are of approximately 150 amino acids whereas cluster II domains harbor approximately 250 amino acids.6 Cluster I domains were identified as PAS, GAF, CHASE, or TarH domains.6 In contrast, around 40% of chemoreceptors possess larger cluster II domains which are mostly un-annotated and in cases an annotation is available it was found to be unreliable.6
The methyl-accepting chemotaxis protein S (McpS) of Pseudomonas putida binds and mediates chemotaxis toward six Krebs cycle intermediates, butyrate, and acetate.7,8 McpS has a cluster II sensor domain and its secondary structure prediction (two long and four short helices) was incompatible with structures of known sensor domains.7 We have recently reported the 3D structure of the McpS sensor domain which is composed of two structural modules each composed of a 4-helix bundle.8 Interestingly, malate and succinate were bound to the membrane proximal module whereas acetate was present on the membrane distal module. In a superimposition of both modules the ligand binding sites were found to overlap. Interestingly, ligand binding to each module causes a chemotactic response and we have proposed a molecular mechanism that is based on a piston-like movement of the final, long helix H6 that forms part of both ligand binding sites. Although the 3D structure of McpS is composed of two 4-helical bundles, its sequence is not recognized by the corresponding domain signature 4HB_MCP.9 We report here the generation of the domain signature of the family McpS-like sensor domains, which we have termed Helical Bi-Modular (HBM) domain.
Results and Discussion
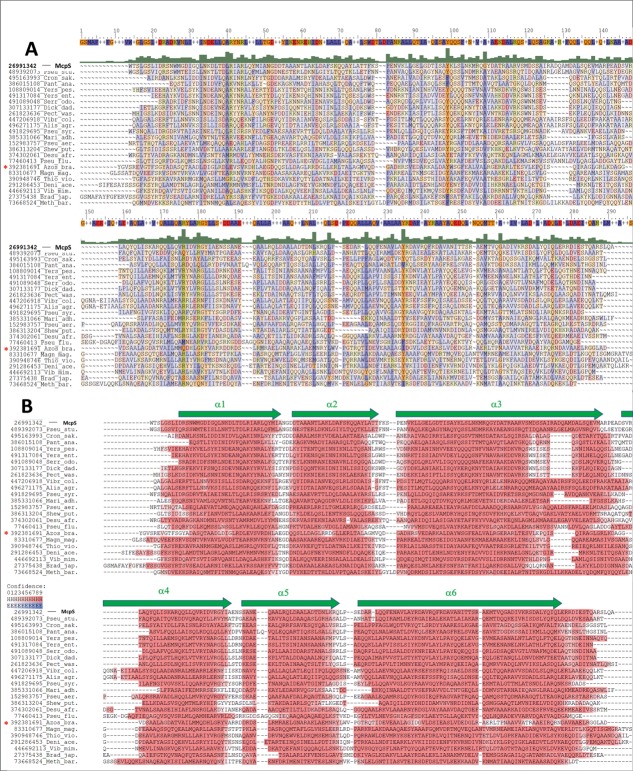
An alignment of a subset of HBM domain sequences representing the major phylogenetic categories is shown in Figure 1(A) and the corresponding secondary structure prediction is provided in Figure 1(B). The HBM domains are predicted to form two short helices followed by a long helix, another pair of small helices, and a final extended helix [Fig. 1(B)], which is in full agreement with the 3D structure of the McpS sensor domain [Fig. 2(A)].8 The profile Hidden Markov Model (HMM) generated as explained in the Materials and Methods section was then used to perform a search in the UniprotKB database, which resulted in the retrieval of the approximately 1200 sequences. The majority of proteins were chemoreceptors, but some proteins contained a Histidine kinase phosphoacceptor domain (HisKA, PF00512) and Histidine kinase like ATPase domain (HATPase_c, PF02518), indicating that HBM domains are also found in histidine kinases. The secondary structure prediction and domain size of HBM domains of histidine kinases was entirely comparable to those of chemoreceptors. The sensor domain of the TorS histidine kinase has a similar structure as compared to McpS,10 but has not been retrieved by the UniprotKB search. TorS binds the protein TorT in complex with the signal molecule. We show below that the conserved residues of the HBM domain are primarily in the two small ligand binding sites. The fact that TorS does not bind small signal molecules is responsible for the lacking sequence conservation which explains why it is not recognized by the HBM domain signature. Using BLAST we have retrieved homologues of the TorS sensor domain and constructed a domain signature, which, however, was incompatible with the HBM signature described here. This is consistent with the notion that there are two protein families, the HBM; domain and the TorS-like sensor domains, which do not share significant sequence similarities but possess similar structures. The family of TorS-like domains was found to be significantly less populated than the HBM family. A sequence alignment of the McpS and TorS sensor domain gave rise to an identity of less than 12%, which is a value very close to the random identity of two unrelated proteins.
Figure 1.
Alignment of a representative subset of HBM domain sequences from chemoreceptors and a sensor kinase. The asterisk marks the sensor kinase, the remaining sequences are from chemoreceptors. Sequences were selected to cover the phylogenetic distribution of species with HBM domains. The GI numbers from the NCBI database are indicated. Alis_agr: Alishewanella agri; Azos_lip: Azospirillum lipoferum; Brad_jap; Bradyrhizobium japonicum; Cron_sak: Cronobacter sakazakii; Deni_ace: Denitrovibrio acetiphilus; Desu_afr: Desulfovibrio africanus; Dick_dad: Dickeya dadantii; Magn_mag: Magnetospirillum magneticum; Mari_adh: Marinobacter adhaerens; Meth_bar: Methanosarcina barkeri; Pan_ana: Pantoea ananas; Pect_was: Pectobacterium wasabiae; Pseu_aer: Pseudomonas aeruginosa; Pseu_flu: Pseudomonas fluorescens; Pseu_stu: Pseudomonas stutzeri; Pseu_syr: Pseudomonas syringae; Serr_odo: Serratia odorifera; Shew_put: Shewanella putrefasciens;Thio_vio: Thiocystis violascens; Vibr_cho: Vibrio cholerae; Vibr_mim: Vibrio mimicus;Yers_ent: Yersinia enterocolitica; Yers_pes: Yersinia pestis (A). ClustalX alignment highlighting conserved residues: blue for hydrophobic residues (ACFILMPVW), red for acidic residues (DE), green-yellow for basic amino acids (HKR) and orange for the remaining residues (GNQSTY). The consensus sequence is shown on top of the alignment. (B). Secondary structure prediction for the same set of sequences, red color indicates alpha-helical regions in ascending intensity according to confidence. Green arrows represent the position of the α helices in the 3D structure of the McpS sensor domain.
Figure 2.
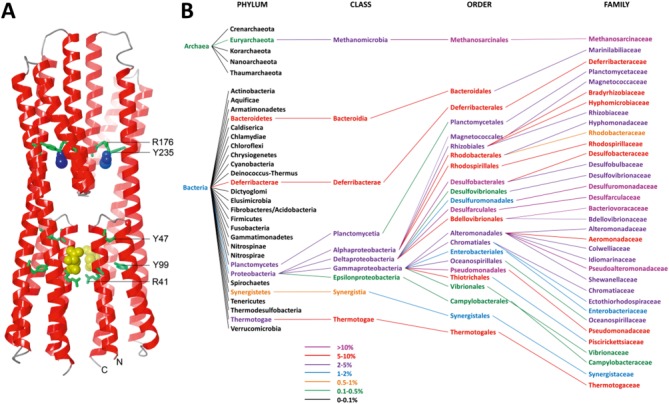
(A). Three dimensional structure of the McpS sensor domain. Bound malate and acetate are shown in yellow and blue, respectively. Conserved amino acids (numbering according to Fig. 1) are depicted in green. (B). Phylogenetic distribution of species with HBM domain containing chemoreceptors. Data were derived from the analysis of the complete set of chemoreceptor sequences that were used as seed for the generation of the HMM. Phyla shown in black contain chemoreceptors (protein that are detected by PS50111), but not those containing HBM domains. The color coding corresponds to the relative abundance of chemoreceptors with an HBM domain over the total number of chemoreceptors.
HBM domains are found in bacteria and archaea but are absent from eukaryotes. HBM domain containing chemoreceptors are primarily found in Proteobacteria (98%) and in particular in Alpha- (16%) and Gammaproteobacteria (76%) [Fig. 2(B)]. This may not be surprising since approximately 75% of all chemoreceptor sequences are found in this phylum. Although most of the remaining chemoreceptors are found in Firmicutes, HBM domains were not detected in this phylum. Apart from Proteobacteria HBM domain containing chemoreceptors were also detected in species like Methanosarcina or Methanolobus that belong to the phylum of Euryarchaeota of the Archaea superkingdom. In addition, receptors with the HBM domain are found in species that belong to the phyla Deferribacterae, Bacteroidia, Thermotogae, or Planctomycetia (≤1%).
Significant differences were identified in the relative abundance of HBM containing chemoreceptors which is illustrated by the color coding in Figure 2(B). HBM containing receptors were found to represent more than 10% of total receptors in a number of families including Desulfomonadaceae, Bacteriovoracaceae, and Pseudoalteromonadaceae and 5–10% of total chemoreceptors in orders like Rhodospirillales, Thiotrichales, and Pseudomonadales.
A major challenge in signal transduction reside in the identification of the signal molecules that are recognized by a given sensor domain. However, the sequence alignments derived from the selected subset of sequences (Fig. 1) as well as from the complete set of 1200 sequences provide interesting clues. We have noted the conservation of residues R41, Y47, and Y99 in the proximal bundle of the McpS sensor domain as well as R176 and Y235 of its membrane distal bundle (numbering according to Fig. 1). These residues are located close to the bound chemo-attractants in the structure of the McpS sensor domain [Fig. 2(A)] and establish in the case of Y235, R176, and R41 direct interactions with bound ligand. The involvement of some of these residues in ligand binding to McpS has been determined experimentally since the mutation of R41 (corresponding to R60 in McpS) abolished malate binding whereas mutation of R176 (R183 in McpS) reduced significantly acetate binding.8 The novelty of McpS sensor domain resides in its bimodular architecture and the fact that ligand binding at each module causes a response. The conservation of residues in both ligand binding sites suggests that the capacity to recognize ligands at the different modules is a characteristic common to the HBM family. McpS recognizes different carboxylic acids and the conservation of binding site residues, in particular the arginine residues, may also indicate that family members recognize similar type of ligands.
Materials and Methods
The sequence of the McpS sensor domain comprising residues 32–291 was used for PSI-BLAST11 searches in the database of non-redundant sequences (as of August 26, 2013). Several iterations were carried out until no further sequence was included into the subset with E-values lower than 0.005 (BLAST default value). After each iteration, the sequences were analyzed and only those with the expected sensor domain size (220–299 amino acids as defined for cluster II domains) were retained.
The sensor domain was defined as the protein fragment flanked by two transmembrane helices as predicted by the DAS algorithm.12 Sequences were selected according to the prediction of their secondary structure (predicted by PSIPRED)13 and only sequences were retained that were predicted to be exclusively composed of helix and turn. Each curated subset was employed for the subsequent iteration. Additional PSI-BLAST searches were done with sequences of low E-values and belonging to phylogenetically distant organisms and the resulting sequences found were included into the set. The final set of sequences was then manually inspected and curated to discard incomplete sequences. Finally, CD-Hit14 was used to eliminate redundancy (100% identical sequences).
The final set of more than 1200 sequences from a wide variety of Bacteria and Archaea was used to generate the domain signature. These sequences were aligned using the multiple sequence alignment (MSA) tool ClustalX15 and their secondary structure was predicted by PSIPRED through the Quick2D tool included in the MPI Bioinformatics Toolkit.16
A profile hidden Markov (profile HMM) model was generated using the HMMBUILD tool from HMMER3 software package17 implemented in the Mobyle portal.18 To test the specificity of this profile HMM, a search with the HMMSearch tool of HMMER was performed in the UniprotKB database19 that produced more than 1200 protein sequences that harbor the HBM domain. To determine the total number of chemoreceptors per bacterial species the UniProtKB knowledgebase was searched for sequences that match Prosite code PS50111. This information was then used to calculate the relative abundance of chemoreceptors with an HBM domain over the total number of receptors.
References
- 1.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lacal J, Garcia-Fontana C, Munoz-Martinez F, Ramos JL, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol. 2010;12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 7.Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos JL, Krell T. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem. 2010;285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pineda-Molina E, Reyes-Darias JA, Lacal J, Ramos JL, García-Ruiz JM, Gavira JA, Krell T. Evidence for chemoreceptors with bimodular ligand binding regions harboring two signal-binding sites. Proc. Acad. Natl. Sci. USA. 2012;109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich LE, Zhulin IB. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics. 2005;21:iii45–iii48. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore JO, Hendrickson WA. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure. 2012;20:729–741. doi: 10.1016/j.str.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 13.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 16.Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 18.Neron B, Menager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magrane M, Consortium tU. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011 doi: 10.1093/database/bar009. 2011:bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]