Abstract
Anti-poly(ADP-ribose)polymerase (PARP) drugs were initially developed as catalytic inhibitors to block the repair of DNA single-strand breaks. We recently reported that several PARP inhibitors have an additional cytotoxic mechanism by trapping PARP-DNA complexes, and that both olaparib and niraparib act as PARP poisons at pharmacological concentrations. Therefore, we have proposed that PARP inhibitors should be evaluated based both on catalytic PARP inhibition and PARP-DNA trapping. Here, we evaluated the novel PARP inhibitor, BMN 673, and compared its effects on PARP1 and PARP2 with two other clinical PARP inhibitors, olaparib and rucaparib, using biochemical and cellular assays in genetically-modified chicken DT40 and human cancer cell lines. Although BMN 673, olaparib and rucaparib are comparable at inhibiting PARP catalytic activity, BMN 673 is ~100-fold more potent at trapping PARP-DNA complexes and more cytotoxic as single agent than olaparib, while olaparib and rucaparib show similar potencies in trapping PARP-DNA complexes. The high level of resistance of PARP1/2 knockout cells to BMN 673 demonstrates the selectivity of BMN 673 for PARP1/2. Moreover, we show that BMN 673 acts by stereospecific binding to PARP1 as its enantiomer, LT674, is several orders of magnitude less efficient. BMN 673 is also ~100-fold more cytotoxic than olaparib and rucaparib in combination with the DNA alkylating agents methyl methane sufonate (MMS) and temozolomide. Our study demonstrates that BMN 673 is the most potent clinical PARP inhibitor tested to date with the highest efficiency at trapping PARP-DNA complexes.
Keywords: PARP inhibitor, poly(ADP-ribosyl)ation, PARP-DNA complex, chemotherapy, homologous recombination
Introduction
Poly(ADP-ribose) polymerase 1 (PARP1) and PARP2 detect DNA damage with great sensitivity (1–5). PARP1 is an abundant nuclear protein that binds damaged DNA through its N-terminal zinc finger motifs, which activates its catalytic C-terminal domain to hydrolyze NAD+ and produce linear and branched PAR chains that can extend over hundreds of ADP-ribose units (1–5). The rapid binding of PARP1 and PARP2 to DNA is critical for the resealing of DNA single-strand breaks (SSBs) during base excision repair (BER) and for the repair of topoisomerase I cleavage complexes (6–11). A large number of SSBs are generated endogenously as well as BER intermediates (12, 13). When replication forks encounter SSBs, they generate double-strand breaks (DSBs) that need to be repaired by homologous recombination (HR) (7, 13–15). Accordingly, PARP inhibition results in the accumulation of recombinogenic substrates marked by RAD51 and γH2AX nuclear foci (16, 17).
Since the discovery of the synthetic lethality of PARP inhibitors in HR deficient cells (15, 18–23), the mechanism by which PARP inhibitors exert their cytotoxicity has been dominantly interpreted as an accumulation of unrepaired SSBs resulting from catalytic PARP inhibition (7, 24). Hence, more highly efficacious PARP catalytic inhibitors with IC50 (inhibitory concentration 50%) values reaching the low nanomolar range have been developed (4, 21, 24–26).
Recently, we showed that, in addition to catalytic inhibition, PARP inhibitors exert their cytotoxicity by trapping PARP1 and PARP2 on SSB sites (27). Such PARP-DNA complexes are more effective at killing cancer cells than unrepaired SSBs caused by the absence of PARP. Since the cytotoxicity is mediated by the presence of PARP1 and PARP2, PARP inhibitors have been proposed to act as “PARP poisons”. PARP trapping is also not merely interpreted as resulting from catalytic PARP inhibition, which prevents dissociation of PARP from DNA and is required for repair completion (28), because the potency to trap PARP-DNA complexes varies widely across the different PARP inhibitors and is not correlated with their PARP catalytic inhibition potency (27). Indeed, veliparib is a highly potent catalytic PARP inhibitor with relatively limited trapping of PARP-DNA complexes in comparison with niraparib and olaparib (27). Therefore, we have proposed that PARP inhibitors should be categorized according to PARP poisoning capability in addition to catalytic PARP inhibition.
In the present study, we examined the ability of three clinical inhibitors, olaparib (AstraZeneca), rucaparib (Clovis) and BMN 673 (BioMarin) (Figure 1A) in terms of catalytic PARP inhibition and PARP poisoning. We also evaluated potential off-target effects of these drugs. To do so, we took advantage of the fact that avian cells genetically lack PARP2 (29). PARP1−/− avian B-lymphoblast DT40 cells are equivalent to PARP1 and PARP2 double-knockout cells, and do not have detectable level of poly(ADP-ribosyl)ation (27, 29). We also compared the cytotoxicity of the three PARP inhibitors as a single agent in the BRCA-deficient DT40 cells, and in human prostate cancer and Ewing’s sarcoma cells, which have been reported to be selectively sensitive to PARP inhibitors (30), in the NCI60 cell line panel, and in combination with the DNA alkylating agents, methyl methane sulfonate (MMS) and temozolomide.
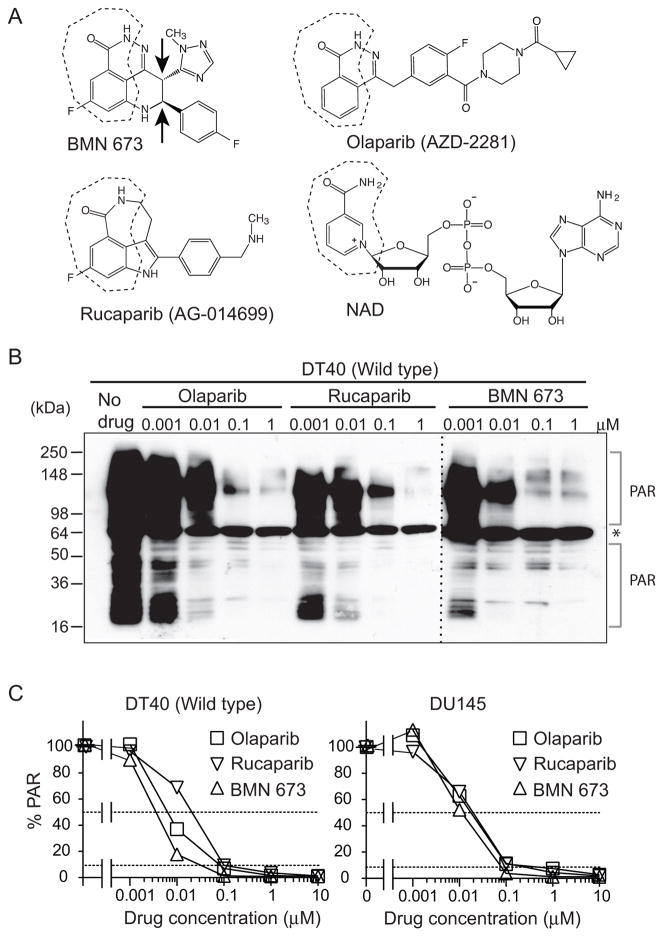
Figure 1. Comparative PARP catalytic inhibition of BMN 673.
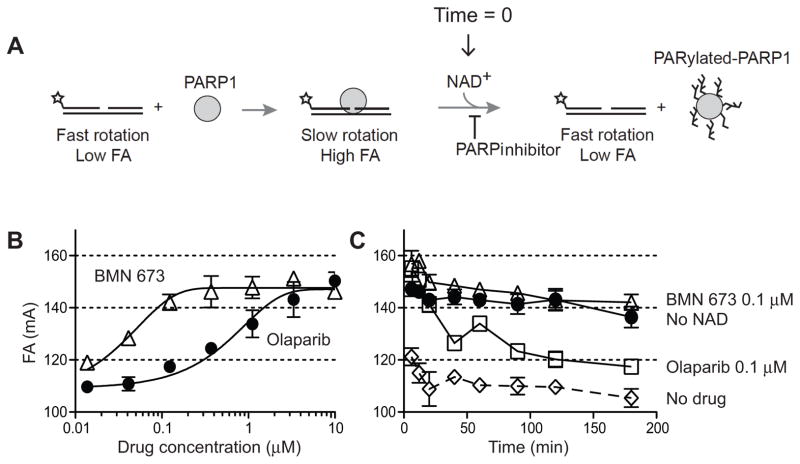
(A) Chemical structures of olaparib (AZD2281), rucaparib (AG-014699), BMN 673 and NAD. The nicotinamide moiety is outlined in dotted lines. Arrows in the BMN 673 structure indicate chiral centers involved in drug activity (see Figure S1) (B) Catalytic PARP inhibition potency of BMN 673 in comparison with olaparib and rucaparib. Total poly(ADP-ribosyl)ation (PAR) levels in wild type DT40 cells were examined by Western blotting against PAR 30 min after the indicated drug treatments. The asterisk represents a nonspecific band. (C) PAR levels in drug-treated wild type DT40 and DU145 cells measured by ELISA. Cells were incubated with the indicated concentrations of PARP inhibitors for 2 hours. PAR levels without drug treatment were set as 100% in each cell line.
Materials and Methods
Cell lines and drug treatments
The DT40 cell lines used in this study were obtained from the Laboratory of Radiation Genetics Graduate School of Medicine in Kyoto University, Japan in 2011–2012. Human prostate cancer cells (DU145) and human breast cancer cells (MDA-MB231) were obtained from the National Cancer Institute Developmental Therapeutics Program (Frederick, USA) in 2011–2012. The human Ewing’s sarcoma cell line (EW8) was a kind gift from Dr. Lee Helman, Pediatric Oncology Branch, NCI, NIH obtained in 2012. We did not authenticate these cells in our laboratory. BMN 673 and LT674 were provided by Dr. Leonard E. Post, BioMarin Pharmaceutical Inc. (San Rafael, CA). Olaparib, rucaparib and temozolomide were obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, DCTD, NCI. Drug stock solutions were made in DMSO at 10 mM. The stock solutions were stored at −20ºC in the dark and diluted in culture medium immediately before use. 1% or 10% MMS was prepared fresh each time from 99% MMS (129925, Sigma-Aldrich) in PBS, and then diluted in culture medium to final concentration.
Immunoblotting
To prepare whole cell lysates, cells were lysed with CelLytic™M lysis reagent (C2978, Sigma-Aldrich, St Louis, MO). After thorough mixing and incubation at 4°C for 30 min, lysates were centrifuged at 15,000 g at 4°C for 10 min, and supernatants were collected. To prepare chromatin bound subcellular fraction, ten million DT40 cells with 10 ml medium in 15 ml tube or semi-confluent human cells with 10 ml medium in 10 cm dish were treated with indicated drugs for 30 min or 4 hours, respectively. Cells were collected and fractionated using a Subcellular Protein Fractionation Kit from Thermo Scientific (78840, Rockford, IL, USA) following the manufacturer’s instructions. Immunoblotting was carried out using standard procedures.
Rabbit polyclonal anti-PARP1 antibody (sc-7150) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit polyclonal anti-histone H3 antibody (07-690) was from Upstate Biotechnology (Lake Placid, NY, USA). Rabbit polyclonal anti-PAR polymer antibody (#4336-BPC-100) was from Trevigen (Gaithersburg, MD, USA). Rabbit polyclonal anti-PARP2 antibody (ab93416) was from Abcam (Cambridge, MA, USA). Secondary antibodies were horseradish peroxidase (HRP)-conjugated antibodies to rabbit IgG (GE Healthcare, UK).
PAR immunoassay
The validated chemiluminescent immunoassay for PAR using commercially available reagents has been described previously (31). The detailed lab procedures can be viewed at the website (32).
Measurement of cellular sensitivity to DNA-damaging agents
To measure the sensitivity of cells to drugs, cells were continuously exposed to various concentrations of the drugs for 72 hours in triplicate. For DT40 cells, 200 cells were seeded into 384-well white plates (#6007680 Perkin Elmer Life Sciences, Waltham, MA) in 40 μl of medium per well. For human cells, 1,500 cells were seeded in 96-well while plates (#6005680 Perkin Elmer Life Sciences) in 100 μl of medium per well. Cell survival was determined using the ATPlite 1-step kits (PerkinElmer). Briefly, 20 μl or 50 μl ATPlite solution for 384-well or 96-well plate respectively, was added to each well. After 5 min, luminescence was measured with an EnVision 2104 Multilabel Reader (PerkinElmer). The ATP level in untreated cells was defined as 100%. Survival of treated cells was defined as ATP treated cells/ATP untreated cells × 100.
The cell viability assays across the NCI60 cell panel were carried out by the NCI/NIH Developmental Therapeutics Program (33) using standard procedures (34–36).
Fluorescence anisotropy DNA binding assay
The fluorescence anisotropy (FA) experiments were carried out with a 30 bp duplex labeled with 5’-Alexa Fluor488. The deoxyoligonucleotide (sequence: 5’-Alexa Fluor 488-ACCCTGCTGTGGGCdUGGAGAACAAGGTGAT) was annealed to its complementary DNA strand in buffer containing 50 mM potassium acetate, 20 mM tris-acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol, pH 7.9. All oligonucleotides were purchased from Integrated DNA Technologies. Uracil-DNA glycosylase and APE1 (New England Biolabs) were added to the annealed DNA sample and incubated at 37°C for 1 hour. The resulting DNA construct contains a DNA nick and a 5’-dRP at the nicked site. The completion of digestion is verified by denaturing PAGE.
For FA measurements, 250 nM recombinant PARP1 (a kind gift from Dr. Valerie Schreiber, University of Strasbourg, France), 1 nM DNA construct, and increasing concentration of PARP inhibitors were combined and incubated for 30 minutes in buffer containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 100 mM NaCl, and 100 ng/μL BSA. 1 mM NAD+ was added to the samples to initiate the experiment and the FA values were measured at indicated time using an EnVision 2104 Multilabel Reader equipped with a FITC FP Label (PerkinElmer). The control samples lack NAD+ or PARP inhibitor. The FA values reported here were average of two independent experiments (each experiment was measured in duplicates).
Results
BMN 673 is a stereospecific PARP catalytic inhibitor at least as potent as olaparib and rucaparib
We first tested the potency of BMN 673 to inhibit total cellular PARylation in comparison with olaparib and rucaparib. Western blotting analyses against PAR using total cell lysates of drug-treated wild type DT40 cells (27) showed that all three PARP inhibitors reduced total cellular PAR levels in a concentration-dependent manner at submicromolar concentrations, with BMN 673 producing full inhibition at 0.1 μM (Figure 1B). Figure 1C and Table 1 show a quantitative analysis using the clinically validated PAR ELISA assay (27, 37) with IC50 (inhibitory concentration 50%) and IC90 (inhibitory concentration 90%) for all three drugs in DT40 cells and human prostate DU145 cancer cells. In DT40 cells, BMN 673 was ~2-fold more potent than olaparib, and rucaparib was ~3-fold less potent than olaparib. In DU145 cells, the three drugs were comparable. We also compared BMN 673 with its isomer, LT674 (Figure S1). LT674 showed no detectable cellular PARylation inhibition by Western blotting and several orders of magnitude less reduction of PAR by ELISA. These results demonstrate that BMN 673, olaparib and rucaparib are highly potent PARP inhibitors at low nanomolar concentrations, and they are indistinguishable above 0.1 μM since PAR levels are almost zero under these conditions.
Table 1.
Summary of the IC50 (inhibitory concentration 50%) and IC90 (inhibitory concentration 90%) PAR level inhibitions for each drug in DT40 and DU145 cells.
| Drug | IC50 (nM) | IC90 (nM) | ||
|---|---|---|---|---|
| DT40 | DU145 | DT40 | DU145 | |
| BMN 673 | 4 | 11 | 30 | 71 |
| Olaparib | 6 | 18 | 80 | 120 |
| Rucaparib | 21 | 18 | 100 | 120 |
BMN 673 produces higher PARP-mediated cytotoxicity than olaparib or rucaparib
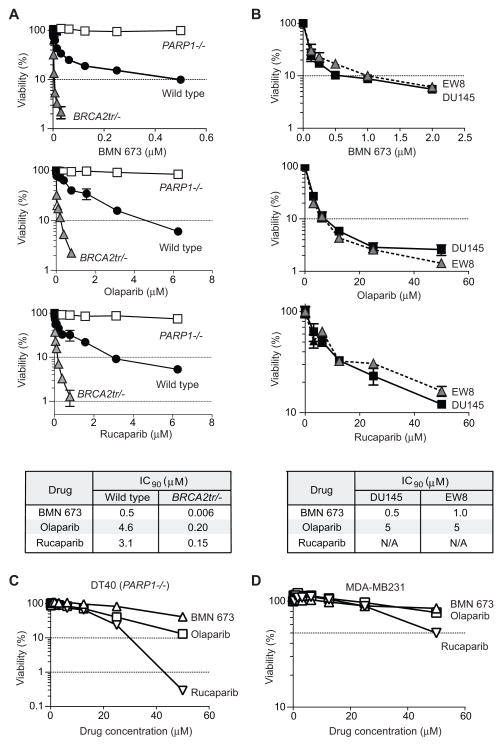
To determine the selective targeting of PARP1 by BMN 673, we examined its single-agent cytotoxicity in wild type, PARP1−/− and BRCA2tr/− [BRCA2 truncated mutant that is deficient in HR (38)] DT40 cells (Figure 2A) (27). We measured ATP concentration to evaluate cellular viability across this study. Since the alternation of NAD+ pool by catalytic PARP inhibition might affect the ATP pool, we checked ATP concentration in treated and untreated cells with BMN 673 in the same conditions as Figure 1B (Figure S2). We confirmed that catalytic PARP inhibition did not alter the total ATP concentration. BMN 673 showed single agent cytotoxicity at nanomolar concentrations. Yet, PARP1−/− cells were immune to BMN 673, indicating a PARP1 requirement for the cytotoxicity of BMN 673, and therefore the selective targeting of PARP1 with no off-target effect for BMN 673. Similar results were observed for olaparib (27) (Figure 2A, 2nd panel from top). Rucaparib also showed PARP-dependent cytotoxicity at low micromolar concentrations (Figure 2A, 3rd panel from top).
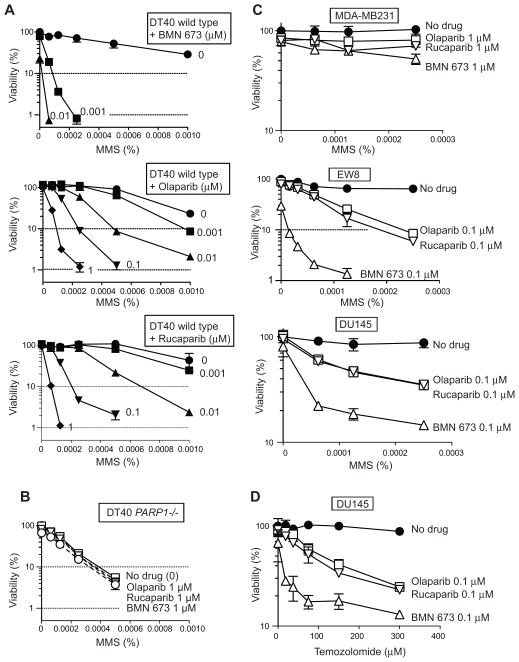
Figure 2. BMN 673 is markedly more cytotoxic than olaparib and rucaparib while requiring PARP-1/2 for activity.
For all experiments, viability curves were derived after continuous treatment for 72 hours with the indicated PARP inhibitors in the indicated cell lines. Cellular ATP concentration was used to measure cell viability. The survival of untreated cells was set as 100%. Error bars represent standard deviation (SD) (n ≥ 3). (A) Survival curves of wild type, PARP1−/−, and BRCA2tr/− DT40 cells. Drug IC90’s are tabulated at the bottom. (B) Survival curves of DU145 (human prostate cancer) and EW8 (Ewing’s sarcoma) cells. Drug IC90’s are tabulated at the bottom. (C) Survival curves of PARP1−/− DT40 cell line to high concentrations of the PARP inhibitors. (D) Survival curves of MDB-MB231 (human breast cancer) cells.
The additional sensitivity of wild type compared to PARP1−/− cells is mediated by PARP1 (27, 39). The IC90 of wild type DT40 cells to BMN 673 was 6–10 times lower than for the other two PARP inhibitors. We also confirmed by flow cytometry analyses the higher cytotoxicity of BMN 673 compared to olaparib and rucaparib (Figure S3A). As expected (20, 27), homologous recombination (HR)-deficient BRCA2tr/− cells showed greater sensitivity than wild type cells to all three drugs. The IC90 of BRCA2tr/− cells toBMN 673 was 25–33 times lower than for olaparib and rucaparib (Figure 2A, bottom panel). Moreover, LT674, the inactive stereoisomer of BMN 673, was markedly less cytotoxic (~100-fold) even in the BRCA2-deficient cells (Figure S4).
We next examined the cytotoxicity of each drug in human cancer cells (Figure 2B). Ewing’s sarcoma cells have recently been identified as being selectively sensitive to olaparib (30), and EW8 is an Ewing’s sarcoma cell line that carries the EWS-FLI1 translocation (40). DU145, a prostate cancer cell line without TMPRSS2-ERG translocation, is among the most sensitive NCI60 cell lines to olaparib and BMN 673. Both cell lines showed comparable sensitivity curves to the three PARP inhibitors (Figure 2B). The IC90 of BMN 673 was 10- and 5-fold lower than that of olaparib in DU145 and EW8 cells, respectively (Figure 2B, bottom panel). Rucaparib was also markedly less cytotoxic than BMN 673 in EW8 and DU145 cells (Figure 2B). Consistently, the flow cytometry analyses revealed higher cytotoxic effect in BMN 673 compared to olaparib and rucaparib (Figure S3B). These results indicate that BMN 673 produces greater PARP-mediated cytotoxicity than olaparib and rucaparib.
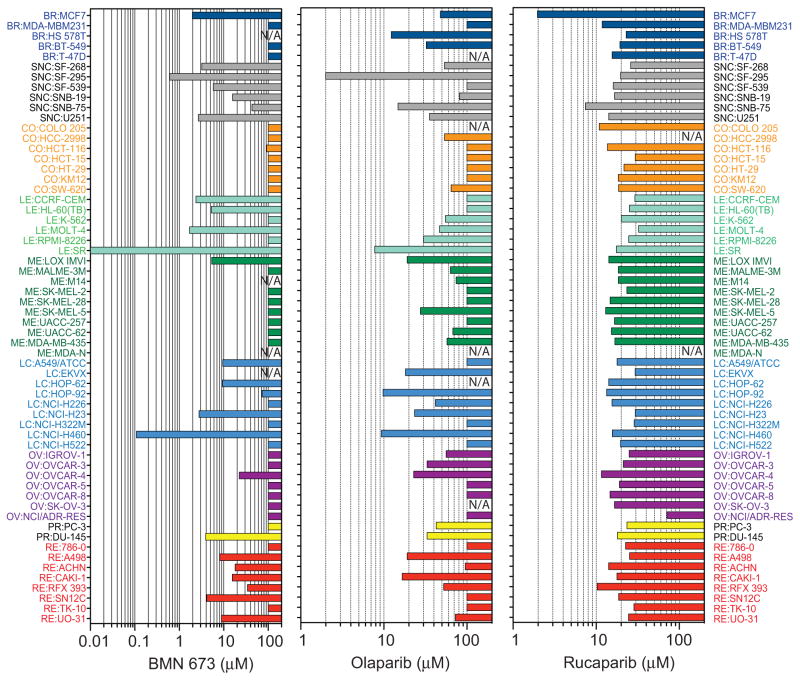
To test whether the three PARP inhibitors have other cellular target(s) beside PARP1 and PARP2, the drugs were tested at high concentrations in PARP1−/− DT40 cells (Figure 2C). While BMN 673 and olaparib remained relatively non-cytotoxic at 50 μM, rucaparib showed marked PARP1/2-independent cytotoxicity. The off-target effect of rucaparib (with respect to PARP-1/2) was also demonstrated in the human MDA-MB231 breast cancer cell line (Figure 2D), one of the NCI60 cell lines (34, 41), that was insensitive to 100 μM olaparib or BMN 673 (Figure 3). Rucaparib decreased MDA-MB231 cells viabilities down to 50% at 50 μM, while olaparib and BMN 673 showed no significant effect (Figure 2D). Moreover the sensitivity data across NCI60 showed that the cytotoxicity profile of rucaparib is not correlated with olaparib and BMN 673, whereas olaparib and BMN 673 are significantly correlated with each other, as expected for drugs with similar target(s) (41) (Figure 3 and Table 2). Notably, the IC50 of BMN 673 was overall lower than that of olaparib in the olaparib-sensitive cell lines, which is consistent with the fact that BMN 673 has higher PARP-mediated cytotoxicity than olaparib.
Figure 3. Comparison of the sensitivity patterns in the three PARP inhibitors across the NCI60 cell lines.
The IC50 (drug concentration that reduces cell survival down to 50%) obtained from the NCI60 databases (35, 41) (http://discover.nci.nih.gov/cellminer) is plotted for each cell line. Cell lines are colored according to tissue of origin (41). NA: data not available.
Table 2.
Pearson’s correlation coefficient analyses of Figure 3 between the drugs.
| BMN 673 | Olaparib | Rucaparib | |
|---|---|---|---|
| BMN 673 | 1.00 | ||
| Olaparib | 0.52 | 1.00 | |
| Rucaparib | 0.04 | 0.16 | 1.00 |
Numbers in Italic indicate highly significant correlations (p< 0.001).
BMN 673 traps PARP-DNA complexes ~100-fold more efficiently than olaparib and rucaparib
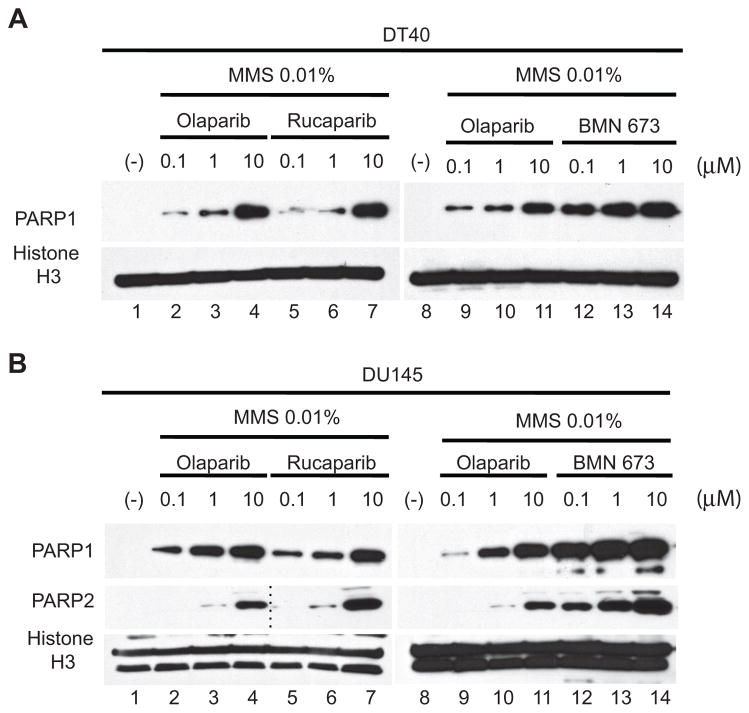
Trapping PARP1 and PARP2 on damaged DNA has recently be proposed as a mechanism accounting for the cytotoxicity of olaparib, niraparib, and to a lesser extent veliparib (27). Using a cellular assay to measure PARP trapping on damaged DNA (27), we examined chromatin-bound PARP1 and PARP2 (Figure 4). Wild type DT40 and prostate cancer DU145 cells were treated with different concentrations of PARP inhibitors in the presence of 0.01 % MMS to produce base damage that recruits PARP-1/2 (Figure 4). DT40 cells only have PARP1 (no PARP2) (Figure 4A) whereas DU145 have both PARP1 and PARP2 (Figure 4B) (27). PARP1 and PARP2 were not detectable in chromatin bound fractions without drug exposure (lanes 1 and 8, Figure 4A and B). While olaparib and rucaparib induced similar amounts of PARP-DNA complexes (lanes 2–7, Figure 4A and B), 0.1 μM BMN 673 induced equivalent levels of PARP–DNA complexes as 10 μM olaparib (lanes 11 and 12, Figure 4A and B), indicating that BMN 673 is ~100-fold more potent than olaparib and rucaparib at trapping PARP1 and PARP2.
Figure 4. Comparative trapping of PARP1- and PARP2-DNA complexes by BMN 673, olaparib and rucaparib.
PARP-DNA complexes were determined by Western blotting analyses of chromatin bound fractions from drug-treated DT40 cells (A) and DU145 cells (B). DT40 and DU145 treatments were for 30 min and 4 hours, respectively. Blots were probed with the indicated antibodies. Histone H3 was used as positive markers for chromatin-bound fractions and as loading control. The blots of lanes 1–4 are identical to the blots of lanes 8–11 for PARP2 in (B). The blots are representative of multiple experiments.
To further investigate the differential trapping of PARP-DNA complexes by BMN 673 at the molecular level, we expanded PARP1-DNA binding using fluorescence anisotropy (27). A nicked oligonucleotide duplex DNA with a single 5’-dRP end at the break site was used as a fluorescent substrate (Figure 5A). Its anisotropy was enhanced upon PARP1 binding to the damaged DNA site. PARylation following addition of NAD+ reduced the fluorescence anisotropy signal by freeing the DNA. Figure 5B shows that both PARP inhibitors enhanced the fluorescence anisotropy signal, which reflects the stabilization of PARP1-DNA complexes. BMN 673 was approximately 40-fold more potent than olaparib. Time-course experiments following NAD+ addition also showed that BMN 673 slowed the dissociation of PARP1-DNA complexes more efficiently than olaparib (Figure 5C). Together, these results demonstrate that BMN 673 is markedly more effective at trapping PARP than olaparib and rucaparib.
Figure 5. Biochemical trapping of PARP1 by BMN 673.
(A) Scheme of the fluorescence anisotropy (FA) binding assay. The star indicates the site labeled on the DNA substrate with Alexa Fluor488. Unbound nicked DNA substrate rotates fast and gives low FA. PARP1 binding to the substrate slows the rotation and gives high FA. Addition of NAD+ leads to PARP1 dissociation from DNA due to autoPARylation. (B) Concentration-dependent PARP1-DNA association in the presence of BMN 673 or olaparib. FA was measured 40 min after adding NAD+. (C) Time-course of PARP1-DNA dissociation in the presence of BMN 673 and olaparib (0.12 μM each). Addition of NAD+ in the absence of PARP inhibitor immediately reduces PARP1-DNA complexes (DMSO control). In the absence of NAD+, PARP1-DNA complexes remain stable for at least 120 min (no NAD+).
BMN 673 is substantially more potent than olaparib and rucaparib in combination with temozolomide and base damaging agents
The lack of detectable off-target effects of BMN 673 (with respect to PARP-1/2) (see Figures 1–2 and S4), and its high potency to trap PARP-DNA complexes (Figures 4–5), led us to test the combinations of BMN 673 with alkylating agents in comparison with olaparib and rucaparib. As expected from the well-established role of PARylation for SSB repair, PARP1−/− cells were hypersensitive to MMS (compare open and closed circles in panels A and B, Figure 6). Consistent with the recently established role of PARP-DNA complexes in the cytotoxicity of PARP inhibitors (27, 39), each drug had no impact on MMS sensitivity in PARP1−/− cells (Figure 6B), confirming the lack of off-target effects of the three PARP inhibitors tested up to 1 μM concentration.
Figure 6. BMN 673 enhances the cytotoxicity of alkylating agents more efficiently than olaparib and rucaparib.
(A) Survival curves of wild type DT40 cells treated with MMS alone (upper curves labeled “0”) or with the indicated concentrations of PARP inhibitors (at the concentration shown beside each curve in micromolar units). The survival of untreated cells was set as 100%. Data are mean ± SD (n ≥ 3). (B) PARP1−/−cells are hypersensitive to MMS (compare with upper curves in panel A) and resistant to the PARP inhibitors. (C) Survival curves of the indicated human cancer cells treated with MMS in combination with the indicated PARP inhibitors (the concentration of each PARP inhibitor is shown beside each curve). The survival of untreated cells was set as 100%. Data are mean ± SD (n ≥ 3). (D) Same as C but using temozolomide instead of MMS in prostate cancer DU145 cells.
On the other hand, the three drugs produced supra-additive effects in wild type DT40 cells treated with MMS in a concentration-dependent manner (Figure 6A, upper three panels). Notably, the MMS sensitivity of wild type cells treated with 0.1 μM olaparib, 0.1 μM rucaparib or 0.001 μM BMN 673 was greater than that of PARP1−/−cells (Figure 6, compare upper three A panels and the B panel). These results indicate that BMN 673 induces PARP-mediated cytotoxicity ~100 times more efficiently than olaparib or rucaparib and that its cytotoxicity is mediated not only by PARP catalytic inhibition but also by trapping PARP-DNA complexes.
Human prostate cancer DU145 and Ewing’s sarcoma EW8 cells also showed supra-sensitization to MMS at submicromolar concentration for all three PARP inhibitors (Figure 6C). However, BMN 673 was clearly more effective than olaparib or rucaparib (Figure 6C, top and middle panels). Notably, breast cancer MDA-MB231 cells, a cell line tolerant to PARP inhibitors (see above), were not markedly sensitized to MMS even with 1μM of each PARP inhibitor (Figure 6C, top panel).
To extend these findings to a clinically relevant combination, we tested temozolomide, an alkylating agent known to act synergistically with other PARP inhibitors (9, 42, 43). Figure 6D shows that all three PARP inhibitors enhanced the cytotoxicity of temozolomide, with BMN 673 being markedly more potent than olaparib and rucaparib. These results demonstrate that BMN 673 is the most potent drug among the three PARP inhibitors tested in combination with temozolomide.
Discussion
In this study, we report that BMN 673 is the most potent PARP trapping drug tested to date. It is approximately 2 orders of magnitude more potent than olaparib both in prostate cancer DU145 and lymphoma DT40 cells for both PARP1 and PARP2 (see Figure 4). We also show that olaparib and rucaparib trap PARP1 and PARP2 with comparable efficiency. The present results complement our recent study (27) revealing PARP-DNA complex trapping, and comparing olaparib with niraparib and veliparib. Veliparib differed from the two other drugs by its much weaker ability to trap PARP-DNA complexes in spite of its remarkable activity as a PARP catalytic inhibitor (27).
Our data indicate that BMN 673 is only slightly more potent than olaparib and rucaparib at inhibiting PARP catalytic activity. The differential potencies of the drugs at trapping PARP vs. inhibiting PARP catalytic activity may possibly be interpreted as resulting from an allosteric effect of the drugs (27). As shown in Figure 1A, the chemical structure of BMN 673 is rigid, whereas olaparib and rucaparib are flexible. This might explain their weaker impact on PARP trapping. The binding of PARP inhibitor to the catalytic pocket of PARP1 and PARP2 may enhance the binding between DNA and the DNA binding domains of PARP, which would be the converse allosteric effect produced by the binding of PARP to DNA, which induces conformational distortions that stimulate the catalytic domain (44). Notably, LT674, the inactive enantiomer of BMN 673, is markedly less active than BMN 673 both at PARP-mediated cytotoxicity and at inhibiting its catalytic activity. This difference between the enantiomers reflects the optimal structure of BMN 673 for PARP binding and the inability of LT674 to fit in the nicotinamide binding pocket (45). We believe that BMN 673 can now be viewed not only as a valuable anticancer agent but also as a molecular tool to elucidate PARP allosteric regulation. For the comprehensive understanding of the mechanism of differential PARP trapping, further studies such as co-crystal structure analysis will be required.
The nanomolar cytotoxicity of BMN 673 is notably greater than that of rucaparib or olaparib (≥10-fold in lymphoma DT40 and prostate cancer DU145 and ≥ 5-fold in Ewing’s sarcoma EW8 cells) (see Figure 2). The potency of BMN 673 as a cytotoxic agent was just reported independently (46). However, these studies did not examine the molecular mechanism of action of BMN 673, especially with respect to PARP trapping. The potency of BMN 673 observed across the NCI60 cell line panel (see Figure 3) showed significant correlation between BMN 673 and olaparib (Table 2). The greater cytotoxic potency of BMN 673 over olaparib and rucaparib can be related to the trapping of PARP-DNA complexes because knocking out PARP1 in lymphoma DT40 cells, which by itself is well-tolerated in spite of the fact that DT40 cells also lack PARP2 (29), conferred extreme resistance to BMN 673 (and olaparib) (see Figure 2). Moreover, the greater cytotoxicity of BMN 673 compared to olaparib is correlated with the greater potency of BMN 673 at trapping PARP (see Figures 4 and 5) while both drugs are equally effective at inhibiting PARP catalytic activity (see Figure 2). The BMN 673 findings reinforce our proposal (27) that PARP inhibitors should be categorized and evaluated based on both PARP inhibition and PARP trapping.
Our study shows that rucaparib exhibits off-target effect with respect to PARP1 and PARP2 (Figure 2C), which fits with a previous report showing that rucaparib has more promiscuous inhibitory activity (extending to PARP1–4 and tankyrases) than olaparib (specific to PARP1–4) (47). The NCI60 data also revealed the differences between rucaparib and BMN 673 or olaparib, and the general cytotoxicity of rucaparib irrespective of cell lines and tissue origin (Figure 3). We speculate that the inhibition of tankyrases may contribute to the broader cytotoxicity of rucaparib as tankyrase-1 RNAi results in mitotic arrest (48).
Our results provide relevant information for the clinical use of PARP inhibitors. As single agent in BRCA-deficient cells, we found that BMN 673 demonstrates ≈ 30-fold greater potency in isogenic BRCA2-deficient lymphoma DT40 cells (see Figure 2). Consistent results have just been reported independently using other cellular systems (45). BMN 673 is also significantly more potent than olaparib in combination with temozolomide or MMS (see Figure 6) (45), which is consistent with the enhanced trapping of PARP by BMN 673 and olaparib in the presence of MMS (see Figure 4). In spite of the fact that BMN 673 is a highly potent drug by inducing PARP-DNA complexes, it is surprising that half of the NCI60 cell lines are resistant even at 100 μM of BMN 673 (see Figure 3). Further studies are warranted to elucidate why some cell lines are tolerant or selectively sensitive to PARP trapping. One possibility is that sensitive cell lines are deficient in post-replication repair, Fanconi anemia pathway, ATM, homologous recombination (27) or PTEN (49). It will also be important to determine whether the resistant cells exhibit preferential homologous recombination by 53BP1 inactivation (50, 51).
Supplementary Material
Acknowledgments
J. Murai is a recipient of a fellowship from the Japan Society for the Promotion of Science (JSPS) and the Kyoto University Foundation. Y. Pommier and J. Murai were supported by the Intramural Program of the National Cancer Institute, Center for Cancer Research (Z01 BC 006150). J. Murai and S. Takeda were supported by JSPS Core-to-Core Program. J. Murai was supported by JSPS KAKENHI Grant Number 25740016.
We wish to thank the Intramural Program of the National Cancer Institute, Center for Cancer Research (CCR), by the Developmental Therapeutics Program (DTP), Division of Cancer Treatment and Diagnosis (DCTD) for sharing the NCI60 data and drugs. We also thank CRADA with BioMarin Pharmaceuticals Inc. for providing BMN 673 and LT674 compounds.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 3.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee S, Kaye SB, Ashworth A. Making the best of PARP inhibitors in ovarian cancer. Nature reviews Clinical oncology. 2010;7:508–19. doi: 10.1038/nrclinonc.2010.116. [DOI] [PubMed] [Google Scholar]
- 6.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 7.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–20. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney CA, Wang LZ, Kyle S, White AW, Calvert AH, Curtin NJ, et al. Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin Cancer Res. 2000;6:2860–7. [PubMed] [Google Scholar]
- 10.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–23. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 11.Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol. 2013;20:347–54. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 13.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 14.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5'-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–87. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5:387–93. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noel G, Godon C, Fernet M, Giocanti N, Megnin-Chanet F, Favaudon V. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1,8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Mol Cancer Ther. 2006;5:564–74. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 17.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–69. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 21.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: Anticancer therapy and beyond. Molecular aspects of medicine. 2013;34:1217–56. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 23.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–90. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 24.Kummar S, Chen A, Parchment RE, Kinders RJ, Ji J, Tomaszewski JE, et al. Advances in using PARP inhibitors to treat cancer. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat. 2012;134:649–59. doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced Killing of Cancer Cells by Poly(ADP-ribose) Polymerase Inhibitors and Topoisomerase I Inhibitors Reflects Poisoning of Both Enzymes. J Biol Chem. 2012;287:4198–210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–8. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 29.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. Embo J. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji J, Kinders RJ, Zhang Y, Rubinstein L, Kummar S, Parchment RE, et al. Modeling pharmacodynamic response to the poly(ADP-Ribose) polymerase inhibitor ABT-888 in human peripheral blood mononuclear cells. PLoS One. 2011;6:e26152. doi: 10.1371/journal.pone.0026152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Division of Cancer Treatment and Diagnosis [Internet] Bethesda (MD): National Cancer Institute, National Institutes of Health (US); [updated 2013 Aug 16; cited 2013 Oct 10]. Available from: http://dctd.cancer.gov/ResearchResources/biomarkers/PolyAdenosylRibose.htm. [Google Scholar]
- 33.NCI-60 DTP Human Tumor Cell Line Screen [Internet] Bethesda (MD): National Cancer Institute, National Institutes of Health (US); [cited 2013 Oct 10]. Available from: http://dtp.nci.nih.gov/branches/btb/ivclsp.html. [Google Scholar]
- 34.Holbeck SL, Collins JM, Doroshow JH. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther. 2010;9:1451–60. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holbeck S, Chang J, Best AM, Bookout AL, Mangelsdorf DJ, Martinez ED. Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol Endocrinol. 2010;24:1287–96. doi: 10.1210/me.2010-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kummar S, Chen HX, Wright J, Holbeck S, Millin MD, Tomaszewski J, et al. Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nature reviews Drug discovery. 2010;9:843–56. doi: 10.1038/nrd3216. [DOI] [PubMed] [Google Scholar]
- 37.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–11. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, et al. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25:1124–34. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettitt SJ, Rehman FL, Bajrami I, Brough R, Wallberg F, Kozarewa I, et al. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PLoS One. 2013;8:e61520. doi: 10.1371/journal.pone.0061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grohar PJ, Griffin LB, Yeung C, Chen QR, Pommier Y, Khanna C, et al. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells. Neoplasia. 2011;13:145–53. doi: 10.1593/neo.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: A Web-Based Suite of Genomic and Pharmacologic Tools to Explore Transcript and Drug Patterns in the NCI-60 Cell Line Set. Cancer Res. 2012;72:3499–511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulton S, Pemberton LC, Porteous JK, Curtin NJ, Griffin RJ, Golding BT, et al. Potentiation of temozolomide-induced cytotoxicity: a comparative study of the biological effects of poly(ADP-ribose) polymerase inhibitors. Br J Cancer. 1995;72:849–56. doi: 10.1038/bjc.1995.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:7917–23. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langelier M-F, Planck JL, Roy S, Pascal JM. Structural Basis for DNA Damage-Dependent Poly(ADP-ribosyl)ation by Human PARP-1. Science. 2012;336:728–32. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, Rehman FL, Feng Y, Boshuizen J, Bajrami I, Elliott R, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clinical Cancer Research. 2013;19:5003–15. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postel-Vinay S, Bajrami I, Friboulet L, Elliott R, Fontebasso Y, Dorvault N, et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene. 2013;32:5377–87. doi: 10.1038/onc.2013.311. [DOI] [PubMed] [Google Scholar]
- 47.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature biotechnology. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 48.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature cell biology. 2005;7:1133–9. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 49.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim J-S, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. Embo Mol Med. 2009;1:315–22. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer discovery. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.