Abstract
Background
Hedgehog signaling plays an important role in embryonic development, organogenesis and cancer. In the adult liver, Hedgehog signaling in non-parenchymal cells has been found to play a role in certain disease states such as fibrosis and cirrhosis. However, whether the Hedgehog pathway is active in mature healthy hepatocytes and is of significance to liver function are controversial.
Findings
Two types of mice with distinct conditional hepatic deletion of the Smoothened gene, an essential co-receptor protein of the Hedgehog pathway, were generated for investigating the role of Hedgehog signaling in mature hepatocytes. The knockout animals (KO) were inconspicuous and healthy with no changes in serum transaminases, but showed a slower weight gain. The liver was smaller, but presented a normal architecture and cellular composition. By quantitative RT-PCR the downregulation of the expression of Indian hedgehog (Ihh) and the Gli3 transcription factor could be demonstrated in healthy mature hepatocytes from these mice, whereas Patched1 was upregulated. Strong alterations in gene expression were also observed for the IGF axis. While expression of Igf1 was downregulated, that of Igfbp1 was upregulated in the livers of both genders. Corresponding changes in the serum levels of both proteins could be detected by ELISA. By activating and inhibiting the transcriptional output of Hedgehog signaling in cultured hepatocytes through siRNAs against Ptch1 and Gli3, respectively, in combination with a ChIP assay evidence was collected indicating that Igf1 expression is directly dependent on the activator function of Gli3. In contrast, the mRNA level of Igfbp1 appears to be controlled through the repressor function of Gli3, while that of Igfbp2 and Igfbp3 did not change. Interestingly, body weight of the transgenic mice correlated well with IGF-I levels in both genders and also with IGFBP-1 levels in females, whereas it did not correlate with serum growth hormone levels.
Conclusions
Our results demonstrate for the first time that Hedgehog signaling is active in healthy mature mouse hepatocytes and that it has considerable importance for IGF-I homeostasis in the circulation. These findings may have various implications for mouse physiology including the regulation of body weight and size, glucose homeostasis and reproductive capacity.
Keywords: Gli3, Growth hormone, Hedgehog signaling, Hepatocytes, IGF-I, IGFBP-1, Liver, MotifMap, Patched1, Smoothened
Lay abstract
Background
The Hedgehog signaling pathway, like that of other morphogens, plays an important role in embryonic development, organogenesis and tissue homeostasis. In the adult stage, its activity is downregulated in most organs, but can be reactivated in physiological and pathological processes such as tissue regeneration and cancer. In the adult liver, Hedgehog signaling is lower in parenchymal cells, the hepatocytes, than in non-parenchymal cells where it has been found to play a fundamental role in certain disease states such as fibrosis and cirrhosis. Whether the Hedgehog pathway is active in mature healthy hepatocytes and is of significance to liver function are controversial.
Findings
In order to approach these questions we used two types of mice with distinct conditional hepatic deletion of the Smoothened gene, an essential co-receptor protein of the Hedgehog pathway. Knockout of this gene interrupted the Hedgehog pathway only in hepatocytes. The knockout mice were healthy, but showed a slower gain of body and liver weight. Alterations in the expression of Hedgehog signaling components revealed the functional activity of this pathway in the liver. Strong alterations in gene expression were also observed for insulin-like growth factor-I (IGF-I) the serum level of which correlated well with body weight.
Conclusions
Our results demonstrate for the first time that Hedgehog signaling is active in mature mouse hepatocytes and that it has considerable importance for IGF-I homeostasis in the circulation. These findings may have various implications for mouse physiology including the regulation of body weight and size, glucose homeostasis and reproductive capacity.
Background
Hedgehog (Hh) signaling is known to play an important role in embryonic development, organogenesis and cancer [1,2]. Secreted Hh ligands (Sonic hedgehog (SHH), Indian hedgehog (IHH) and Desert hedgehog (DHH)) influence on target cells via autocrine, paracrine or even endocrine-like ways. They bind to PTCH1 and PTCH2 receptors and alleviate Patched-mediated suppression of Smoothened (SMO), a putative seven-transmembrane protein with features of a G-protein coupled receptor [3,4]. Activated Smo induces a number of downstream reactions, finally leading to stabilization of Gli transcriptional factors, their translocation into nucleus and transcription of Hh-dependent genes. In vertebrates three Gli genes are present. GLI1 is acting predominantly as a transcriptional activator of Hh target genes, while GLI2 and GLI3 may act as both activators and repressors depending on Hh activity [5,6].
In adult tissues Hh signaling is commonly downregulated and may be reactivated only in certain types of cancer [7,8]. Like for other morphogens, however, Hh signaling in adult tissues frequently shows activity gradients that are involved in maintenance of tissue homeostasis. Well studied examples are the maintenance of the specific phenotype of gastric and intestinal epithelium [9,10] and the fate of adipose tissue [11,12]. In the adult liver, active Hh signaling has been observed in some non-parenchymal cells namely hepatic stellate cells (HSC) and cholangiocytes [13,14], where it plays a fundamental role in maintaining the normal phenotype of these cells but also during their activation in certain disease states such as fibrosis and cirrhosis [14]. Under diseased conditions also stressed or dying hepatocytes (e.g. when ballooning in severe liver steatosis) were described to secret high amounts of Shh which appears to act as a major signal for inducing alternative regenerative pathways via stem cell populations [15-17]. However, whether the Hh signaling pathway is active in healthy, mature hepatocytes and relevant to normal liver function remains controversial [13,18]. On the one hand, a significant contribution of Hh signaling to the regulation of liver function has been denied because of the low expression level of the respective signaling components in mature hepatocytes compared with certain types of liver cancer cells or even non-parenchymal cells such as HSC or cholangiocytes [13,14]. On the other hand, theoretical considerations based on analogy with other tissues considerably support the hypothesis that Hh signaling might indeed play a fundamental role in controlling liver function [18].
In order to clarify the important question of whether Hh signaling is active in healthy, mature hepatocytes, we have chosen two independent types of transgenic mice with conditional hepatocellular ablation of Smoothened, a bottleneck component of the Hh pathway [19], to interrupt Hh signaling in hepatocytes. These mice should be optimally suited for investigating the functional relevance, if any, of low Hh signaling in these cells. To exclude any aberrant findings due to damaged or immature hepatocytes we carefully checked the histological integrity of the livers after knockout of Smo including the integrity and phenotype of the hepatocytes in situ. A second method for gaining insight into the functional relevance of Hh signaling in hepatocytes was to use RNAi experiments in vitro for modulating (i. e. inhibiting or activating) Hh signaling and monitoring the altered transcriptional and functional output. For that purpose a high purity of the cultured hepatocytes was guaranteed. Using these approaches and various analytical techniques we were able to demonstrate for the first time to our knowledge that Hh signaling is active in mature hepatocytes and contributes to the regulation of IGF-I and IGFBP-1 protein serum levels in mice.
Results
Generation and phenotypic characteristics of Smo-KO mice
For investigating the influence of the Hedgehog signaling pathway on liver function, hepatocyte-specific ablation of Smo, a bottleneck component of Hh signaling [19], seemed a suitable approach. In the first type of transgenic mice, conditional ablation of Smo in liver epithelial cells was induced by mating Smoflox/flox and Alfp-Cre mouse lines (Additional file 1: Figure S1A-C). The resulting double transgenic homozygous Smo-/- mice are abbreviated as SAC-KO mice. Alfp-Cre mice [20] have been successfully used for liver-specific knockout studies [21]. In agreement with these studies, Cre-recombinase could be detected by immunohistochemistry in all liver epithelial cells (hepatocytes and cholangiocytes) (Additional file 1: Figure S1D), and only the recombined Smo alleles could be detected in the livers of homozygous mice after 12 weeks (Additional file 1: Figure S1E). As expected, significant loss of Smo expression was found in the liver and in isolated hepatocytes, and no changes were found in the heart, lung, spleen, adipose tissue or kidney (Additional file 1: Figure S1F).
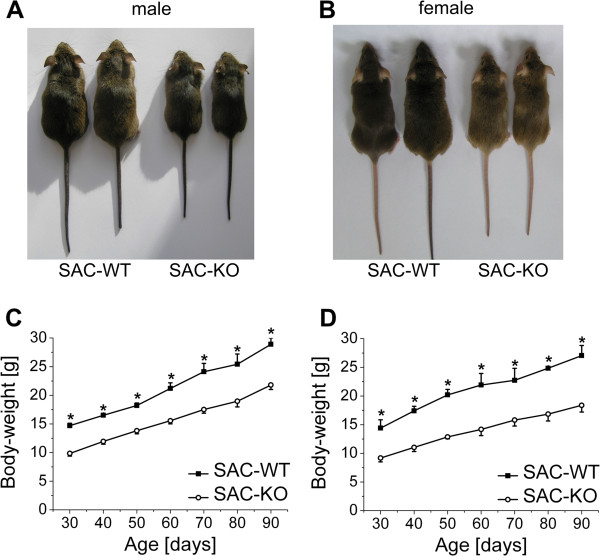
The first visible phenotype of the SAC-KO mice was evident immediately after birth; the mice were small in size compared to their WT littermates (Figure 1A, B). At 30 days after birth, when the offspring were separated from their dams, body weight was significantly reduced in both male and female homozygous SAC-KO mice compared to gender-matched WT mice (Figure 1C, D). Weight gain in the transgenic males paralleled that in WT males up to the age of 12 weeks, after which it appeared to progress at a slightly lower pace in transgenic female mice during the same period of time (Figure 1D). Interestingly, the weight of the heterozygous SAC-KO mice (Smo+/-) was significantly different from that of both, the WT littermates and the homozygous mice (Additional file 2: Figure S2A,B) indicating that the loss of each Smo allel caused a distinct drop of body weight. The age of 12 weeks was chosen for further investigations because the animals were fully mature at this time point, which allowed the proper isolation of the mature hepatocytes essential for this study.
Figure 1.
Phenotypic features induced by hepatocyte-specific ablation of Smoothened in transgenic SAC mice. Photographs of (A): male SAC mice and (B): female SAC mice show two WT mice (left) and two KO mice (right). Comparison of body weight of (C): male SAC-WT (black squares) (n = 4) and SAC-KO (open circles) (n = 5) mice and (D): female SAC-WT (black squares) (n = 3) and SAC-KO (open circles) (n = 4) mice. Values are presented as the means ± SEM; *, p < 0.05.
Generally, the SAC-KO mice were healthy and their behavior was inconspicuous. They showed slightly reduced blood glucose levels in the postprandial state and had significantly lower plasma insulin levels (Table 1).
Table 1.
Liver parameters and serum activities of male and female SAC-WT and SAC-KO mice
| Male mice | SAC-WT | n | SAC-KO | n | p value (MWU-test) |
|---|---|---|---|---|---|
| Liver weight (g) |
1.65 ± 0.09 |
6 |
0.95 ± 0.03 |
14 |
0.001** |
| Liver-body-weight ratio (%) |
5.58 ± 0.16 |
6 |
4.38 ± 0.14 |
14 |
0.001** |
| ALAT (μkat/l) |
1.64 ± 0.48 |
3 |
2.47 ± 0.37 |
4 |
0.228 |
| ASAT (μkat/l) |
8.07 ± 3.97 |
3 |
6.82 ± 1.83 |
4 |
1.000 |
| GLDH (μkat/l) |
0.35 ± 0.08 |
3 |
1.93 ± 0.58 |
4 |
0.057 |
| Insulin (pmol/l) |
160.53 ± 43.84 |
7 |
44.86 ± 11.97 |
5 |
0.0101* |
| Glucose (mMol/dl) |
8.12 ± 0.48 |
5 |
7.22 ± 0.40 |
7 |
0.34 |
|
Female mice |
|
|
|
|
|
| Liver weight (g) |
1.29 ± 0.06 |
17 |
1.10 ± 0.06 |
8 |
0.724 |
| Liver-body-weight ratio (%) |
5.82 ± 0.18 |
17 |
4.91 ± 0.16 |
8 |
0.001** |
| ALAT (μkat/l) |
1.48 ± 0.50 |
6 |
2.47 ± 0.37 |
6 |
0.228 |
| ASAT (μkat/l) |
4.80 ± 0.44 |
6 |
7.07 ± 1.10 |
6 |
1.000 |
| GLDH (μkat/l) |
0.54 ± 0.10 |
6 |
1.04 ± 0.28 |
6 |
0.057 |
| Insulin (pmol/l) |
98.24 ± 10.68 |
10 |
56.73 ± 5.89 |
8 |
0.0021** |
| Glucose (mMol/dl) | 7.19 ± 0.24 | 9 | 6.22 ± 0.40 | 10 | 0.079 |
Determination of body weight and liver parameters at the age of 12 weeks. Values represent means ± SEM; *, p < 0.05; **, p < 0.01.
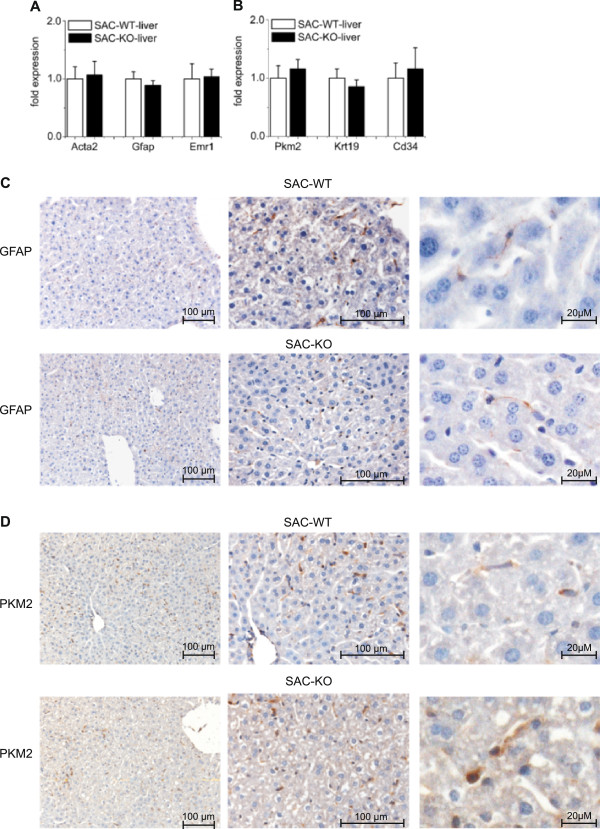
Histological examination of the liver revealed a normal lobular architecture, indicating that Smo ablation did not influence the correct development of this organ despite leading to a smaller size. Liver weight was significantly reduced by approx. 40% in male SAC-KO mice (Table 1), and the liver weight/body weight ratio dropped from 5.6% in WT mice to 4.4% in SAC-KO mice. In female mice, similar alterations were observed, except for a smaller difference in total liver weight (Table 1). Apart from the slower liver weight/body weight ratio compared to SAC-WT mice, there were no indications of overt liver damage in the SAC-KO mice in both genders. Accordingly, no significant differences could be observed in serum activities of ASAT, ALAT and GLDH between SAC-WT and SAC-KO mice (Table 1). Furthermore, comparing the expression levels of different marker genes for hepatic stellate cells (HSC) and myofibroblasts (Gfap, Acta2) or Kupffer cells (Emr1), in liver tissue from SAC-WT mice and SAC-KO mice revealed no major changes in the cellular composition (Figure 2A, B). This could be confirmed by immunohistochemistry as exemplified by the distribution of GFAP which was found only in HSC and cholangiocytes (Figure 2C). Only the general marker of non-parenchymal cells, PKM2 [22] seemed to be slightly induced in hepatocytes from SAC-KO mice (Figure 2D).
Figure 2.
Expression of markers for non-parenchymal cells in transgenic SAC mice. qRT-PCR analyses of (A): Acta2, Gfap and Emr1; (B): Pkm2, Krt19 and Cd34 in liver tissue of 12 weeks old male SAC-WT (white bars) (n = 13) and SAC-KO (black bars) (n = 11) mice. Values are presented as the means ± SEM. Immunohistochemistry in liver sections from male SAC-WT and SAC-KO mice of (C): GFAP (present only in HSC’s), (D): PKM2 (Is present in all non-parenchymal cell types. Hepatocytes in SAC-KO mice show a very light cytoplasmic staining). Bar: 100 μm and 20 μM.
Influence of Smoothened ablation on Hedgehog pathway components in hepatocytes in situ and ex vivo
To analyze hepatocellular Hh signaling, the expression of several components involved in the pathway was determined by qRT-PCR in freshly isolated hepatocytes (Figure 3). Purity of the hepatocyte preparation, prepared as described in Materials and Methods, was carefully checked using the markers for hepatocytes (Arg1, Gck, Gls2, Ldha, Krt8, Krt18) (Additional file 3: Figure S3A,B). For non-parenchymal cells like hepatic stellate cells, myofibroblasts and Kupffer cells we used the markers shown above. Furthermore we used markers for cholangiocytes (Krt19) and endothelial cells (Cd34) (Additional file 3: Figure S3C,D). In particular, contamination of hepatocyte preparations by cholangiocytes was excluded as described previously [23].
Figure 3.
Expression changes of Hedgehog signaling components. qRT-PCR analyses of hedgehog members in hepatocytes freshly isolated from 12-week-old male SAC-WT (white bars) (n = 8-17) and SAC-KO (black bars) (n = 8-13) mice: (A): Smo, Ihh and Shh; (B): Boc, Cdo and Ptch1; (C): Hhip1, Fu and Sufu, and the transcription factors (D): Gli1, Gli2 and Gli3. Values are presented as relative means ± SEM; *, p < 0.05, **, p < 0.01. Immunohistochemical staining of (E): Indian Hedgehog and (F): GLI3 in liver sections from male transgenic SAC-WT and SAC-KO mice at the age of 12 weeks. Scale bars: 100 μM; 50 μM and 25 μM.
Apart from the expected loss of Smo expression in hepatocytes, the mRNA level of Ihh was significantly downregulated, whereas Boc, Cdo and Ptch1 were significantly upregulated (Figure 3A, B). The expression levels of Shh, the inhibitor Hhip1 and components acting further downstream, such as Fused and Sufu, were not significantly affected (Figure 3C). Of the three members of the Gli family of zinc-finger transcription factors, Gli1 and Gli3 were strongly downregulated (p < 0.05, n = 7), whereas Gli2 appeared to be less affected (Figure 3D). These results were comparable in female mice (not shown) and clearly indicate that Hh signaling is active in mature hepatocytes. These findings are in close agreement with those in other cells challenged by Smo knockout [24]. Notably, Ihh expression appears to be characteristic of healthy hepatocytes, whereas Shh is expressed by damaged hepatocytes [16].
By immunohistochemistry it was possible to demonstrate that indeed hepatocytes in situ stained positive for IHH and GLI3 protein in the cytoplasm and the nuclei, respectively (Figure 3E, F). Interestingly, staining for IHH showed a pronounced pericentral distribution. In livers of SAC-KO mice both proteins were downregulated resulting in a weaker cytoplasmic staining (IHH) or a loss of nuclear staining (GLI3) (Figure 3F).
Ablation of Smo in hepatocytes alters serum levels of proteins of the IGF axis
Hepatocytes are the major source of circulating IGF-I in many species including mice [25]. Because we have recently hypothesized that IGF-I might be a possible target of Hh signaling in these cells [18], we set out to evaluate the expression of IGF-I and other members of the IGF axis in hepatocytes freshly isolated from WT and SAC-KO mice.
The expression levels of Igf1 mRNA and Igfbp1 mRNA were measured by qRT-PCR. As shown in Figure 4A and B, males and females demonstrated a significant downregulation of Igf1 by approximately 80% and 60%, respectively. Igfbp1 mRNA was upregulated by approximately 3-fold in males and 8-fold in females (Figure 4C, D). Neither Igfbp2 nor Igfbp3 changed significantly (Figure 4C, D). Determination of the levels of IGF-I protein in serum revealed strong downregulation in males and females (Figure 4E, F), whereas the upregulation of IGFBP-1 was significant only in female mice.
Figure 4.
Changes of IGF axis members in the liver and serum. Quantitative RT-PCR analyses of (A, B): Igf1 and (C, D): Igfbp1, Igfbp2 and Igfbp3 mRNA levels in hepatocytes isolated from 12-week-old male SAC-WT (white bars) (n = 7) and SAC-KO (black bars) (n = 7) (left diagrams) and female SAC-WT (white bars) (n = 7) and SAC-KO (black bars) (n = 5) (right diagrams) mice relative to that of β-actin. Values are presented as the means ± SEM; *, p < 0.05. (E, F): Serum analyses of circulating IGF-I and IGFBP-1 in male SAC-WT (white bars) (n = 9) and SAC-KO (black bars) (n = 8) mice and female SAC-WT (white bars) (n = 11) and SAC-KO (black bars) (n = 13) mice. Values are presented as the means ± SEM; *, p < 0.05.
Completely similar results were obtained with a second transgenic mouse model, the SLC-KO mice (Additional file 4: Figure S4). Deletion of Smo in these mice is inducible by transient exposure to Doxycycline at any desired age of the animals. When the Smo knockout was induced at 8 weeks of age, the SLC-KO mice show an immediate reduction in weight gain during the next 5 weeks, the same significant alterations of Gli3, Igf1 and Igfbp1 expression levels in isolated hepatocytes, and the corresponding changes in IGF-I and IGFBP-1 serum protein concentrations (Additional file 4: Figure S4). These findings convincingly demonstrate that the consequences of hepatocellular deletion of Smo are independent upon the specific mechanisms for conditional expression of Cre recombinase and other characteristics of the two types of transgenic mice.
Gli3 is a transcriptional activator of Igf1
To gain insight into the mechanism through which Hh signaling might control the expression of Igf1 and Igfbp1, RNA interference experiments were performed in cultured hepatocytes from C75BL/6-N mice. Because Gli1 and Gli3 were significantly down-regulated in SAC-KO mice, we wanted to know which Gli-factor is the predominant one responding immediately to the loss of Smo. As shown in Figure 5A, downregulation of Smo by Smo siRNA resulted in the significant decrease of Gli3 mRNA level within 48 h, while that of Gli1 was decreased only by trend at this time. Therefore, we focused on GLI3 in subsequent in vitro experiments. First, we asked whether the downregulation of Gli3 might be sufficient to account for the observed changes in the expression of Igf1. As expected, transfection of cultured hepatocytes with Gli3 siRNA depleted the Gli3 mRNA level by 80% (Figure 5C). The decrease in Gli3 expression was paralleled by a significant decrease in Igf1 mRNA (Figure 5C), which was in perfect agreement with the results obtained in SAC-KO mice. Moreover, the knockdown lead to a significant decrease in IGF-I protein determined by ELISA in the culture medium after 72 h (Figure 5D).
Figure 5.
RNA interference, ELISA and ChIP experiments. (A): Relative expression of Smo, Gli1, Gli2 and Gli3 determined by qRT-PCR analyses in cultured hepatocytes of male C57BL/6-N mice in response to transfection of Smo siRNA (black bars) (n = 8) compared to nonsense transfection (white bars) (n = 8) after 48 h of incubation. Values are presented as relative means ± SEM; *, p < 0.05. (B): Relative expression of Ptch1, Gli3, Igf1 and Igfbp1 determined by qRT-PCR analyses in cultured hepatocytes of male C57BL/6-N mice in response to transfection with Ptch1 siRNA (black bars) compared to nonsense transfection (white bars) after 48 h (n = 10-12) and 72 h (n = 5-6) of incubation. Values are presented as relative means ± SEM; *, p < 0.05; **, p < 0.01; ***, p < 0.001. (C): Relative expression of Gli3 determined by qRT-PCR analyses in cultured hepatocytes of male C57BL/6-N mice in response to transfection of Gli3 siRNA (black bars) (n = 11) compared to nonsense transfection (white bars) (n = 11) after 72 h of incubation and relative expression of Igf1 and Igfbp1 in C57BL/6 N-hepatocytes after 72 h of incubation with Gli3 siRNA (black bars) (n = 8) compared to nonsense transfection (white bars) (n = 8). Values are presented as relative means ± SEM; *, p < 0.05; **, p < 0.01. (D): Supernatant analyses of secreted IGF-I and IGFBP-1 in cultured hepatocytes of male C57BL/6-N mice in response to transfection of Gli3 siRNA (black bars) (n = 6) compared to nonsense transfection (white bars) (n = 6) after 72 h of incubation. Values are presented as the means ± SEM; *, p < 0.05. (E): GLI3 binding site at -405 bp of the Igf1 promoter according to MotifMap. (F): qRT-PCR analyses of precipitated DNA. DNA immunoprecipitated with GLI3 Antibody (black), Histone H3 Antibody as positive control (grey), and IgG as a negative control (white) calculated relative to input.
Second, we were interested in whether the upregulation of Hh signaling causes the opposite regulatory response by the Igf1 gene. In line with other studies [19], the siRNA-mediated downregulation of Ptch1 gene expression was chosen to activate Hh signaling. Hepatocyte transfection with Ptch1 siRNA resulted in significant and selective downregulation of Ptch1 expression and significant upregulation of Gli3 after 48 h (Figure 5B), reflecting the known activation of the Hh pathway through the loss of Ptch1[26]. In addition, upregulation of the pathway was also indicated by the upregulation of Gli1, Gli2 and Hhip1 (Additional file 5: Figure S5B,D). Other Hh components (e.g. Smo, Ihh, Shh, Fu and Sufu) showed no changes in expression, when the pathway was activated via Ptch1 siRNA (Additional file 5: Figure S5A,C). When Igf1 expression was evaluated in these experiments, a significant upregulation of Igf1 mRNA levels was observed that persisted till 72 h (Figure 5B). Together, these results suggest that Igf1 is a direct transcriptional target of the activator function of GLI3. Data mining by MotifMap [27] provides three different possible binding sites for Gli3 within the range of -5000 bp upstream to 5000 bp downstream of the transcription start site (Additional file 6: Table S5). One of these located at -405 bp upstream with the sequence TGTGTGGTC shows the most promising features (Figure 5E) and, thus, was selected for a chromatin immunoprecipitation (ChIP) analysis. As shown in Figure 5F GLI3 binding to the selected sequence could definitively be proven by the ChIP assay. Both, positive and negative controls showed the expected results.
Collectively, our results from hepatocytes ex vivo (SAC-KO mice) and in vitro clearly show that the activity of the Hh signaling pathway in the liver is a major determinant of circulating IGF-I levels. To date, hepatic Igf1 expression is considered to be primarily regulated by growth hormone via STAT5 [28,29]. Interestingly, in our SAC mice no correlation was found between the serum levels of growth hormone (GH) and IGF-I (correlation coefficient = 0.2488; p = 0.4124) indicating that GH is not the dominant regulator of IGF-I serum levels. Whether Hh signaling acts independently of or interferes with GH signaling in hepatocytes remains to be established.
Gli3 represses the expression of Igfbp1 but does not affect that of Igfbp2 and 3
In contrast to Igf1, the mRNA level of Igfbp1 was upregulated in livers of SAC-KO mice (Figure 4C). Therefore, we wanted to know whether siRNA-mediated knockdown of Gli3 in cultured hepatocytes also results in an inverse response of Igfbp1 mRNA in vitro. Indeed, an inverse picture was found, characterized by the selective upregulation of Igfbp1 expression in response to Gli3 knockdown (Figure 5C). Likewise, the concentration of IGFBP-1 protein secreted into the culture medium increased significantly after a 72 h cultivation period (Figure 5D).
Conversely, activation of the Hh pathway by Ptch1 knockdown resulted in significant downregulation of Igfbp1 mRNA within 72 h (Figure 5B). Apparently, the response here is slightly delayed, because at 48 h after Ptch1 knockdown the effect is seen only by trend. Again, there is a nice correspondence to the presence of several binding sites for GLI3 in the Igfbp1 promoter region (Additional file 6: Table S5). In contrast, modulation of Hh signaling in vivo or of Gli3 activity in vitro did not show any response at all on the mRNA level of Igfbp2 and 3, for which no GLI3 binding sites could be identified using MotifMap (data not shown).
Discussion
The results obtained with newly generated transgenic mice with hepatocyte-specific conditional knockout of Smo reported herein replenished by RNAi experiments using primary hepatocyte cultures demonstrate for the first time that Hedgehog signaling is active in healthy mature liver parenchymal cells and has considerable impact for liver function, and – via modulating liver-derived endocrine factors – for the entire organism. Even though we agree with published data [30] that the levels of Hh signaling components are low in hepatocytes, complete ablation of Smo at early or late stages of development as in both, the SAC-KO and the SLC-KO mice, leads to several characteristic phenotypic alterations: a) an obvious growth retardation resulting in lower body size and weight gain, b) a reduced liver/body weight ratio, and c) altered levels of production of IGFI and IGFBP-1 by the liver. Furthermore, changes in the expression of some Hh components, particularly of Patched1, were noted indicating the expected response to the interruption of the Hh pathway. Likewise, the Gli transcription factors Gli1 and Gli3 were significantly downregulated in vivo at the mRNA level. The fact that knockdown of Smo in vitro resulted only in significant downregulation of Gli3 mRNA is compatible with the well-known finding that Gli1 is a target gene of GLI3 [31]. These findings provide a first proof of principle for our recent hypothesis concerning the existence of a measurable activity of Hh signaling in mature hepatocytes [18].
Since Hh signaling in hepatocytes is currently considered to be mainly associated with hepatocellular damage and resulting regenerative phenomena [17], we carefully investigated molecular markers for damage and precursors of hepatocytes, but found no overt signs of cell death, of activation or relative shifts of non-parenchymal cell populations, and of other changes of liver architecture in the transgenic mice. Thus, there is certainly an effective Hh signaling that is not associated with hepatocyte toxicity and/or replacement. Instead, zonation of the liver seemed to be affected which is of utmost importance for liver function [32,33]. For example, the production of IHH was found to be restricted to pericentral hepatocytes in control livers and was downregulated in SAC-KO livers to an even smaller pericentral area (c.f. Figure 3E). These findings are in line with reports that IHH is a target gene of Wnt/beta-catenin signaling [34] and indicate that the knockout of Smo affects Wnt/beta-catenin signaling as predicted recently [18]. Details of this crosstalk between the two morphogen signaling pathways are currently investigated.
Another major result of this study is the unequivocal evidence that hepatocellular Hh signaling is involved in controlling the IGF-I axis in the mouse. The observed decreased levels of Igf1 mRNA in hepatocytes and of IGF-I protein in serum are in line with the fact that liver-derived IGF-I is the principle source of this hormone in blood [35]. Mechanistically, our results from RNAi experiments on cultured hepatocytes in vitro encompassing downregulation of Hh signaling by Gli siRNAs and, conversely, upregulation by Ptch1 siRNA strongly suggest that Hh signaling acts via the activator function of GLI3 in the transcriptional control of Igf1. Indeed, two active GLI3 binding sites in the promoter region of the Igf1 gene have already been identified by McMahon and co-workers [36], while data mining by MotifMap [27] provides even three different possible binding sites within the range of -5000 bp upstream to 5000 bp downstream of the transcription start site (Additional file 6: Table S5). The sequence TGTGTGGTC located at -405 bp upstream of the transcription start site which we selected for a ChIP analysis nicely fits with the consensus sequence for GLI3 binding sites (Figure 5E). Binding of GLI3 to this sequence could definitively be proven by the ChIP assay, while both positive and negative controls showed the expected results (Figure 5F).
The contribution of Hh signaling to Igf1 transcription in liver and to the control of serum levels of IGF-I is a novel finding reported for the first time. Hitherto, transcription of the Igf1 gene was thought to be mainly controlled by growth hormone mediated by dispersed STAT5b-binding elements [28,29]. Our finding that in the SAC mice the IGF-I serum level does not correlate with growth hormone levels argues against an exclusive role of growth hormone. Thus, it seems an important challenge for endocrinologists to elucidate the interplay between these two regulatory mechanisms influencing IGF-I levels. In particular, it remains to be determined whether regulation of Igf1 expression by GLI3 affects its regulation by STAT5.
Further, we provide strong evidence that IGFBP-1 which is also mainly produced in the liver [37,38] is also influenced by hepatocyte Hh signaling via a Gli3-mediated mechanism. So far, IGFBP-1 is known to be inversely regulated by insulin [39]. Though plasma insulin levels are lower in SAC mice of both genders and, thus might contribute to the changes in IGFBP-1 in vivo, our in vitro results clearly show that the influence by Hh is independent of insulin. Since IGF-I and IGFBP-1 were found to be inversely regulated by Smo knockdown it is tempting to speculate that in the case of IGFBP-1 GLI3 may act in a repressive manner, since GLI binding sites were predicted in the promoter region of the IGFBP-1 gene (Additional file 6: Table S5). This would be compatible with the known fact that GLI3 has both repression and activation domains depending on the activation status [8]. Thus, GLI3 may suppress Igfbp1 expression following knockdown of Gli3 when GLI3 is truncated to the repressor form. However, the situation is not as clear as for the activating role in the case of IGF-I for two reasons: a) there are no suitable antibodies available that recognize solely the truncated repressor form of GLI3, and b) at present it cannot be excluded that GLI3 acts via repression of an as yet unknown activator of Igfbp1 expression (unpublished observation). Nonetheless, our results suggest that GLI3 is an important mediator in the regulation of Igf1 and Igfbp1 expression by Hh signaling in mouse hepatocytes.
Since IGF-I is long known as an important growth factor with high impact on skeletal growth and body size [40], it is tempting to ask whether the observed changes in the IGF-I axis found in the SAC-KO mice are responsible for the observed changes in body size and weight gain of these mice. Although there is general agreement that liver-derived IGF-I is the principle source of this hormone in blood, its role for postnatal growth in mice remains controversial [35,41,42]. Interestingly, the data obtained in this study shows a clear correlation between IGF-I levels in serum and body weight for male (correlation coefficient = 0.8422; p = 0.0003) and for female (correlation coefficient = 0.6814; p = 0.0026) mice (Additional file 7: Figure S6A,B). Likewise, the correlation between IGFBP-1 and body weight is highly significant also in female mice (correlation coefficient = -0.4834; p = 0.0493) confirming earlier results obtained with mice overexpressing IGFBP-1 [43]. To our surprise, however, there is no such correlation for GH in both genders (correlation coefficient = 0.3356; p = 0.2623 in males and correlation coefficient = 0.2747; p = 0.4136 in females) suggesting that the dominant factors for determining body weight and size in our mice are members of the IGF axis rather than GH.
Apart from contributing to body size, the physiological consequences of the modulation of the IGF axis in SAC-KO mice may be manifold. Liver-derived IGF-I was found to be important for normal carbohydrate and lipid metabolism [41]. Particularly, IGF-I contributes to maintenance of normal glucose homeostasis and is necessary for normal insulin sensitivity [44]. Likewise, IGFBP-1 is recognized as an important regulator of glucose levels and a potential marker for the metabolic syndrome [45]. Our findings that plasma insulin levels are decreased in SAC-KO mice, while refeed glucose levels were slightly decreased rather than increased generally reflect an important influence of hepatic Hh signaling on glucose homeostasis mediated, at least in part, by modulation of the IGF axis. However, since the effects of IGF-I and IGFBP-1 are opposite in nature [44,45], it is hard to draw conclusions about the individual contribution of these factors in the context of our transgenic mice.
Another important consequence of the altered hepatic Hh signaling may concern the reproductive capacity. Both, IGF-I and IGFBP-1 were found to interfere with reproductive functions particularly in females [46,47]. Most interestingly, low IGF-I serum levels and/or upregulated levels of IGFBP-1 are associated with considerably impaired fertilization or implantation [37]. Indeed female SAC-KO mice show a strongly reduced reproductive capacity in line with the effects ascribed to the alterations in the IGF-I axis rather than with GH. The anatomical and physiological aspects of this effect are currently investigated in detail.
Conclusions
In conclusion, the results of this study, obtained using different transgenic mice with conditional hepatic knockout of Smo, demonstrate for the first time that Hh signaling is active in healthy mature hepatocytes and has considerable impact on endocrine liver function. The consequences are manifold and comprise phenomena as different as body size and weight, glucose metabolism and homeostasis or female fertility.
Materials and methods
Generation of transgenic SAC mice
Transgenic mice with hepatocyte-specific knockout of Smoothend (Smo) were obtained by crossing Smoflox/flox and Alfp-Cre lines (Additional file 1: Figure S1A-C). After establishing the animals the general breading protocol was as follows: male mice: Smoflox/flox + Cre-recombinase mated with female Smoflox/flox - Cre recombinase. In agreement with the results of Kellendonk et al. [20], Cre-recombinase could be detected by immunohistochemistry in all hepatocytes and cholangiocytes (Additional file 1: Figure S1D).
Genotypes of the transgenic mice were identified by PCR using DNA from tailtips. After birth mice were screened for expressing the Smo wildtyp (Smo WT), the Smo floxed (Smo flx.) allele and the Cre-recombinase by PCR. All primers are listed in Additional file 6: Table S1.
Generation of transgenic SLC mice
Triple transgenic mice allowing conditional hepatocyte-specific ablation of Smo in response to exposure to Doxycycline (abbreviated SLC mice) were generated by crossing the Smotm2Amc/J mice (Jackson Laboratories) possessing loxP sites on either side of exon 1 of the Smo gene [48] with double transgenic LC-1/rTALAP -1 mice [49] (generously provided by Hermann Bujard). For inducing ablation of Smo, SLC mice were grown up for 8 weeks under normal conditions. Then, expression of Cre-recombinase was induced by adding Doxycycline hydrochloride (2 mg/ml) to the drinking water for a period of 10 days.
Maintenance of mice and feeding
Mice were maintained according to the German guidelines and local regulations for the care and safe use of experimental animals in a pathogen-free facility in a 12:12 h LD cycle (permission numbers: TVV11/08; N05/12; A19/11). Animals had free access to regular chow (ssniff® M-Z V1124-0 composed of 22.0% protein, 50.1% carbohydrate, 4.5% fat; usable energy: 13.7 kJ/g; ssniff® Spezialdiäten GmbH, Soest, Germany) and tap water throughout life. Before sacrifice (between 9 and 11 am), mice were starved for 24 h and re-fed with regular chow for 12 h, in order to obtain a synchronized feeding state.
Measurements in blood samples and cell culture-supernatant
Blood samples were taken from the beating heart of anesthesized mice. Serum activities of ASAT, ALAT, and GLDH were measured with an automated analyser (Roche modular) using standardized assays (Roche, Mannheim, Germany). Blood glucose levels were determined using a Blood Glucose Meter (Freestyle Mini, Abbot, Wiesbaden).
Insulin was detected in serum with an Elisa- Kit from DRG Instruments (Marburg, Germany) (EIA 3439). IGF-I, IGFBP-1 and GH, were detected in serum with Elisa-Kits from Mediagnost (Reutlingen, Germany and Cell Applications (San Diego, USA).
To determine IGF-I and IGFBP-I in cell culture supernatant we used Elisa-Kits from Boster Biological Technology (Fremont, CA, USA).
Isolation and cultivation of primary hepatocytes
Primary hepatocytes from SAC mice were isolated by collagenase perfusion of the liver as described [50]. Cell suspension was carefully cleared from non-parenchymal cells by differential centrifugation. Finally, hepatocytes were suspended in Williams Medium E containing 10% fetal calf serum and further additions as described [50,51], and were plated onto 12-well plates precoated with collagen type 1. After 2 h, serum-free medium was used throughout cultivation.
To give an overview of the purity of our primary hepatocyte isolation we looked for the expression of typical markers of mature hepatocytes like Arg1 (Arginase 1), Gck (Glucokinase), Gls2 (Glutaminase 2 (liver, mitochondrial)), Ldha (lactate dehydrogenase A chain), Krt18 (Keratin 18) and Krt8 (Keratin 8). For the non-parenchymal cell population (e.g. hepatic stellate cells, vascular smooth muscle cells kupffer cells and cholangiocytes) we analyzed Gfap (glial fibrillary acidic protein), Acta2 (actin, alpha 2, smooth muscle, aorta), Emr1 (EGF-like module containing, mucin-like, hormone receptor-like sequence 1), and Krt19 (Keratin 19). All genes were analyzed in SAC-WT and SAC-KO material (Additional file 3: Figure S3A,B,C,D).
RNA preparation and quantitative real-time PCR (qRT-PCR)
Total RNA from hepatocytes, liver tissue and other organs was extracted using TRIzol (peqlab, Erlangen, Germay). RNA was reverse transcribed using oligo(dt) primers and Proto Script M-MuLV First Strand cDNA Synthesis Kit (New England Biolabs, Frankfurt am Main, Germany). The resulting cDNA samples were then quantified for each test gene using target gene-specific primers designed using the online tools Universal Probe Library software, Perl Primer and Primer 3. Primers are listed in Additional file 6: Table S2. The levels of all mRNA transcripts were determined in duplicate by qRT-PCR using the Light Cycler® 2.0 Instrument and the Light Cycler® Fast Start DNA Masterplus SYBR Green I (Roche, Mannheim, Germany). Using the standard curve method, the absolute quantification of specific PCR products for each primer set was performed. For normalization, β-actin was amplified from each sample.
RNA-interference
Smo-, Ptch1- and Gli3-specific siRNAs and respective nonsense control siRNAs were purchased from Invitrogen, Darmstadt, Germany. Freshly isolated hepatocytes were seeded at a density of 100000 cells per well of 12-well plates in normal culture medium containing 10% FCS. After 4 h, serum-free medium was used and chemically synthesized siRNAs (10 nmol for Smo and Ptch1, 25 nmol for Gli3) were transfected with Interferin from peqlab, Erlangen, Germany, according to the manufacturer’s instructions. Sequences for siRNA primers are listen in Additional file 6: Table S3. Twenty-four hours after transfection, the medium was changed and fresh medium without siRNA was added. Changes in gene expression were analysed by qRT-PCR at 48 h and 72 h post-transfection.
Chromatin Immunoprecipitation (ChIP)
To determine the binding of the transcription factor GLI3 to the Igf1 promoter region we used the SimpleChIP Plus Enzymatic Chromatin IP Kit (Agarose Beads) (Cell Signaling, Erlangen, Germany) according to the manufacturer’s instructions. Freshly isolated hepatocytes from three male C57BL/6 mice were pooled, washed and cross-linked with 37% formaldehyde. As positive experimental control the Histone H3 (D2B12) XP® Rabbit mAb (#2729) was used whereas normal Rabbit IgG served as negative control. The cross-linked GLI3 DNA complex was precipitated with the goat anti mouse GLI3 antibody (20 μg) (R&D Wiesbaden-Nordenstadt, Germany). The quantification analysis was performed using qRT-PCR with 5 μl of each DNA sample and specific primers listed in Additional file 6: Table S4. Primer pairs for the putative GLI3 binding site in the Igf1 promoter region were designed using Primer-BLAST of NCBI [52]. The range of interest in the promoter region (~ -405 bp) was obtained by the MotifMap [27] analysis. The applicability of the primer pair was tested using the UCSC In silico PCR analysis (http://genome.csdb.cn/cgi-bin/hgPcr). The desired Igf1 amplicon resulted in a 283 bp fragment. The primers for β–actin were used to analyse the unspecific DNA-Antibody-binding. Rpl30 primers (included in the SimpleChIP Plus Enzymatic Chromatin IP Kit) were used as a positive control for the histone H3 antibody precipitation.
Immunohistochemistry
Immunohistochemistry on paraffin sections (3 μm) was performed similarly as described [53]. After dewaxing and rehydration sections for Cre-recombinase, IHH, GLI3 and PKM2 staining were microwaved (3 × 5 min) in citrate buffer (0.01 M sodium citrate, pH 6.0) for antigen retrieval. For GFAP staining the slides were incubated for 4 min with 0.1% Proteinase K and 0.1% Calcium for antigen retrieval. Endogenous peroxidase was destroyed by incubation with 3% hydrogen peroxide in TBS (0.01 M Tris-base, 0.9% NaCl, pH 7.6) for 10 min. To block unspecific binding, slides for Cre-Recombinase, GFAP, IHH and GLI3 staining were incubated for 30 min in 5% goat serum (Sigma, München, Germany) and slides for PKM2 staining were incubated for 30 min in 10% horse serum (Sigma, München, Germany). Primary antibodies were rabbit anti Cre-recombinase (1:4000; Abcam, Cambridge, UK), rabbit anti Indian Hedgehog (1:250, Abcam, Cambridge, UK), rabbit anti GLI3 (1:250, GeneTex, Irvine, USA), rabbit anti Gfap (1:1000, Dako, Hamburg, Germany) rabbit anti Pyruvate Kinase muscle (1:500, Biomol, Hamburg, Germany). To visualize IHH and GLI3 proteins the Dako REAL™ EnVision™ Detection System, Peroxidase/DAB+,Rabbit Mouse (Dako, Hamburg, Germany) was used according to the manufacturer’s protocol. Secondary antibody for Cre Recombinase, PKM2 and GFAP staining was biotinylated goat anti-rabbit IgG (Millipore, Schwalbach, Germany) used together with Extravidine (Sigma, München, Germany). Diaminobenzidine and Aminotriazole were used for POD staining as described [53]. Sections were counterstaining with hematoxylin.
Statistical analysis
Values are expressed as means ± standard deviation of the mean (SEM) and sample size (n) is given in each figure ore table. Statistical evaluation was performed using the Mann–Whitney-U-Test for fresh tissue and hepatocytes and the wilcoxon matched pairs test for the siRNA experiments in cultured hepatocytes. The null hypothesis was rejected at the p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***) levels. The Pearson correlation coefficients and the related p-values for Pearson’s correlation were calculated using the Statistics toolbox of MATLAB (The MathWorks Inc., Natick, MA, USA).
Availability of supporting data
The data sets supporting the results of this article are included within the article (and its additional files).
Abbreviations
Acta2: actin, alpha 2, smooth muscle, aorta; ALAT: Alanine aminotransferase; ASAT: Aspartate aminotransferase; Arg1: arginase 1; BBLS: Bayesian branch length score; BLS: Branch length score; Boc: brother of Cdo; Cd34: cd34 antigen; Cdo: cell adhesion molecule-related/downregulated by oncogenes; Dhh: dessert hedgehog; Emr1: EGF-like module containing, mucin-like, hormone receptor-like sequence 1; FDR: False discovery rate; Gck: glucokinase; GLDH: Glutamate dehydrogenase; Gls2: glutaminase 2; Gfap: glial fibrillary acidic protein; Gli1: glioma-associated oncogene family zinc finger 1; Gli2: glioma-associated oncogene family zinc finger 2; Gli3: glioma-associated oncogene family zinc finger 3; Hh: Hedgehog; IGFBP-1: Insulin-like growth factor binding protein 1; IGFBP-2: Insulin-like growth factor binding protein 2; IGFBP-3: Insulin-like growth factor binding protein 3; IGF-I: Insulin-like growth factor 1; Ihh: Indian hedgehog; Krt8: keratin8; Krt18: keratin18; Krt19: keratin 19; Ldha: lactate dehydrogenase A; NLOD: Normalized log-odds score; Pkm2: pyruvate kinase, muscle; Ptch1: patched1; Ptch2: patched2; Shh: sonic hedgehog; Smo: smoothened.
Competing interests
GR is listed as the inventor on a patent application filed by the University of Leipzig (PCT/EP2013/051425, Therapeutic use of activators of zinc finger protein GLI3).
Authors’ contributions
MSM designed and conducted most experiments, analyzed the data and wrote the manuscript. AS, BJ, KJ, AK and ME designed and conducted the experiments and analyzed the data. SHW analyzed the data. GR conceived the project, analyzed the data, and wrote and corrected the manuscript. All authors read and approved the final version of the manuscript.
Supplementary Material
Generation of the transgenic mouse model with hepatocyte-specific deletion of Smoothened. (A): Structure of the Alfp-Cre vector construct. (B): The Smo locus with loxP sites on either side of exon 1in the absence of Cre-recombinase activity. (C): The floxed Smo locus lacking exon 1 in the presence of Cre-recombinase activity. (D): Immunohistochemical detection of Cre-recombinase in liver sections of SAC-WT and SAC-KO mice. Brown colour indicating Cre-recombinase is present in hepatocyte and cholangiocyte nuclei (strong staining) and cytoplasm (weak staining). Bar: 50 μm. (E): PCR analysis, using DNA extracted from liver tissue of SAC-WT and SAC-KO mice yields a 600-bp amplicon of wild-type Smo in different tissues SAC-WT (black bars) (n = 7-20) and SAC-KO (white bars) (n = 7-20) mice determined by qRT-PCR. Significant decrease of Smo mRNA relative to β-actin is detected only in liver. Values are presented as means ± SEM; *, p<0.05.
Phenotypic features induced by hepatocyte-specific ablation of Smoothened in transgenic SAC mice. Photographs of (A): male SAC wildtype SAC-WT (Smo+/+), heterozygous SAC-KO (Smo+/-) and homozygous SAC-KO (Smo-/-) mouse. (B): Comparison of body weight of 4, 8 and 12 weeks old male SAC-WT (Smo+/+) (white bars) (n = 4-9), heterozygous SAC-KO (Smo+/-) (gray bars) (n = 8-9) and homozygous SAC-KO (Smo-/-) (black bars) (n = 8-21) mice. Values are presented as the means ± SEM; *, p<0.05; **, p<0.01; ***, p<0,001.
Expression of markers for hepatocytes and non-parenchymal cells in liver sections and isolated hepatocytes from male transgenic SAC mice. qRT-PCR analyses of the expression of markers Arg1 Gck, Gls2, Ldha, Krt8 and Krt18 in male (A): SAC-WT liver (white bars) (n = 8) versus SAC-WT hepatocytes (hatched bars) (n = 7); (B) SAC-KO liver (white bars) (n = 8) versus SAC-KO hepatocytes (hatched bars) (n = 8). Non-parenchymal cell markers Acta2, Gfap, Emr1, and Krt19 in male; (C): SAC-WT liver (black bars) (n = 8) versus SAC-WT hepatocytes (checkered bars) (n = 8); (D): SAC-KO liver (black bars) (n = 8) versus SAC-KO hepatocytes (chekered bars) (n = 8). *, p<0.05;***, p<0.01; ***,p<0.001. Values are presented as the means ± SEM.
Physiological and molecular effect of hepatocellular deletion of Smo in transgenic SLC mice. (A): Comparison of body weight of female SLC-WT mice without Doxycycline (Dox.) (white bars) (n = 17), SLC-WT mice with Dox. (gray bars) (n = 4) and SLC-KO mice with Dox. (black bars) (n = 12) at 5 weeks after deletion of Smo. Doxycycline was given at the age of 8 weeks for 10 days to induce the Cre-recombinase expression as described in Materials and methods. (B): Comparison of body weight of female SLC-WT mice (black squares) (n = 8-14) and SLC-KO mice (open circles) (n = 8-10). Deletion of Smo was induced at the age of 8 weeks. (C): qRT-PCR analyses of the expression of Smo, Gli1, Gli2 and Gli3 in isolated hepatocytes from female SLC-WT mice (white bars) (n = 5-15) and SLC-KO mice (black bars) (n = 5-16) at 5 weeks after deletion of Smo. (D): qRT-PCR analyses of the expression of Igf1 and Igfbp1 in isolated hepatocytes from female SLC-WT mice (white bars) (n = 7) and SLC-KO mice (black bars) (n = 4) at k weeks after deletion of Smo. *, p<0.05; **, p<0.01; ***, p<0.001. Values are presented as the means ± SEM.
Response of selected Hh pathway genes to siRNA mediated knockdown of Ptch1. qRT-PCR analyses of (A): Smo, lhh and Shh; (B): Ptch2 and Hhip1, (C): Fu and Sufu and (D): Gli1 and Gli2 in cultured hepatocytes of male C57BL/6N mice in response to transfection with Ptch1 siRNA (black bars) (n = 7-8) compared to nonsense transfection (white bars) (n = 7-8) after 48 h of incubation. Values are presented as relative means ± SEM.
Primers for PCR genotyping of SAC mice. Table S2: Primers for qRT-PCR analyses. Table S3: Primers for RNA interference experiments. Table S4: Primers for Chromatin IP. Table S5: Binding sites of Gli factors and Gli factor-dependent transcription factors to the promoter region of Igf1 and Igfbp-1 in mouse determined by MotifMap.
Correlation analyses of IGF-I serum concentrations and body weight from male and female transgenic SAC mice. Correlation analyses of IGF-I serum concentrations and body weight from (A): male SAC-WT (black circles) (n = 7) mice and SAC-KO (open circles) (n = 6) mice and (B): female SAC-WT (black circles) (n = 9) mice and SAC-KO (open circles) (n = 8) mice. c = correlation coefficient; p = significance level.
Contributor Information
Madlen Matz-Soja, Email: Madlen.matz@medizin.uni-leipzig.de.
Susanne Aleithe, Email: Susanne.Aleithe@medizin.uni-leipzig.de.
Eugenia Marbach, Email: Eugenia.Marbach@medizin.uni-leipzig.de.
Jan Böttger, Email: Jan.Boettger@medizin.uni-leipzig.de.
Katrin Arnold, Email: Katrin.Arnold@medizin.uni-leipzig.de.
Wolfgang Schmidt-Heck, Email: wolfgang.schmidt-heck@hki-jena.de.
Jürgen Kratzsch, Email: juergen.kratzsch@medizin.uni-leipzig.de.
Rolf Gebhardt, Email: rgebhardt@medizin.uni-leipzig.de.
Acknowledgements
We are grateful to Dr. Sebastian Zellmer for partaking in valuable discussions. We cordially thank Kerstin Heise, Doris Mahn, Frank Struck, Fatina Siwczak and Vivien Karrasch for providing excellent technical assistance.
Further, we would like to thank Ute Krügel for the possibility to breed the SAC mice in the animal facility of the Rudolf-Böhm-Institute and Petra Fink-Sterba, Sigrid Weisheit, Sandra Richter and Manuela Liebig from the MEZ of the Faculty of Medicine for taking excellent care of the transgenic mice. We thank Prof. Dr. Daniel Teupser and Franziska Jeromin for the measurement of ALAT, ASAT and GLDH.
This work was supported in part by the Medical Faculty of the University of Leipzig and by a grant from the Bundesministerium für Forschung und Technologie (BMBF) in the framework of the Systems Biology initiative “Virtual Liver Network” (grant: 0315735). We acknow-ledge support from the German Research Foundation (DFG) and Leipzig University within the program of Open Access Publishing.
References
- Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends in Molecular Medicine. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Ayers KL, Therond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol. 2010;20:287–298. doi: 10.1016/j.tcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137:2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz I, Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Di Pasca MM, Hebrok M. Hedgehog signaling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Ruiz I, Altaba A. Gli proteins and Hedgehog signaling: development and cancer. Trends Genet. 1999;15:418–425. doi: 10.1016/S0168-9525(99)01840-5. [DOI] [PubMed] [Google Scholar]
- van den Brink GR. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- Wang LC, Nassir F, Liu ZY, Ling L, Luo F, Crowell T, Olson D, Davidson NO, Burkly LC. Disruption of hedgehog signaling reveals a novel role in intestinal morphogenesis and intestinal-specific lipid metabolism in mice. Gastroenterology. 2002;122:469–482. doi: 10.1053/gast.2002.31102. [DOI] [PubMed] [Google Scholar]
- Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G, Dalgaard K, Selvaraj M, Gaster M, Lee-Young RS, Febbraio MA, Knauf C, Cani PD, Aberger F, Penninger JM, Pospisilik JA, Esterbauer H. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Pospisilik JA, Schramek D, Schnidar H, Cronin SJF, Nehme NT, Zhang X, Knauf C, Cani PD, Aumayr K, Todoric J, Bayer M, Haschemi A, Puviindran V, Tar K, Orthofer M, Neely GG, Dietzl G, Manoukian A, Funovics M, Prager G, Wagner O, Ferrandon D, Aberger F, Hui C, Esterbauer H, Penninger JM. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010;140:148–160. doi: 10.1016/j.cell.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Sicklick JK, Li Y, Choi SS, Qi Y, Chen W, Bustamante M, Huang J, Zdanowicz M, Camp T, Torbenson MS, Rojkind M, Diehl AM. Role for hedgehog signaling in hepatic stellate cell activation and viability. Lab Invest. 2005;85:1368–1380. doi: 10.1038/labinvest.3700349. [DOI] [PubMed] [Google Scholar]
- Omenetti A, Diehl AM. Hedgehog signaling in cholangiocytes. Curr Opin Gastroenterol. 2011;27:268–275. doi: 10.1097/MOG.0b013e32834550b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Witek RP, Syn WK, Choi SS, Omenetti A, Premont R, Guy CD, Diehl AM. Signals from dying hepatocytes trigger growth of liver progenitors. Gut. 2010;59:655–665. doi: 10.1136/gut.2009.204354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala F, Guy CD, Lu J, Suzuki A, Burchette JL, Abdelmalek MF, Chen W, Diehl AM. Increased production of sonic hedgehog by ballooned hepatocytes. J. Pathol. 2011;224:401–410. doi: 10.1002/path.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54:366–373. doi: 10.1016/j.jhep.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz-Soja M, Hovhannisyan A, Gebhardt R. Hedgehog signaling pathway in adult liver: a major new player in hepatocyte metabolism and zonation? Med Hypotheses. 2013;80:589–594. doi: 10.1016/j.mehy.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Opherk C, Anlag K, Schutz G, Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151–153. doi: 10.1002/(SICI)1526-968X(200002)26:2<151::AID-GENE17>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Blaas L, Kornfeld J, Schramek D, Musteanu M, Zollner G, Gumhold J, van Zijl F, Schneller D, Esterbauer H, Egger G, Mair M, Kenner L, Mikulits W, Eferl R, Moriggl R, Penninger J, Trauner M, Casanova E. Disruption of the growth hormone-Signal transducer and activator of transcription 5-Insulinlike growth factor 1 axis severely aggravates liver fibrosis in a mouse model of cholestasis. Hepatology. 2010;51:1319–1326. doi: 10.1002/hep.23469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueberham E, Bottger J, Ueberham U, Grosche J, Gebhardt R. Response of sinusoidal mouse liver cells to choline-deficient ethionine-supplemented diet. Comp Hepatol. 2010;9:8. doi: 10.1186/1476-5926-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann-Tappe I, Wizigmann S, Gebhardt R. Glutamate uptake in primary cultures of biliary epithelial cells from normal rat liver. Cell Biol Toxicol. 1991;7:315–325. doi: 10.1007/BF00124068. [DOI] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von KD, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Skrtic S, Wallenius K, Sjogren K, Isaksson OG, Ohlsson C, Jansson JO. Possible roles of insulin-like growth factor in regulation of physiological and pathophysiological liver growth. Horm Res. 2001;55(Suppl 1):1–6. doi: 10.1159/000063454. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics. 2011;12:495. doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DJ, Varco-Merth B, Rotwein P. Dispersed Chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J Biol Chem. 2010;285:17636–17647. doi: 10.1074/jbc.M110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicklick JK, Li Y, Melhem A, Schmelzer E, Zdanowicz M, Huang J, Caballero M, Fair JH, Ludlow JW, McClelland RE, Reid LM, Diehl AM. Hedgehog signaling maintains resident hepatic progenitors throughout life. Am J Physiol Gastrointest Liver Physiol. 2006;290:G859–70. doi: 10.1152/ajpgi.00456.2005. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Gebhardt R. Metabolic zonation of the liver: regulation and implications for liver function. Pharmacol Ther. 1992;53:275–354. doi: 10.1016/0163-7258(92)90055-5. [DOI] [PubMed] [Google Scholar]
- Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: the role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- Reed KR, Athineos D, Meniel VS, Wilkins JA, Ridgway RA, Burke ZD, Muncan V, Clarke AR, Sansom OJ. B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc Natl Acad Sci USA. 2008;105:18919–23. doi: 10.1073/pnas.0805778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson OG, Jansson JO, Ohlsson C. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci USA. 1999;96:7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay E, Seurin D, Babajko S, Doublier S, Cazillis M, Binoux M. Liver-specific expression of human insulin-like growth factor binding protein-1 in transgenic mice: repercussions on reproduction, ante- and perinatal mortality and postnatal growth. Endocrinology. 1997;138:2937–2947. doi: 10.1210/endo.138.7.5282. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Lahm H, Wu M, Hoeflich A, Wolf E. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. FASEB J. 2000;14:629–640. doi: 10.1096/fasebj.14.5.629. [DOI] [PubMed] [Google Scholar]
- Lee PD, Conover CA, Powell DR. Regulation and function of insulin-like growth factor-binding protein-1. Proc Soc Exp Biol Med. 1993;204:4–29. doi: 10.3181/00379727-204-43630. [DOI] [PubMed] [Google Scholar]
- Camacho-Hubner C. Assessment of growth hormone status in acromegaly: what biochemical markers to measure and how? Growth Horm IGF Res. 2000;10:S125–9. doi: 10.1016/s1096-6374(00)80024-6. [DOI] [PubMed] [Google Scholar]
- Sjogren K, Wallenius K, Liu JL, Bohlooly-Y M, Pacini G, Svensson L, Tornell J, Isaksson OG, Ahren B, Jansson JO, Ohlsson C. Liver-derived IGF-I is of importance for normal carbohydrate and lipid metabolism. Diabetes. 2001;50:1539–1545. doi: 10.2337/diabetes.50.7.1539. [DOI] [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LJ. Overexpression of insulin-like growth factor binding protein-1 in transgenic mice. Pediatr Nephrol. 2000;14:567–571. doi: 10.1007/s004670000347. [DOI] [PubMed] [Google Scholar]
- Clemmons DR. Role of insulin-like growth factor in maintaining normal glucose homeostasis. Horm Res. 2004;62(Suppl 1):77–82. doi: 10.1159/000080763. [DOI] [PubMed] [Google Scholar]
- Ruan W, Lai M. Insulin-like growth factor binding protein: a possible marker for the metabolic syndrome? Acta Diabetol. 2010;47:5–14. doi: 10.1007/s00592-009-0142-3. [DOI] [PubMed] [Google Scholar]
- Froment P, Seurin D, Hembert S, Levine JE, Pisselet C, Monniaux D, Binoux M, Monget P. Reproductive abnormalities in human IGF binding protein-1 transgenic female mice. Endocrinology. 2002;143:1801–1808. doi: 10.1210/endo.143.5.8815. [DOI] [PubMed] [Google Scholar]
- Yuan R, Meng Q, Nautiyal J, Flurkey K, Tsaih S, Krier R, Parker MG, Harrison DE, Paigen B. Genetic coregulation of age of female sexual maturation and lifespan through circulating IGF1 among inbred mouse strains. Proc Natl Acad Sci USA. 2012;109:8224–8229. doi: 10.1073/pnas.1121113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- Schonig K, Schwenk F, Rajewsky K, Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002;30:e134. doi: 10.1093/nar/gnf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt R, Lerche KS, Gotschel F, Gunther R, Kolander J, Teich L, Zellmer S, Hofmann H, Eger K, Hecht A, Gaunitz F. 4-Aminoethylamino-emodin–a novel potent inhibitor of GSK-3beta–acts as an insulin-sensitizer avoiding downstream effects of activated beta-catenin. J Cell Mol Med. 2010;14:1276–1293. doi: 10.1111/j.1582-4934.2009.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingmuller U, Bauer A, Bohl S, Nickel PJ, Breitkopf K, Dooley S, Zellmer S, Kern C, Merfort I, Sparna T, Donauer J, Walz G, Geyer M, Kreutz C, Hermes M, Gotschel F, Hecht A, Walter D, Egger L, Neubert K, Borner C, Brulport M, Schormann W, Sauer C, Baumann F, Preiss R, MacNelly S, Godoy P, Wiercinska E, Ciuclan L. et al. Primary mouse hepatocytes for systems biology approaches: a standardized in vitro system for modelling of signal transduction pathways. Syst Biol (Stevenage) 2006;153:433–447. doi: 10.1049/ip-syb:20050067. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellmer S, Sickinger S, Schmidt-Heck W, Guthke R, Gebhardt R. Heterogeneous expression of suppressor of cytokine signaling 2 (SOCS-2) in liver tissue. J Anat. 2009;215:176–183. doi: 10.1111/j.1469-7580.2009.01085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of the transgenic mouse model with hepatocyte-specific deletion of Smoothened. (A): Structure of the Alfp-Cre vector construct. (B): The Smo locus with loxP sites on either side of exon 1in the absence of Cre-recombinase activity. (C): The floxed Smo locus lacking exon 1 in the presence of Cre-recombinase activity. (D): Immunohistochemical detection of Cre-recombinase in liver sections of SAC-WT and SAC-KO mice. Brown colour indicating Cre-recombinase is present in hepatocyte and cholangiocyte nuclei (strong staining) and cytoplasm (weak staining). Bar: 50 μm. (E): PCR analysis, using DNA extracted from liver tissue of SAC-WT and SAC-KO mice yields a 600-bp amplicon of wild-type Smo in different tissues SAC-WT (black bars) (n = 7-20) and SAC-KO (white bars) (n = 7-20) mice determined by qRT-PCR. Significant decrease of Smo mRNA relative to β-actin is detected only in liver. Values are presented as means ± SEM; *, p<0.05.
Phenotypic features induced by hepatocyte-specific ablation of Smoothened in transgenic SAC mice. Photographs of (A): male SAC wildtype SAC-WT (Smo+/+), heterozygous SAC-KO (Smo+/-) and homozygous SAC-KO (Smo-/-) mouse. (B): Comparison of body weight of 4, 8 and 12 weeks old male SAC-WT (Smo+/+) (white bars) (n = 4-9), heterozygous SAC-KO (Smo+/-) (gray bars) (n = 8-9) and homozygous SAC-KO (Smo-/-) (black bars) (n = 8-21) mice. Values are presented as the means ± SEM; *, p<0.05; **, p<0.01; ***, p<0,001.
Expression of markers for hepatocytes and non-parenchymal cells in liver sections and isolated hepatocytes from male transgenic SAC mice. qRT-PCR analyses of the expression of markers Arg1 Gck, Gls2, Ldha, Krt8 and Krt18 in male (A): SAC-WT liver (white bars) (n = 8) versus SAC-WT hepatocytes (hatched bars) (n = 7); (B) SAC-KO liver (white bars) (n = 8) versus SAC-KO hepatocytes (hatched bars) (n = 8). Non-parenchymal cell markers Acta2, Gfap, Emr1, and Krt19 in male; (C): SAC-WT liver (black bars) (n = 8) versus SAC-WT hepatocytes (checkered bars) (n = 8); (D): SAC-KO liver (black bars) (n = 8) versus SAC-KO hepatocytes (chekered bars) (n = 8). *, p<0.05;***, p<0.01; ***,p<0.001. Values are presented as the means ± SEM.
Physiological and molecular effect of hepatocellular deletion of Smo in transgenic SLC mice. (A): Comparison of body weight of female SLC-WT mice without Doxycycline (Dox.) (white bars) (n = 17), SLC-WT mice with Dox. (gray bars) (n = 4) and SLC-KO mice with Dox. (black bars) (n = 12) at 5 weeks after deletion of Smo. Doxycycline was given at the age of 8 weeks for 10 days to induce the Cre-recombinase expression as described in Materials and methods. (B): Comparison of body weight of female SLC-WT mice (black squares) (n = 8-14) and SLC-KO mice (open circles) (n = 8-10). Deletion of Smo was induced at the age of 8 weeks. (C): qRT-PCR analyses of the expression of Smo, Gli1, Gli2 and Gli3 in isolated hepatocytes from female SLC-WT mice (white bars) (n = 5-15) and SLC-KO mice (black bars) (n = 5-16) at 5 weeks after deletion of Smo. (D): qRT-PCR analyses of the expression of Igf1 and Igfbp1 in isolated hepatocytes from female SLC-WT mice (white bars) (n = 7) and SLC-KO mice (black bars) (n = 4) at k weeks after deletion of Smo. *, p<0.05; **, p<0.01; ***, p<0.001. Values are presented as the means ± SEM.
Response of selected Hh pathway genes to siRNA mediated knockdown of Ptch1. qRT-PCR analyses of (A): Smo, lhh and Shh; (B): Ptch2 and Hhip1, (C): Fu and Sufu and (D): Gli1 and Gli2 in cultured hepatocytes of male C57BL/6N mice in response to transfection with Ptch1 siRNA (black bars) (n = 7-8) compared to nonsense transfection (white bars) (n = 7-8) after 48 h of incubation. Values are presented as relative means ± SEM.
Primers for PCR genotyping of SAC mice. Table S2: Primers for qRT-PCR analyses. Table S3: Primers for RNA interference experiments. Table S4: Primers for Chromatin IP. Table S5: Binding sites of Gli factors and Gli factor-dependent transcription factors to the promoter region of Igf1 and Igfbp-1 in mouse determined by MotifMap.
Correlation analyses of IGF-I serum concentrations and body weight from male and female transgenic SAC mice. Correlation analyses of IGF-I serum concentrations and body weight from (A): male SAC-WT (black circles) (n = 7) mice and SAC-KO (open circles) (n = 6) mice and (B): female SAC-WT (black circles) (n = 9) mice and SAC-KO (open circles) (n = 8) mice. c = correlation coefficient; p = significance level.