Abstract
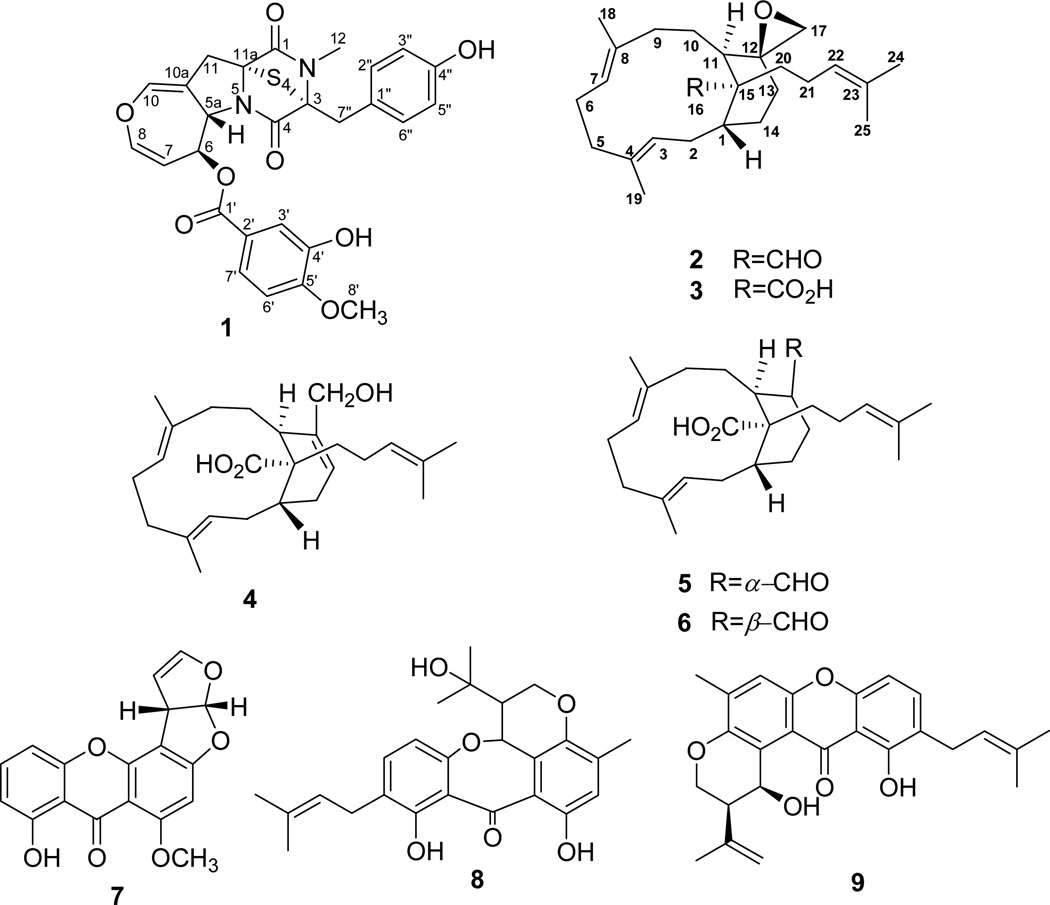
A new epitetrathiodioxopiperizine, secoemestrin D (1), five sesterterpenoids bearing a new carbon skeleton, emericellenes A–E (2–6), together with previously known fungal metabolites, sterigmatocystin (7), arugosin C (8), and epiisoshamixanthone (9), were obtained from the endophytic fungal strain Emericella sp. AST0036 isolated from a healthy leaf tissue of Astragalus lentiginosus. The planar structures and relative configurations of the new metabolites 1–6 were elucidated using MS, 1D and 2D NMR spectroscopic data. All compounds were evaluated for their potential anticancer activity using a panel of six tumor cell lines and normal human fibroblast cells. Only metabolites 1 and 7 showed cytotoxic activity. More importantly, secoemestrin D (1) exhibited significant cytotoxicity with IC50s ranging from 0.06–0.24 µM and moderate selectivity to human glioma (SF-268) and metastatic breast adenocarcinoma (MDA-MB-231) cell lines.
Fungal endophytes that colonize internal tissues of healthy plants represent one of the largest but least-explored sources of small-molecule natural products.2–4 These fungi are a fundamental feature of plant biology in biomes ranging from Arctic tundra to tropical rainforests and hot deserts.4 Although interactions between endophytes with their hosts are not fully understood in most cases, many endophytes produce bioactive small-molecule natural products that may protect hosts from herbivores, plant pathogens, and abiotic stressors such as drought.5 Despite the fact that the existence of endophytes were first observed over a century ago, this group of microorganisms did not receive significant attention until the recent realization of their ecological relevance4 and the potential to yield metabolites with diverse structures and biological functions.6 In the course of our ongoing studies directed towards the discovery of potential anticancer agents1,7 and isolation of compounds new to the NIH Molecular Libraries Small-Molecule Repository (MLSMR), we have investigated a large number of endophytic fungi among which Emericella sp. AST0036 was found to be one of those promising. Emericella species are the perfect states of Aspergillus, a common genus of Ascomycota (Trichocomaceae, Eurotiales, Eurotiomycetes, Pezizomycotina),8 and it is noteworthy that both Emericella and Aspergillus species are known to produce carcinogenic mycotoxins.9
An EtOAc extract derived from a solid (potato dextrose agar, PDA) culture of the endophytic fungal strain, Emericella sp. AST0036, isolated from a healthy leaf tissue of Astragalus lentiginosus (spotted locoweed, Fabaceae) was found to be active in the resazurin (alamarBlue®) cell viability assay for cell proliferation/survival.7 Bioactivity-guided fractionation of this extract provided a new epitetrathiodioxopiperizine, named secoemestrin D (1), and five new sesterterpenes, emericellenes A–E (2–6), together with sterigmatocystin (7),10 arugosin C (8),11 and epiisoshamixanthone (9)12 of which 1 and 7 were found to be cytotoxic. Metabolites 2–6 contain a hitherto unprecedented bicarbocyclic sesterterpene molecular scaffold which we have named as emericellane skeleton. Herein we report the isolation, characterization, and biological evaluation of 1–9 from Emericella sp. AST0036. Previous investigations of Emericella species have led to the identification of mycotoxins,9 xanthones,13 epithiodiketopiperazines,14 variecolin-type sesterterpenoids,15 and steroids.16
RESULTS AND DISCUSSION
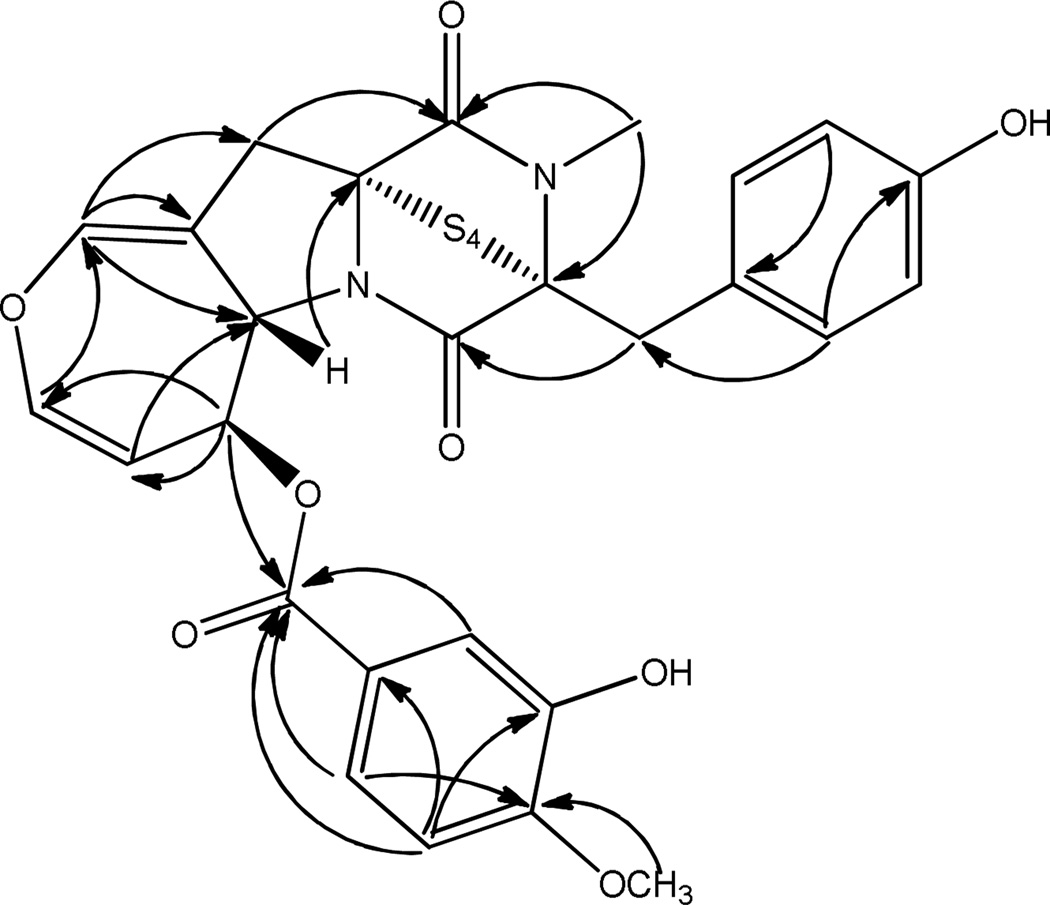
Secoemestrin D (1) was obtained as an off-white amorphous solid that analyzed for C27H24N2O8S4 by a combination of HRESIMS and NMR data and indicated 17 degrees of unsaturation. The positive APCI-LRMS ion at m/z 505 [M – S4]+ (base peak) suggested the presence of a tetrasulfide moiety in 1 and IR absorption bands at 1720 (sh), 1682, and 1663 cm−1 indicated that it contained an ester and two amide groups. The 13C NMR spectrum of 1 displayed 27 signals consisting of 2 methyl, 2 methylene, 12 methine (of which 10 were aromatic/olefinic), and 11 quaternary (of which 3 were carbonyls and 6 were aromatic/olefinic) carbons as judged by the DEPT spectrum. The signals due to carbonyl groups at δc 168.3 and 164.0 and those at δc 78.7 and 74.2 due to quaternary carbons bearing the sulfide moiety were typical of epipolythiodioxopiperazines.17 The presence of signals due to a dihydrooxepine moiety [δH 6.58 (1H, brd, J = 1.6 Hz), δc 138.6 (CH); δH 6.28 (1H, dd, J = 2.4, 8.0 Hz), δc 139.4 (CH); δH 4.82 (1H, dd, J = 2.0, 8.4 Hz), δc 106.6 (CH); δH 5.46 (1H, ddd, J = 2.0, 2.4, 8.4 Hz), δc 71.5 (CH); δH 5.33 (1H, dd, J = 2.0, 8.4 Hz), δc 61.7 (CH); and δc 108.2 (C)], and a prominent peak at m/z 465 (C19H17N2O4S4) in the HREIMS due to the facile loss of the hydroxy and methoxy-substituted benzoic acid moiety from [M + H]+ suggested that the structure of 1 closely resembled that of secoemestrin C1.18 The 1H NMR spectrum of 1 (Table 1) also exhibited signals due to a 1,3,4-trisubstituted benzene ring [δ 7.81 (1H, d, J = 2.0 Hz), 6.98 (1H, d, J = 8.8 Hz), and 7.85 (1H, dd, J = 8.4, 2.0 Hz)], a 1,4-disubstituted benzene ring [δ 6.74 (2H, d, J = 8.4 Hz), 7.08 (1H, d, J = 8.4 Hz)], a benzylic methylene attached to a chiral center [δ 3.23 and 3.96 (1H each, d, J = 14.8 Hz)], an allylic methylene [δ 3.12 (m)], an OMe (δ 4.01) and an NMe (δ 3.19) groups. The dihydrooxepine and epitetrathiodioxopiperazine partial structures and all the above moieties were joined together with the help of HMBC correlations (Figure 2) suggesting that secoemestrin D is an epitetrathiodioxopiperizine analogue with the gross structure as depicted in 1. The coupling constant (8.4 Hz) for protons at C-5a and C-6 indicated that their relative configurations in secoemestrin D (1) to be β similar to those reported for emestrin.17
Table 1.
1H (400 MHz, CDCl3) and 13C NMR Data (100 MHz, CDCl3) for Secoemestrin D (1)
| position | δc, type | δH (J in Hz) |
|---|---|---|
| 1 | 168.3, C | |
| 3 | 78.7, C | |
| 4 | 164.0, C | |
| 5a | 61.7, CH | 5.33, dd (2.0, 8.4) |
| 6 | 71.5, CH | 5.46, ddd (2.0, 2.4, 8.4) |
| 7 | 106.6, CH | 4.82, dd (2.0, 8.4) |
| 8 | 139.4, CH | 6.28, dd (2.4, 8.0) |
| 10 | 138.6, CH | 6.58, brd (1.6) |
| 10a | 108.2, C | |
| 11 | 41.3, CH2 | 3.12, m |
| 11a | 74.2, C | |
| 1' | 166.2, C | |
| 2' | 123.0, C | |
| 3' | 116.4, CH | 7.81, d (2.0) |
| 4' | 145.2, C | |
| 5' | 150.8, C | |
| 6' | 110.0, CH | 6.98, d (8.8) |
| 7' | 123.8, CH | 7.85, dd (2.0, 8.4) |
| 8' | 56.0, CH3 | 4.01, s |
| 1" | 126.2, C | |
| 2", 6" | 130.7, CH | 7.08, dd (8.4) |
| 3", 5" | 115.7, CH | 6.74, d (8.4) |
| 4" | 155.1, C | |
| 7" | 38.5, CH2 | 3.23, d (14.8) |
| 3.96, d (14.8) |
Figure 2.
Key HMBC correlations for secoemestrin D (1)
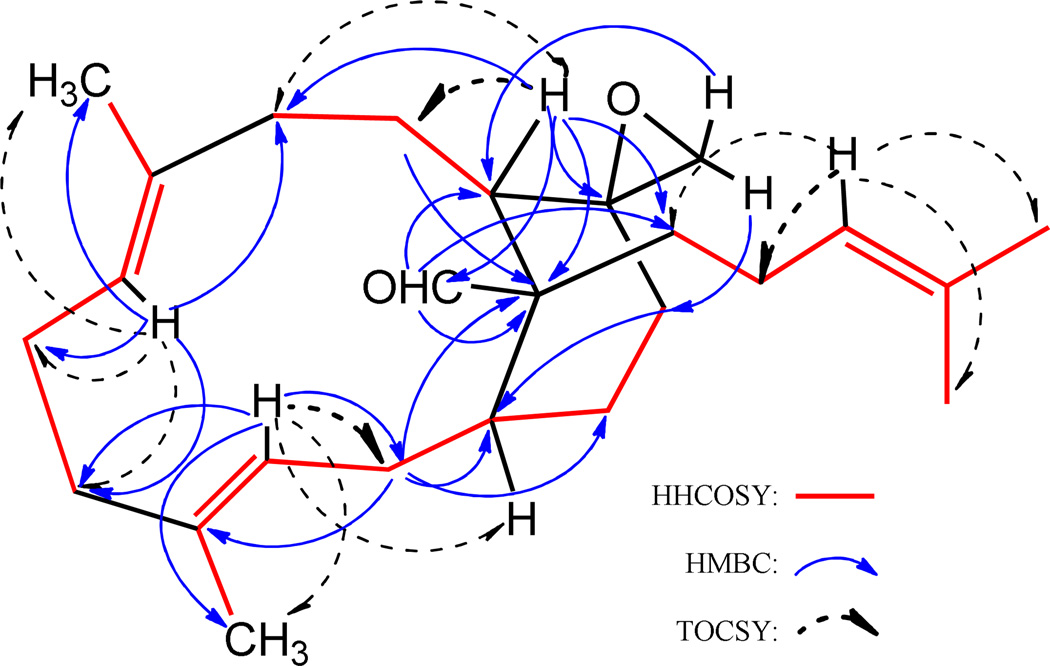
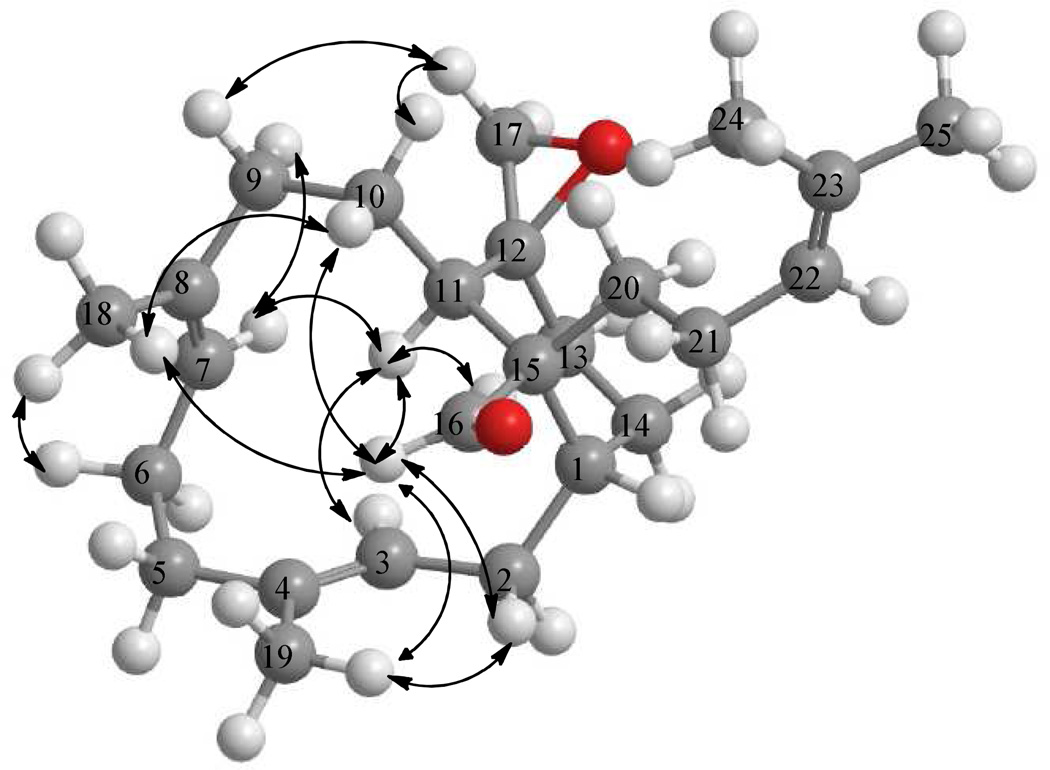
Emericellene A (2), obtained as a colorless amorphous solid, was determined to have the molecular formula C25H38O2 by a combination of HRESIMS and NMR data and indicated seven units of unsaturation. Its 1H NMR spectrum (Table 2) exhibited signals due to three olefinic protons [δ 5.60 (1H, d, J = 12.6 Hz), 4.85 (1H, d, J = 11.2 Hz), and 5.10 (1H, brt, J = 6.8 Hz)], two terminal oxirane protons [δ 2.67 (1H, d, J = 4.4 Hz) and 2.41 (1H, d, J = 4.4 Hz)], four vinylic methyl singlets (δ 1.63, 1.60, 1.50, and 1.48), and an aldehyde proton [δ 9.28 (s)]. The 13C NMR spectrum of 2 (Table 2) which displayed 25 carbon signals, when analyzed by DEPT and HSQC data indicated the presence of four CH3, ten CH2, six CH (including the CHO at δ 209.8 and three olefinic carbons at δ 130.8, 126.0, and 125.0), and five quaternary carbons (including three olefinic carbons at δ 135.0, 132.4, and 131.2). These data suggested that 2 is a bicyclic sesterterpenoid bearing a CHO attached to a tertiary carbon. Analysis of 1H–1H COSY and TOCSY spectra of 2 suggested the presence of the spin systems CH3–C=CH–CH2–CH–CH2–CH2 (A), CH3–C=CH–CH2–CH2 (B), CH2–CH2–CH (C), and (CH3)2C=CH–CH2–CH2 (D). These were linked together via quaternary carbons with the help of its HMBC data (Figure 2). Of significance were the HMBC correlations of H-3 and H-7 to C-5 linking spin systems A and B via C-4, those of H-7 to C-6 and C-9 connecting spin systems B and C via C-8, and those of H-2 and H-10 to C-15 linking C-1 of A with C-11 of C via C-15 suggesting the presence of a 12-membered carbocyclic ring system in 2. The attachments of C-20 of the spin system D and the CHO moiety to C-15 were inferred from the HMBC correlations of H-11/C-20, CHO/C-15, CHO/C-20, and CHO/C-11. The spin systems A–D, CHO, and the quaternary carbon C-15 accounted for C23H26O and five units of unsaturation and the remaining C2H2O and two units of unsaturation suggested the presence of another cyclic moiety in addition to the terminal oxirane ring (see above). The HMBC correlations of each of the CH2 protons of this terminal oxirane moiety to C-11 and C-13 and H-11 to the remaining carbon (C-12) of this moiety suggested that the terminal CH2 of the spin system A is connected to the CH terminal of the spin system C via C-12 generating a [9,3,1] bicarbocyclic ring similar to that of verticillane-type diterpenenoids. However, the presence of the spin system D at C-15 suggested that 2 is a sesterterpenoid containing a new emericellane-type skeleton. The relative stereochemistry including the disposition of CHO and terminal oxirane groups in 2 were determined by the application of NOE data to the MM2 energy-minimized conformation for the confirmed E-configuration of the C3–C4 and C7–C8 double bonds. The CHO showed strong NOE to CH3-18 and CH3-19 suggesting that these groups lie on the same side of the molecule and that CHO at C-15 has equatorial α-orientation. Strong NOE correlations observed between one of the CH2 protons of the terminal oxirane and H-9α, H-9α with H-7α, and H-7α with H-11α suggested that this oxirane is of β-orientation. Emericellene A was thus identified as (+)-12β,17-epoxy-emericella-3E,7E,22-trien-16-al (2).
Table 2.
1H (400 MHz, CDCl3) and 13C NMR Data (100 MHz, CDCl3) for Emericellenes A–E (2–6)*
| 2 | 3 | 4 | 5 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| position | δH (J in Hz) | δc, type | δH (J in Hz) | δc, type | δH (J in Hz) | δc, type | δH (J in Hz) | δc, type | δH (J in Hz) | δc, type |
| 1 | 2.07, m | 39.3, CH | 2.08, m | 43.7, CH | 2.13, m | 39.3, CH | 2.06, m | 42.3, CH | 1.97, m | 43.9, CH |
| 2 | 2.63, ddd (5.4, 14.2, 14.2) 2.03, m |
33.7, CH2 | 2.55, ddd (4.4, 14.2, 14.2) 2.04, m |
34.3, CH2 | 2.50, m 1.95, m |
34.4, CH2 | 2.58, brt (14.4) 2.03, m |
34.2, CH2 | 2.47, m 2.02, m |
34.0, CH2 |
| 3 | 5.60, brd (12.6) |
126.0, CH | 5.47, brd (11.6) |
124.0, CH | 5.17, brd (11.6) |
121.9, CH | 5.49, brd (11.6) |
124.9, CH | 5.50, brd (10.4) |
124.9, CH |
| 4 | 135.0, C | 135.6, C | 135.5, C | 135.5, C | 135.2, C | |||||
| 5 | 2.22, m 2.06, m |
40.9, CH2 | 2.17, m 2.01, m |
41.1, CH2 | 2.13, m 1.91, m |
41.0, CH2 | 2.19, m 2.05, m |
41.3, CH2 | 2.18, m 2.01, m |
41.0, CH2 |
| 6 | 2.42, m 2.04, m |
26.4, CH2 | 2.46, m 1.99, m |
26.3, CH2 | 2.47, m 1.97, m |
26.7, CH2 | 2.46, m 2.00, m |
26.4, CH2 | 2.49, m 2.02, m |
26.2, CH2 |
| 7 | 4.85, brd (11.2) |
130.8, CH | 4.66, brd (10.8) |
128.7, CH | 4.81, brd (11.2) |
128.9, CH | 4.69, brd (10.8) |
129.0, CH | 4.70, brd (10.8) |
128.1, CH |
| 8 | 132.4, C | 133.6, C | 133.0, C | 133.3, C | 133.4, C | |||||
| 9 | 2.03, m 1.70, m |
39.6, CH2 | 2.00, m 1.72, m |
39.8, CH2 | 2.17, m 2.08, m |
38.6, CH2 | 1.97, m 1.67, m |
37.9, CH2 | 2.10, m 2.03, m |
37.1, CH2 |
| 10 | 1.13, m 0.80, ddd (2.8, 12.8, 12.8) |
21.5, CH2 | 1.53, m 0.97, brdd (10.2, 13.8) |
21.4, CH2 | 1.57, m | 24.8, CH2 | 1.81, m 1.24, m |
27.5, CH2 | 1.92, m 1.88, m |
24.1, CH2 |
| 11 | 3.26, d (9.2) |
34.3, CH | 3.16, d (9.6) |
34.0, CH | 3.76, brs | 34.4, CH | 3.17, dd (9.2, 11.6) |
32.9, CH | 3.27, dd (5.2, 10.0) |
34.6, CH |
| 12 | 56.6, C | 57.6, C | 139.1, C | 2.44, m | 53.7, CH | 2.52, m | 45.2, CH | |||
| 13 | 2.31, m 1.28, m |
33.3, CH2 | 2.31, m 1.20, m |
33.6, CH2 | 5.83, brd (2.4) |
124.1, CH | 1.99, m 1.56, m |
23.4, CH2 | 2.27, m 1.79, m |
21.7, CH2 |
| 14 | 2.25, m 1.63, m |
27.5, CH2 | 2.32, m 1.67, m |
25.9, CH2 | 2.54, m 2.05, m |
29.4, CH2 | 2.04, m 1.75, m |
26.2, CH2 | 1.99, m 1.59, m |
24.6, CH2 |
| 15 | 57.3, C | 56.2, C | 54.6, C | 54.0, C | 54.0, C | |||||
| 16 | 9.28, s | 209.8, CH | 181.7, C | 182.2, C | 180.4, C | 181.2, C | ||||
| 17 | 2.67, d (4.4) 2.41, d (4.4) |
49.0, CH2 | 2.74, d (4.0) 2.41, d (4.0) |
50.2, CH2 | 4.21, s | 65.4, CH2 | 9.77, d (4.8 Hz) |
204.8, CH | 10.14, s | 205.7, CH |
| 18 | 1.48, s | 15.3, CH3 | 1.57, s | 16.4, CH3 | 1.57, s | 16.7, CH3 | 1.54, s | 16.9, CH3 | 1.63, s | 16.4, CH3 |
| 19 | 1.50, s | 14.9, CH3 | 1.49, s | 15.5, CH3 | 1.50, s | 15.6, CH3 | 1.47, s | 15.4, CH3 | 1.45, s | 15.4, CH3 |
| 20 | 1.76, m 1.69, m |
31.8, CH2 | 2.38, m 1.57, m |
37.1, CH2 | 1.69, m 1.45, m |
33.8, CH2 | 1.96, m 1.58, m |
34.5, CH2 | 1.58, m | 37.4, CH2 |
| 21 | 2.08, m 1.75, m |
24.1, CH2 | 2.10, m 1.77, m |
25.9, CH2 | 2.22, m 1.95, m |
25.0, CH2 | 2.13, m 1.89, m |
25.1, CH2 | 2.08, m 1.90, m |
25.6, CH2 |
| 22 | 5.10, brt (6.8) |
125.0, CH | 5.11, brt (7.2) |
124.8, CH | 5.04, brt (7.2) |
124.9, CH | 5.06, brt (6.8) |
124.1, CH | 5.00, ddd (1.2, 6.0, 6.0) |
123.8, CH |
| 23 | 131.2, C | 131.3, C | 131.4, C | 132.0, C | 132.0, C | |||||
| 24 | 1.63, s | 25.7, CH3 | 1.64, s | 25.7, CH3 | 1.65, s | 25.7, CH3 | 1.66, s | 25.7, CH3 | 1.64, s | 25.6, CH3 |
| 25 | 1.60, s | 17.4, CH3 | 1.57, s | 17.5, CH3 | 1.57, s | 17.6, CH3 | 1.57, s | 17.6, CH3 | 1.55, s | 17.6, CH3 |
Signals were assigned by the analysis of 1H–1H COSY, HSQC, and HMBC spectra; coupling constants in Hz are in parenthesis.
Emericellenes B–E (3–6) were also obtained as colorless amorphous solids and were determined to have the common molecular formula, C25H38O3, on the basis of their HRESIMS and NMR data indicating seven units of unsaturation and the presence of an additional oxygen atom compared to emericellene A (2). Comparison of 1H and 13C NMR data of 3–6 (Table 2) with those of 2 indicated that they all contained the basic emericella-3E,7E,22-triene carbon skeleton, additional evidence for which was obtained by the analysis of their HMBC and NOESY data (see Supporting Information, S25 and S26, respectively). The most significant difference observed for emericellene B (3) was the absence of signals at δH 9.28 (s) and δC 209.8 (CH) but the presence of a signal due to a quaternary carbon at δC 181.7 suggesting that the aldehyde group in 2 is oxidized to a carboxylic acid leading to 3. The relative configurations of the C3–C4 and C7–C8 double bonds and the chiral centers at C-1, C-11 and C-12 in 3 were shown to be the same as those of 2 by its NOESY data (Supporting Information, S26). Considering the possible biosynthetic origin of 3 from 2, the configuration at C15 of 3 was assumed to be the same as that of 2. The foregoing led to the identification of emericellene B as (+)-12β,17-epoxy-emericella-3E,7E,22-trien-16-oic acid (3).
Comparison of 1H and 13C NMR data (Table 2) of emericellene C (4) with those of 3 suggested that in 4 the terminal oxirane moiety of 3 [δH 2.74 and 2.41 (each 1H, d, J = 4.0 Hz); δC 50.2 (CH2) and 57.6 (C)] is replaced by a CH2OH moiety on a double bond [δH 4.21 (2H, s) and 5.83 (1H, br d, J = 2.4 Hz); δC 65.4 (CH2), 139.1 (C), and 121.4 (CH)] suggesting that the 12,17-oxirane in 3 has undergone a ring opening reaction with concomitant rearrangement to a 12,13-en-17-ol moiety giving rise to 4 (for probable biosynthetic pathway from 3 to 4, see Supporting Information, S27). The presence of this moiety in 4 was confirmed by the HMBC data which showed correlations of H-17 (δ 4.21) with C-11 (δ 34.4), C-12 (δ 139.1), and C-13 (δ 124.1) (Supporting Information, S25). The strong NOESY observed for H-13 and H-17 suggested Z configuration for the C12–C13 double bond. Thus, emericellene C was identified as (+)-17-hydroxy-emericella-3E,7E,12Z,22-tetraen-16-oic acid (4).
The 1H and 13C NMR spectroscopic data of emericellene D (5) (Table 2) and comparison of these data with those of 3 revealed absence of the signals due to the terminal oxirane moiety, but the presence of a CH–CHO moiety [δH 9.77 (1H, d, J = 4.8 Hz); δC 204.8 (CH) and δH 2.44 (m); δC 53.7 (CH)] suggesting that the 12,17-epoxide moiety in 3 has undergone a concomitant ring opening and rearrangement to an aldehyde moiety (for probable biosynthetic pathway from 3 to 5, see Supporting Information, S27). The orientation of the CHO group at C-11 of 5 was determined as α by the strong NOESY observed between CHO and H-7 (Supporting Information, S26). These data led to the identification of emericellene D as (+)-emericella-3E,7E,22-trien-17α-al-16-oic acid (5).
Comparison of the 1H and 13C NMR data (Table 2) of emericellene E (6) with those of 5 suggested that they have similar structures except for the configuration of the CHO group at C-12 which was evident from the chemical shift and multiplicity of the NMR signals for CHO [δH 9.77 (1H, d, J = 4.8 Hz); δC 204.8 (d) for 5, and δH 10.14 (br s); δC 205.7 (d) for 6], and H-12 [δH 2.44 (m); δC 53.7 (CH) for 5, and δH 2.52 (m); δC 45.2 (CH) for 6]. The orientation of the CHO group at C-11 of 6 was confirmed to be β by the strong NOEs of CHO with H-20, and H-12 with H-11 (Supporting Information, S26). These data led to the identification of emericellene E as (+)-emericella-3E,7E,22-trien-17β-al-16-oic acid (6). Although the purified epimeric aldehydes 5 and 6 did not undergo any interconversion, it cannot be ruled out that only one of these is the genuine natural product and the other is an artifact formed as a result of epimerization during the isolation process (see Supporting Information, S27). Compounds 7–9 were identified as sterigmatocystin (7),10 arugosin C (8),11 and epiisoshamixathone (9)12 respectively, by comparison of their low-resolution MS and NMR spectroscopic data with those reported.
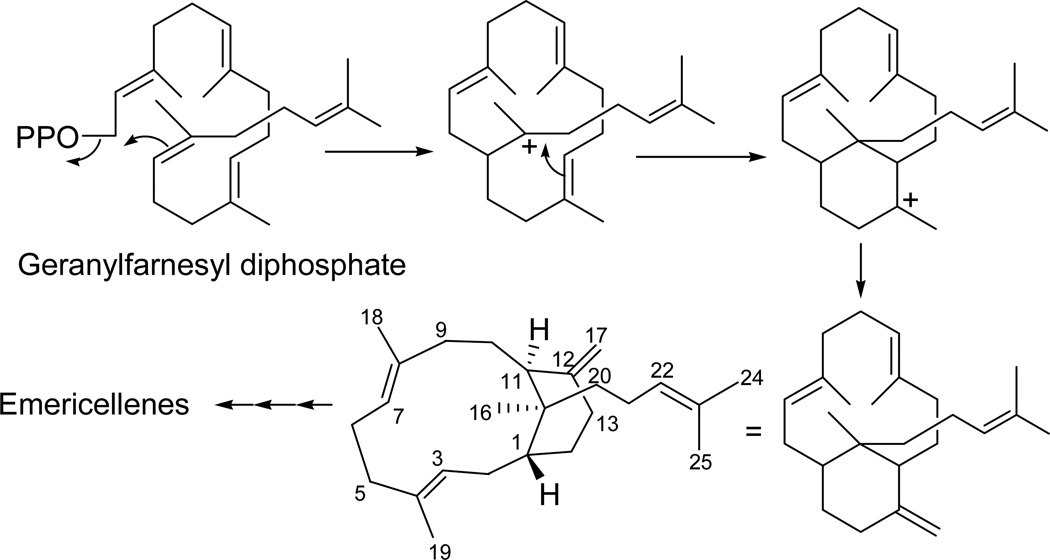
All compounds were evaluated for their potential anticancer activity using a panel of six tumor cell lines, NCI-H460 (human non-small cell lung cancer), SF-268 (human CNS cancer; glioma), MCF-7 (human breast cancer), PC-3M (metastatic human prostate adenocarcinoma), MDA-MB-231 (human breast adenocarcinoma), CHP-100 (human neuroblastoma), and normal human fibroblast cells, WI-38. Of those tested, secoemestrin D (1) exhibited strong cytotoxic activity with some selectivity against the six cancer cell lines compared to normal human fibroblast cells; among the six cell lines used, a moderate selectivity was observed for SF-268 and MDA-MB-231 (Table 3). Sterigmatocystin (7) showed moderate cytotoxicity with no apparent selectivity (Table 3). Metabolites 2–6 and 8–9 were found to be devoid of any cytotoxic activity up to a concentration of 5.0 µM. It is noteworthy that epipolythiodioxopiperazines have previously been studied for their antitumor activity and their mechanism of cytotoxicity has been suggested to be related to conjugation to the targets with susceptible thiol residues and subsequent inactivation or due to the generation of reactive oxygen species involved in redox cycling.19 The mycotoxin, sterigmatocystin (7), has been reported to be cytotoxic to cultured Chinese hamster cells,20 and to have 80-fold higher toxicity to the A-549 lung cancer cell line compared to Hep-G2 liver cells.21 The structures of emericellenes A–E (2–6) represent a unique class of sesterterpenoid metabolites bearing a novel emericellane-type bicarbocyclic ring system. Biosynthetically, emericellenes may be formed from geranylfarnesyl diphosphate by a cyclization pathway (Figure 5) similar to the formation of verticillane-type diterpenoids from geranylgeranyl diphosphate.22
Table 3.
Cytotoxicity Data for Secoemestrin D (1) and Sterigmatocystin (7)a
| Compound | Cell linesb | ||||||
|---|---|---|---|---|---|---|---|
| NCI-H460 | SF-268 | MCF-7 | PC-3M | MDA-MB-231 | CHP-100 | WI-38 | |
| 1 | 0.15±0.01 | 0.06±0.01 | 0.14±0.01 | 0.17±0.01 | 0.06±0.01 | 0.10±0.01 | 0.24±0.01 |
| 7 | 3.41±0.55 | 2.96±0.36 | 3.63±0.93 | 3.23±0.40 | 2.75±0.46 | NT | NT |
| Doxorubicin | 0.05±0.01 | 0.31±0.04 | 0.35±0.08 | 0.27±0.03 | 0.68±0.0.05 | 0.79±0.10 | NT |
Results are expressed as IC50 value ± standard deviation in µM. Doxorubicin and DMSO were used as positive and negative controls. NT = Not tested.
Key: NCI-H460 = human non-small cell lung cancer; SF-268 = human CNS cancer (glioma); MCF-7 = human breast cancer; PC-3M = metastatic human prostate adenocarcinoma; MDA-MB-231 = human breast adenocarcinoma; CHP-100 = human neuroblastoma; WI-38 = human lung fibroblast
Figure 5.
Possible biosynthetic pathway to emericellane-type sesterterpenoids
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured with a Jasco Dip-370 polarimeter using MeOH as the solvent. UV spectra were recorded with Shimadzu UV 2601 spectrophotometer. IR spectra were obtained on a Shimadzu FT IR-8300 spectrophotometer. 1D and 2D NMR spectra were recorded with a Bruker Avance III 400 NMR instrument at 400 MHz for 1H NMR and 100 MHz for 13C NMR. Chemical shift values (δ) are given in parts per million (ppm) and the coupling constants are in Hz. Low-resolution and high-resolution MS were recorded on Shimadzu LCMS-DQ8000α and JEOL HX110A spectrometers, respectively. HPLC purifications were carried out on a 10 × 250 mm Phenomenex Luna 5µ C18 (2) column with Waters Delta Prep system consisting of a PDA 996 detector. MM2 energy minimizations of possible conformations of emericellenes were performed using CambridgeSoft Chembio3D Ultra.
Fungal Isolation and Identification
In June 2008, a healthy individual of Astragalus lentiginosus (spotted locoweed, Fabaceae) was collected from an open, disturbed area in the Verde Valley of central Arizona (34°34'5''N, 111°51'25", 965 m.a.s.l.). Healthy leaves were washed in tap water and cut into ca. 2mm2 segments that were surface-sterilized by agitating sequentially in 95% EtOH for 30 sec, 0.5 % NaOCl for 2 min, and 70% EtOH for 2 min.23 A total of 32 tissue segments was surface-dried under sterile conditions and then placed onto 2% malt extract agar in 100mm Petri plates. Plates were sealed with Parafilm and incubated under ambient light/dark condition at room temperature (ca. 21.5 °C) for 4 months. Emergent fungi were isolated into pure culture on 2% MEA, vouchered in sterile water, and deposited as living vouchers at the Robert L. Gilbertson Mycological Herbarium at the University of Arizona. One fungus of interest was used for the present study: isolate AST0036, which has been accessioned at the Robert L. Gilbertson Mycological Herbarium (accession AST0036). Total genomic DNA was isolated from fresh mycelium23 and the nuclear ribosomal internal transcribed spacers and 5.8s gene (ITS rDNA; ca. 600 base pairs [bp]) was amplified as single fragment by PCR.23 Positive amplicons were sequenced bidirectionally as described previously.23 A consensus sequence was assembled and basecalls were made by phred24 and phrap25 with orchestration by Mesquite,26 followed by manual editing in Sequencher (Gene Codes Corp.). Because the isolate did not produce diagnostic fruiting structures in culture, we compared the entire sequence against the GenBank database using BLAST.27 The top BLAST matches were primarily to cultured and uncultured Emericella. To clarify the phylogenetic placement and taxonomic assignment of AST0036, we downloaded the top 100 BLAST matches from GenBank and aligned AST0036 and the resulting data set automatically using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) with default parameters. The alignment was trimmed so that starting and ending points were generally consistent with the sequence length for AST0036 and adjusted manually in MacClade28 prior to analysis. The type species of Penicillium (P. expansum) was chosen as the outgroup taxon. The final data set consisted of 39 sequences and 561 characters. The data set was analyzed using maximum likelihood in GARLI29 using the GTR+I+G model of evolution as determined by ModelTest,30 followed by a bootstrap analysis with 1000 replicates. The analysis unequivocally placed the sequence with strong support within Emericella (see Supporting Information, S28). Several species of Emericella shared the same ITSrDNA sequence with one another, but AST0036 was not 100% similar to any sequence in the analysis nor to any sequence available in GenBank. Therefore, we designate it as Emericella sp. AST0036, pending morphological description.
Cultivation and Isolation of Metabolites of Emericella sp
The fungus was cultured in 40 T-flasks (800 mL), each containing 135 mL of PDA coated on five sides of the flasks, maximizing the surface area for fungal growth (total surface area/flask ca. 400 cm2). After incubation for 14 days at 28 °C, MeOH (250 mL/T-flask) was added and the flasks were shaken in an ultrasonic bath for 1 h at 25 °C, and the resulting extract was filtered through a layer of Celite 545. The filtrate was concentrated to about one-third of its volume in vacuo below 40 °C and was extracted with EtOAc (3 × 1500 mL). The EtOAc extract was concentrated to afford the crude extract (1.49 g). This extract which showed cytotoxicity activity was fractionated on a reversed-phase (RP) C-18 (40µ ; 100.0 g) open column. The column was eluted sequentially with 80% aq. MeOH (1.0 L), 90% aq. MeOH (1.0 L), and 100% MeOH (500 mL) and the resulting fractions were combined based on their TLC (SiO2; CHCl3–MeOH, 95:5) profiles to afford eight combined fractions A–H. Of these, fractions B and D were found to be cytotoxic. Fraction B (144.7 mg) was subjected to chromatographic separation on a column of Si gel (70 g) and eluted with CHCl3–MeOH (98:2) to afford 1 (3.5 mg). Fraction D (69.0 mg) was further purified by Si gel column chromatography (50 g) and eluted with CHCl3–MeOH (98:2) yielding 7 (62.1 mg). Fractions E (50.7 mg) and F (96.7 mg) were combined and purified by Si gel (50 g) column chromatography and elution with CHCl3–MeOH (98:2) followed by RP-HPLC purification (C-18; 90% aq. MeOH) gave 4 (16.6 mg, tR =10.0 min). Further fractionation of G (350 mg) by Si gel (50 g) column chromatography and elution with CHCl3–MeOH (99:1) followed by purification by HPLC (C-18; 90% aq. MeOH) afforded 3 (26.7 mg, tR =22.5 min), 5 (5.6 mg, , tR =18.5 min), 6 (8.6 mg, tR =20.0 min), and 8 (25.9 mg, tR =21 min). Fraction H (285 mg) on purification by Si gel (20 g) column chromatography and elution with CHCl3–MeOH (99:1) yielded 2 (10.9 mg) and 9 (10.2 mg).
Secoemestrin D (1): off-white amorphous solid; [α]25 D –230.9 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 210 (4.37), 260.5 (3.93), 295.5 (3.67) nm; FT-IR (KBr) νmax 1720 (sh), 1682, 1663 cm−1; 1H NMR and 13C NMR data, see Table 1; Positive HRESIMS m/z 655.0310 (calcd for C27H24N2O8S4Na, 655.0313).
Emericellene A (2): colorless amorphous solid; [α]25 D 15.7 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 201 (3.78) nm; FT-IR (KBr) νmax 1718 cm−1; 1H NMR and 13C NMR data, see Table 2; Positive HRESIMS m/z 370.2878 (calcd for C25H38O2, 370.2872).
Emericellene B (3): colorless amorphous solid; [α]25 D 55.9 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 203 (4.11) nm; FT-IR (KBr) νmax 1705 cm−1; 1H NMR and 13C NMR data, see Table 2; Positive HRESIMS m/z 387.2893 (calcd for C25H38O3+H, 387.2899).
Emericellene C (4): colorless amorphous solid; [α]25 D 106.9 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 206.5 (4.22) nm; FT-IR (KBr) νmax 1701 cm−1; 1H NMR and 13C NMR data, see Table 2; Positive HRESIMS m/z 369.2787 (calcd for C25H39O3–H2O+H, 369.2794).
Emericellene D (5): colorless amorphous solid; [α]25 D 59.0 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 202.5 (3.94) nm; FT-IR (KBr) νmax 1713 (br.) cm−1; 1H NMR and 13C NMR data, see Table 2; Positive HRESIMS m/z 387.2905 (calcd for C25H39O3, 387.2899).
Emericellene E (6): colorless amorphous solid; [α]25 D 24.3 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 201 (3.77) nm; FT-IR (KBr) νmax 1708 (br.) cm−1; 1H NMR and 13C NMR data, see Table 2; Positive HRESIMS m/z 387.2896 (calcd for C25H39O3, 387.2899).
Cytotoxicity Assay
The resazurin-based colorometric (alamarBlue) assay7 was used for evaluating in vitro cytotoxicity of samples against human non-small cell lung (NCI-H460), human CNS glioma (SF-268), human breast (MCF-7), human metastatic breast adenocarcinoma (MDA-MB-231), human prostate adenocarcinoma (PC-3), metastatic prostate adenocarcinoma (PC-3M), human neuroblastoma (CHP-100) cancer cell lines, and human lung fibroblast (WI-38) cells. Doxorubicin and DMSO were used as positive and negative controls, respectively.
Supplementary Material
Figure 1.
Structures of metabolites 1–9 from Emericella sp. AST0036
Figure 3.
Significant HMBC and TOCSY correlations for emericellene A (2)
Figure 4.
Key NOE correlations for emericellene A (2)
ACKNOWLEDGMENTS
Financial support for this work was provided by Grant R01 CA090265 funded by the National Cancer Institute (NCI), Grant P41 GM094060 funded by National Institute of General Medical Sciences (NIGMS), and Grant DEB-1045766 (NSF).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The related supporting information is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Studies on Arid Lands Plants and Microorganisms, Part 25, For Part 24, see: Wanigasekara WMAP, Wijeratne EMK, Arnold AE, Gunatilaka AAL. Nat. Prod. Commun. 2013;8:601–604.
- 2.Strobel G, Daisy B. Microbiol Mol Biol Rev. 2003;67(4):491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusari S, Spiteller M. Nat. Prod. Rep. 2011;28:1203–1207. doi: 10.1039/c1np00030f. [DOI] [PubMed] [Google Scholar]
- 4.(a) Arnold AE. Fungal Biol. Rev. 2007;21:51–66. [Google Scholar]; (b) Rodriguez R, White J, Arnold AE, Redman R. New Phytologist. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 5.Clay K, Holah J. Science. 1999;285(5434):1742–1744. doi: 10.1126/science.285.5434.1742. [DOI] [PubMed] [Google Scholar]
- 6.(a) Schulz B, Boyle C, Draeger S, Römmert A-K, Krohn K. Mycol. Res. 2002;106(9):996–1004. [Google Scholar]; (b) Gunatilaka AAL. J. Nat. Prod. 2006;69:509–526. doi: 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijeratne EMK, Bashyal BP, Liu MX, Rocha DD, Gunaherath GMKB, U’Ren JM, Gunatilaka MK, Arnold AE, Whitesell L, Gunatilaka AAL. J. Nat. Prod. 2012;75(3):361–369. doi: 10.1021/np200769q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawksworth DL. Med. Mycol. 2011;49(Suppl. 1):S70–S76. doi: 10.3109/13693786.2010.504753. [DOI] [PubMed] [Google Scholar]
- 9.Rank C, Nielsena KF, Larsena TO, Vargab J, Samsonc RA, Frisvad JC. Fungal Biol. 2011;115(4–5):406–420. doi: 10.1016/j.funbio.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Fremlin LJ, Piggott AM, Lacey E, Capon RJ. J. Nat. Prod. 2009;72(4):666–670. doi: 10.1021/np800777f. [DOI] [PubMed] [Google Scholar]
- 11.Ballantine JA, Ferrito V, Hassall CH, Jenkins ML. J. Chem. Soc. Perkin Trans. I. 1973;17:1825–1830. doi: 10.1039/p19730001825. [DOI] [PubMed] [Google Scholar]
- 12.Chexal KK, Holker JSE, Simpson T. J. Chem. Soc. Perkin Trans. I. 1975;6:549–554. [PubMed] [Google Scholar]
- 13.(a) Pornpakakul S, Liangsakul J, Ngamrojanavanich N, Roengsumran S, Sihanonth P, Piapukiew J, Sangvichien E, Puthong S, Petsom A. Arch. Pharm. Res. 2006;29(2):140–144. doi: 10.1007/BF02974275. [DOI] [PubMed] [Google Scholar]; (b) Fujimoto H, Asai T, Kim Y-P, Ishibashi M. Chem. Pharm. Bull. 2006;54(4):550–553. doi: 10.1248/cpb.54.550. [DOI] [PubMed] [Google Scholar]; (c) Moosophon P, Kanokmedhakul S, Kanokmedhakul K, Soytong K. J. Nat. Prod. 2009;72(8):1442–1446. doi: 10.1021/np800805f. [DOI] [PubMed] [Google Scholar]
- 14.(a) Kawai K. Hoshi Yakka Daigaku Kiyo. 1996;38:1–8. [Google Scholar]; (b) Ooike M, Nozawa K, Kawai K. Phytochemistry. 1997;46(1):123–126. [Google Scholar]
- 15.Fujimoto H, Nakamura E, Okuyama E, Ishibashi M. Chem. Pharm. Bull. 2000;48(10):1436–1441. doi: 10.1248/cpb.48.1436. [DOI] [PubMed] [Google Scholar]
- 16.Hosoe T, Sameshima T, Dobashi K, Kawai K. Chem. Pharm. Bull. 1998;46(5):850–852. [Google Scholar]
- 17.Seya H, Nozawa K, Nakajima S, Kawai K, Udagawa S. J. Chem. Soc. Perkin Trans. I. 1986:109–116. [Google Scholar]
- 18.Herath KB, Jayasuriya H, Ondeyka JG, Polishook JD, Bills GF, Dombrowski AW, Cabello A, Vicario PP, Zweerink H, Guan Z, Singh SB. J. Antibiot. 2005;58:686–694. doi: 10.1038/ja.2005.94. [DOI] [PubMed] [Google Scholar]
- 19.Gardiner DM, Waring P, Howlett B. J. Microbiol. 2005;151:1021–1032. doi: 10.1099/mic.0.27847-0. [DOI] [PubMed] [Google Scholar]
- 20.Noda K, Umeda M, Ueno Y. Carcinogenesis. 1981;2:945–949. doi: 10.1093/carcin/2.10.945. [DOI] [PubMed] [Google Scholar]
- 21.Bünger J, Westphal G, Monnich A, Hinnendahl B, Hallier E, Müller M. Toxicol. 2004;202:199–211. doi: 10.1016/j.tox.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Seigler DS. Plant Secondary Metabolism. Kluwer: Boston; 1998. p. 401. [Google Scholar]
- 23.U'Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Am. J. Bot. 2012;99:898–914. doi: 10.3732/ajb.1100459. [DOI] [PubMed] [Google Scholar]
- 24.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 25.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 26.Maddison WP, Maddison DR. Mesquite. 2011 www.mesquiteproject.org.
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Maddison DR, Maddison WP. MacClade v. 4.08a. 2005 http://macclade.org.
- 29.Zwickl DJ. Ph.D. Thesis. Austin, TX: The University of Texas at Austin; 2008. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion; pp. 1–114. [Google Scholar]
- 30.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.