Abstract
Graphene-based materials are useful reinforcing agents to modify the mechanical properties of hydrogels. Here, we present an approach to covalently incorporate graphene oxide (GO) into hydrogels via radical copolymerization to enhance the dispersion and conjugation of GO sheets within the hydrogels. GO is chemically modified to present surface-grafted methacrylate groups (MeGO). In comparison to GO, higher concentrations of MeGO can be stably dispersed in a pre-gel solution containing methacrylated gelatin (GelMA) without aggregation or significant increase in viscosity. In addition, the resulting MeGO-GelMA hydrogels demonstrate a significant increase in fracture strength with increasing MeGO concentration. Interestingly, the rigidity of the hydrogels is not significantly affected by the covalently incorporated GO. Therefore, our approach can be used to enhance the structural integrity and resistance to fracture of the hydrogels without inadvertently affecting their rigidity, which is known to affect the behavior of encapsulated cells. The biocompatibility of MeGO-GelMA hydrogels is confirmed by measuring the viability and proliferation of the encapsulated fibroblasts. Overall, this study highlights the advantage of covalently incorporating GO into a hydrogel system, and improves the quality of cell-laden hydrogels.
Keywords: methacrylated graphene oxide (MeGO), methacrylated gelatin (GelMA), hydrogel, toughness, cell encapsulation
1. Introduction
Hydrogels are widely used as extracellular matrix (ECM)-mimicking materials to provide suitable cellular microenvironments in various biomedical applications, because the elastic polymeric network of hydrogels can successfully mimic certain traits of the natural ECM structure.[1, 2] Hydrogels can be designed to exhibit various chemical and physical factors to optimize cell survival and induce specific cell behaviors.[2] For example, hydrogels are often modified with cell recognition domains, such as Arg-Gly-Asp (‘RGD peptide’) to promote cell adhesion and survival, and matrix metalloproteinase recognition domains to allow enzymatic degradation of the hydrogel.[3] Recently, extensive research efforts have been focused on studying the effect of rigidity of hydrogel on the cells, as the mechanical signals imparted by the ECM influence a diverse array of cell phenotypes as well as the differentiation fate of stem cells.[4]
Hydrogel rigidity is most commonly modulated by controlling the crosslinking density of the polymer network via adjustments of monomer concentration and the ratio of monomer to crosslinker.[5] However, varying the crosslinking density inadvertently affects the hydrogel toughness, i.e. the ability to withstand applied mechanical energy without fracture, due to the correlation between rigidity and toughness of polymeric networks. Increasing the crosslinking density to enhance rigidity often results in brittleness, while decreasing the crosslinking density to reduce the rigidity leads to structural weakness.[6] Thus, it is challenging to improve the toughness of hydrogel while maintaining rigidity.
It has been previously shown that incorporating nanostructures with characteristic physical properties into a hydrogel plays a significant role in determining the mechanical properties of the overall hydrogel structure.[7, 8] Graphene is a highly robust yet flexible macromolecular nanomaterial, composed of sp2-carbon atoms in a single two-dimensional layer.[9] Owing to its favorable physical properties (e.g. electrical and optical propeties, high mechanical strength, and biocompatibility), graphene-based materials are increasingly used in biomedical applications.[10] Graphene oxide (GO), readily prepared from the oxidation of graphite, has abundant hydrophilic functional groups on the graphene layer, which allows for dispersion in aqueous media and chemical modifications, and thus has been commonly used in biological applications over pure graphene.[9, 11] Recent research efforts on engineering GO-composite hydrogels with improved mechanical strength have been reported.[12] It is suggested that incorporating GO into hydrogels would significantly enhance the toughness of hydrogels. However, the solubility of GO in biological buffers and pre-gel solutions is rather limited, which impedes the homogeneous incorporation of GO within the polymeric network especially at high concentrations.
Here, we present an approach to chemically modify GO to introduce methacrylate groups on the GO surface, termed methacrylated graphene oxide (MeGO), for the covalent incorporation of GO into a hydrogel system via radical copolymerization. Mechanical properties and the biodegradation rates of the resulting MeGO-linked hydrogels were compared with those made with unmodified GO to evaluate the effects of covalent conjugation. In addition, spectroscopic and microscopic methods were employed to analyze the dispersion of MeGO within the pre-gel solution and hydrogel network. Finally, the biocompatibility of MeGO-linked hydrogels was evaluated by measuring the viability and proliferation of encapsulated fibroblasts.
2. Results and Discussion
2.1. Synthesis and Characterization of Methacrylated Graphene Oxide (MeGO)
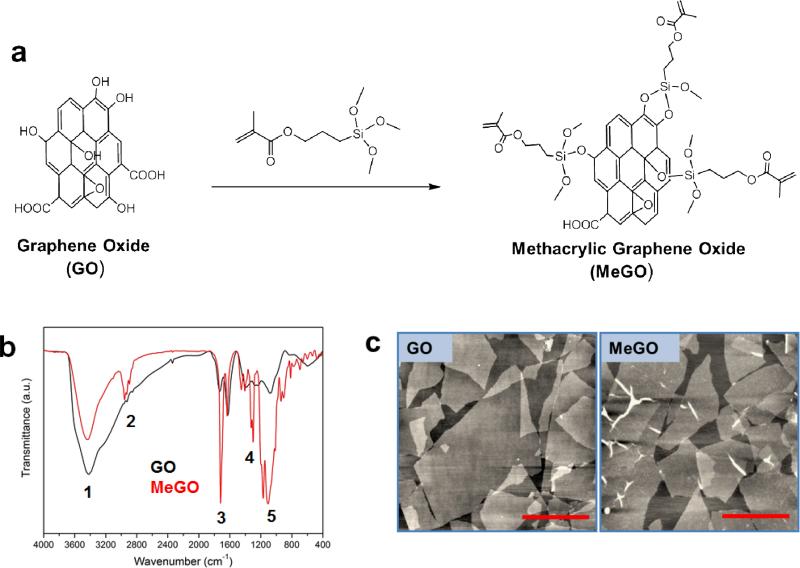
Methacrylate groups were conjugated onto GO by reaction with 3-(trimethoxysilyl)propyl methacrylate (TMSPMA) to prepare methacrylic graphene oxide (MeGO) (Figure 1a). A large number of hydroxyl functional groups on GO were converted to methacrylic groups via silanization, as evidenced by the FT-IR spectroscopy of MeGO; the presence of characteristic vibrational spectral peaks corresponding to siloxyl, silyl and methacrylate groups of TMSPMA (1108 cm−1 (νSi-O), 1300 cm−1 (νSi-C), 1719 cm−1 (νC=O)), and the decrease in hydroxyl peak (3419 cm−1 (νO-H)) due to the reaction between hydroxyl groups and TMSPMA (Figure 1b). The atomic force microscopic (AFM) images of GO and MeGO showed that the chemical reaction did not alter the sheet structure of GO and induce aggregation of multiple GO sheets (Figure 1c).
Figure 1.
(a) Surface functionalization of graphene oxide (GO) with methacrylate via silanization to prepare methacrylated graphene oxide (MeGO). (b) FT-IR spectra of GO (black) and MeGO (red). Characteristic peaks are noted in numbers. 1: 3419 cm−1 (vs(O-H)), 2: 2957 cm−1 (vs(C-C)), 3: 1719 cm−1 (vs(C=O)), 4: 1300 cm−1 (vs(Si-C)), 5: 1108 cm−1 (vs(Si-O)). (c) AFM images of GO (left) and MeGO (right).(Scale bar: 1 μm)
2.2. Dispersion of MeGO in GelMA Solution
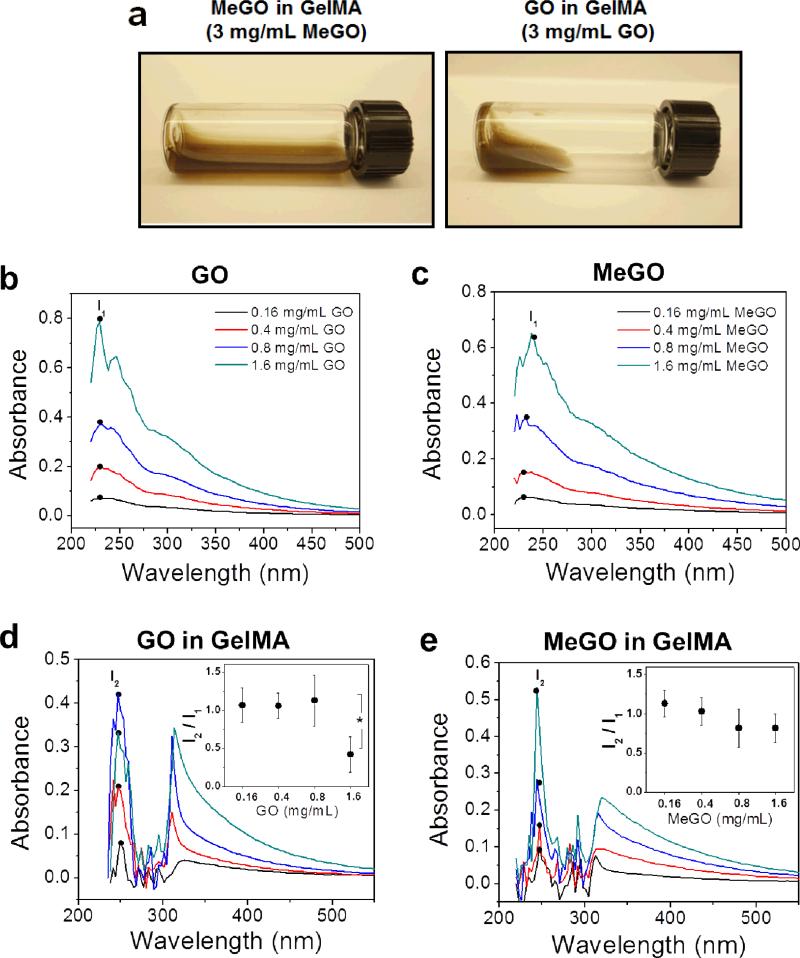
Inducing proper dispersion of nanoparticles within a polymer system is critical for imparting reinforcing effects of nanoparticles to the composite material.[13] Therefore, we first examined the dispersion of MeGO in pre-gel solutions to evaluate the effect of surface methacrylic groups on the dispersibility of GO sheets. Here, methacrylated gelatin (GelMA) was chosen as a model photocrosslinkable polymer system.[14, 15] First, varying amounts of unmodified GO or MeGO up to 3 mg mL−1 were added to pre-gel solutions consisting of 8 wt% GelMA and sonicated to induce dispersion. GO dispersed readily up to 0.8 mg mL−1. However, large aggregations of GO began to appear in the pre-gel solution above 1 mg mL−1, which only disappeared after high-temperature treatment (80 °C for 1 hour). Above 1.6 mg mL−1, the pre-gel solution became viscous with highly diminished fluid mobility and contained large aggregates which could not be disassociated by high-temperature treatment (Figure 2a). Previous studies also reported similar limits of GO dispersion in polymeric solutions, due to extensive physical interaction between polymers and GO, and the propensity of GO sheets to aggregate due to limited solubility.[16] In contrast, MeGO was well dispersed in GelMA solution without aggregation or increase in viscosity up to 3 mg mL−1 (Figure 2a).
Figure 2.
(a) Photographs of 3 mg mL−1 of MeGO (left) or GO (right) dispersed in GelMA solution. UV-vis absorption spectra of (b) GO, (c) MeGO, (d) GO in GelMA, and (e) MeGO in GelMA. The concentration of GO or MeGO was varied from 0.16 to 1.6 mg mL−1. The legends in (b) and (c) are the same for (d) and (e), respectively. Inset graphs in (d) and (e) represent the ratio of characteristic peaks of GO-GelMA or MeGO-GelMA at 254 nm (I2) to that of GO or MeGO at 231 nm (I1).(*p<0.05)
UV-vis spectroscopy was used to further analyze the dispersion of MeGO in GelMA solution. GO displays a characteristic absorption peak at 231 nm, which corresponds to π → π* transition and therefore identifies the dispersion of GO layers (denoted as I1, Figure 2b).[17] MeGO showed similar characteristic absorption spectra as GO, which demonstrated that dispersibility of GO layers were not affected by the presence of methacrylic groups (Figure 2c). When GO or MeGO was incorporated within GelMA solution, the characteristic peak was red-shifted to 254 nm (denoted as I2, Figure 2d and 2e), which is associated with the interaction between GO and polymers.[18] The ratio of I2 to I1 (I2/I1), which measures the change in GO dispersion, significantly decreased (by 60 %) when the concentration of GO was increased to 1.6 mg mL−1, suggesting there was significant aggregation of GO (inset in Figure 2d). However, there was only a small decrease in I2/I1 values (by 15 %), when the concentration of MeGO was increased to 1.6 mg mL−1, demonstrating that MeGO remained effectively dispersed in GelMA solution at a higher concentration than GO (inset in Figure 2e). It should be noted that UV-vis spectra of GO or MeGO in GelMA at 3 mg mL−1 could not be obtained because high concentration of graphene oxide layers absorbed much of UV-vis irradiation. These results demonstrated that the presence of methacrylate groups on GO could effectively prevent aggregation between GO layers, and induce better dispersion within polymer solution.
2.3. Mechanical Properties of MeGO-GelMA Hydrogels
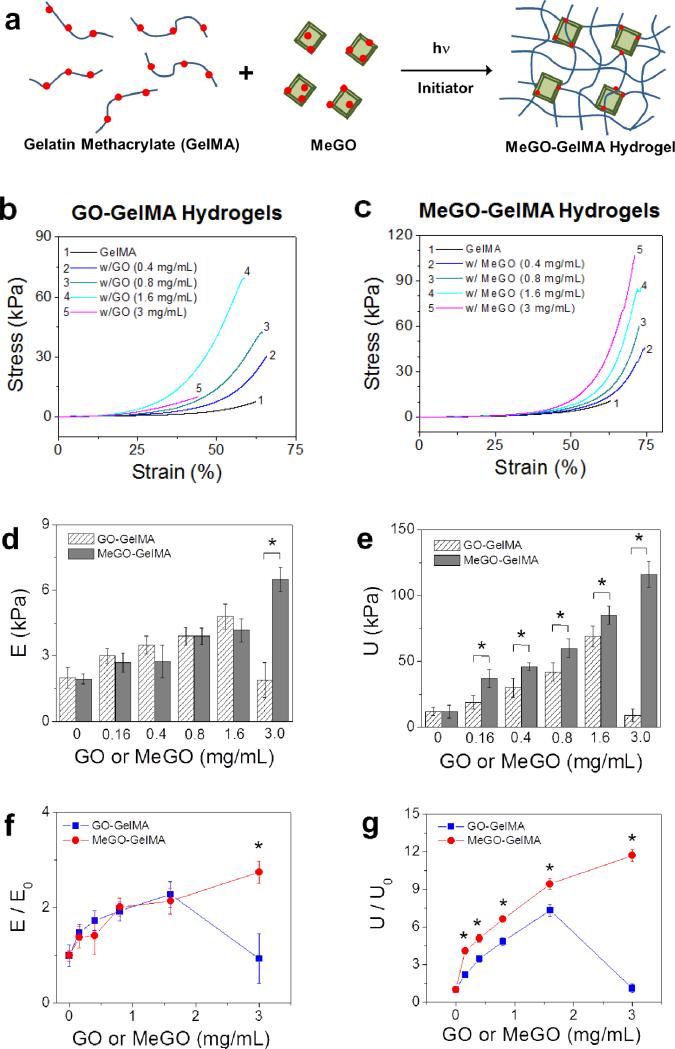
GelMA hydrogels incorporated with varying amounts of GO (‘GO-GelMA hydrogels’) or MeGO (‘MeGO-GelMA hydrogels’) were fabricated by photoinitiated radical copolymerization (Figure 3a). The hydrogels became darker with increasing amount of GO or MeGO (Figure S1a in Supporting Information). Microscopic observation of the hydrogels showed that micron-sized agglomerates began to appear in GO-GelMA hydrogels with GO concentration above 1 mg mL−1, whereas no such agglomerates were observed in MeGO-GelMA hydrogels (Figure S1b in Supporting Information).
Figure 3.
(a) MeGO-GelMA hydrogel is prepared by photoinitiated radical copolymerization of GelMA and MeGO. Stress-strain curves of GelMA hydrogels with varying amounts of (b) GO or (c) MeGO measured from uniaxial compression. (d) Elastic modulus (E) and (e) ultimate stress (U) of GO-GelMA hydrogels and MeGO-GelMA hydrogels. f) Normalized elastic modulus (E/E0) and (g) normalized fracture energy (U/U0) of GelMA hydrogels incorporated with GO or MeGO. The values are normalized with respect to those of pure GelMA hydrogel (E0, U0).(*p<0.05 at the same concentrations of GO and MeGO)
Mechanical properties of the MeGO-GelMA hydrogels were evaluated by uniaxial compression (Figure 3b and 3c). Elastic modulus, determined by the slope of the elastic region of the stress-strain curves, i.e. the initial linear portion of the curves, increased 2.7-fold when the concentration of MeGO was increased up to 3 mg mL−1 (Figure 3d and 3f). On the other hand, the presence of MeGO had a more profound effect on the toughness of the hydrogels, as the stress values began to increase significantly at strains above 50 %. There was an 11-fold increase in the ultimate stress of the MeGO-GelMA hydrogels when MeGO was increased to 3 mg mL−1 (Figure 3e, 3g, and 4a).
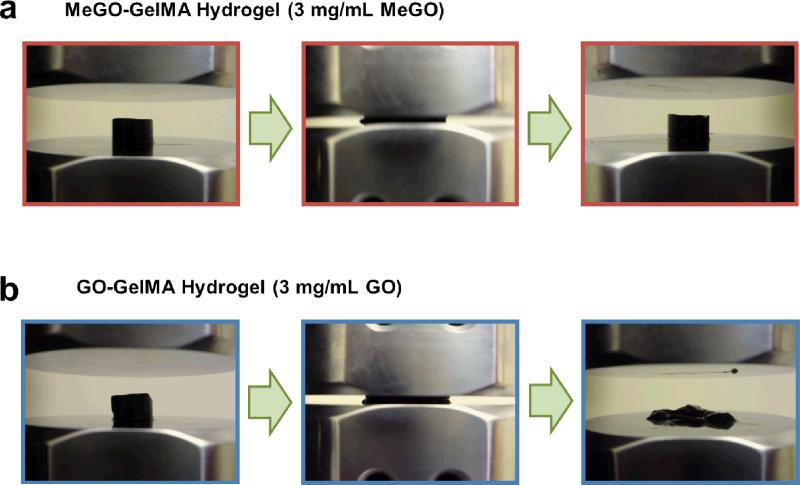
Figure 4.
GelMA hydrogel incorporated with (a) GO or (b) MeGO at 3 mg mL−1 subjected to uniaxial compression. GO-GelMA hydrogel became easily fractured, whereas MeGO-GelMA hydrogel demonstrated resistance to fracture at high strain (70 %).
Elastic moduli and ultimate stress values of GO-GelMA hydrogel were similar to those of MeGO-GelMA hydrogels up to 1.6 mg mL−1. However, the elastic modulus and ultimate stress of GO-GelMA hydrogel at 3 mg mL−1 dramatically decreased (Figure 3d, 3e, 3g, and 4b). This result is in line with the highly limited dispersibility of GO in GelMA solution at 3 mg mL−1 as presented above, which suggests that a large amount of agglomerates prevented proper hydrogel formation. As a result, these agglomerates within the hydrogels acted as structural defects, and led to structural deterioration even at lower strain.
These results also demonstrated that incorporating GO, regardless of the mode of incorporation, had greater influence on toughness than rigidity of the hydrogels. These findings are in contrast with previous studies incorporating other types of carbon-based nanostructures, such as carbon nanotubes (CNTs) and nanodiamonds (NDs), to reinforce hydrogels where both rigidity and toughness were significantly influenced. For example, Shin et al. demonstrated that incorporating CNTs into GelMA hydrogel system resulted in a significant increase in modulus (3-fold increase at 0.5 mg mL−1 GO), while minimally affecting the ultimate stress of the hydrogels.[19] Furthermore, Yildirim et al. showed increases in both elastic modulus and tensile strength of CNT-alginate composite hydrogel.[20] In both studies, however, the brittleness of the hydrogel was also increased with CNT, as evidenced by the decrease in ultimate strain. Behler et el. created ND-polyacrylonitrile composite film which showed 4-fold increase in modulus and 2-fold increase in scratch hardness when the concentration of NDs was increased up to 20 wt%.[21] In other words, CNT or ND-incorporated hydrogels behave like a typical composite system, in which stiffer composites are generally more brittle. In comparison, GO-GelMA and MeGO-GelMA hydrogels deviate from this typical behavior with a significant increase in ultimate stress (11-fold) and a less pronounced increase in stiffness (2.7-fold). It is therefore suggested that characteristic material properties of GO played a critical role in determining the mechanical properties of the overall hydrogel structure. The highly flexible macromolecular sheet structure of GO could effectively dissipate energy applied to the hydrogel through highly dynamic conformational changes, and therefore had a more profound effect on the hydrogel toughness, whereas CNTs and NDs that do not possess such conformational flexibility also had a significant effect on the rigidity of the hydrogel. Therefore, incorporating MeGO into hydrogels could be highly useful for improving their mechanical toughness, without significantly affecting their rigidity which is a known regulator of cellular behavior.
2.4. Morphological Evaluation of MeGO-GelMA Hydrogels
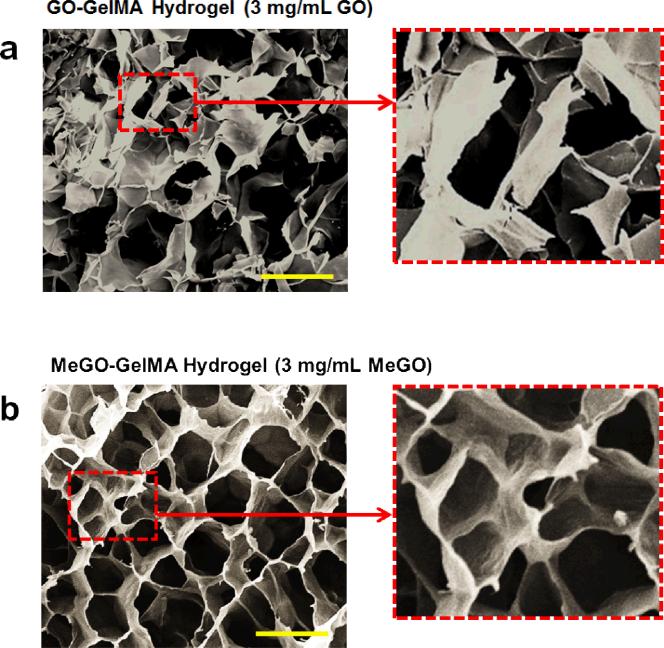
The stark difference in mechanical properties between GO-GelMA hydrogel and MeGO-GelMA hydrogels at high GO or MeGO content (3 mg mL−1), as shown in Figures 3 and 4, suggest that the presence of methacrylic groups on GO sheets facilitated their integration into hydrogels even at high concentrations. To gain further insight into the effect of covalently incorporating GO into GelMA hydrogel, scanning electron microscopy (SEM) was used to visualize the detailed structural features of GelMA hydrogels incorporated with high concentrations of GO or MeGO, at 3 mg mL−1. The GO-GelMA hydrogel displayed highly irregular porous structure, with significant portions of the wall structure being fractured (Figure 5a). In addition, GO was not well distributed within the hydrogel network, as evidenced by the uneven distribution of highly wrinkled and rough surface, which is caused by the presence of GO (inset in Figure 5a). Such structural irregularities were not observed with GO-GelMA at low GO concentration (0.8 mg mL−1, Figure S2a in Supporting Information). It has been shown that the presence of GO can distort the polymeric matrices, resulting in wrinkled structures.[22] Here, high GO content in localized areas of the network without proper dispersion likely weakened the strength of the polymeric network, and led to fracture during the lyophilization process for sample preparation. These findings support the significant decrease in mechanical properties of GelMA hydrogels incorporated with high concentration of GO as shown in Figure 3.
Figure 5.
Scanning electron microscopic (SEM) images of the cross sections of (a) GO-GelMA (3 mg mL−1 GO) hydrogel and (b) MeGO-GelMA (3 mg mL−1 MeGO) hydrogel. The images on right show magnified views of designated area. (Scale bar: 200 μm)
On the other hand, MeGO-GelMA hydrogels, regardless of the concentration of MeGO, showed highly ordered porous structure, without any fractured areas (Figure 5b, Figure S2b in Supporting Information). In addition, the entire surface of the hydrogel network was evenly wrinkled, which indicates that GO was well distributed throughout the hydrogel (inset in Figure 5b). These observations suggested that the covalent conjugation of GO effectively prevented aggregations, and allowed stable dispersion of the GO sheets within the hydrogels even at high concentrations. It is well known that there is enhanced entropy-driven depletion attraction between nanoparticles during polymeric network formation, because it is energetically unfavorable for the polymers to form networks surrounding the nanoparticles.[13, 23] This, coupled with the attractive interaction between GO sheets, makes GO more susceptible for aggregation or phase separation within the polymeric network. However, the covalent linkage between GO and polymer during the polymerization reaction likely stabilized the dispersion and incorporation of GO within the hydrogel network. Furthermore, flexible sheet structures are known to increase the fracture resistance of the composite materials by reducing their Poisson ratio.[8, 24] Therefore, the significant increase in toughness of MeGO-GelMA hydrogel could also be attributed to the presence of MeGO within the polymeric network allowing the material to expand in response to external force, thus effectively dissipating the applied energy without weakening the structure.
2.5. Biodegradation of MeGO-GelMA Hydrogels
GelMA hydrogels have been shown to undergo enzymatic degradation, as gelatin contains functional sequences recognized by collagenase.[14, 25] Thus, we explored the effect of covalent conjugation of GO to the GelMA hydrogels on the enzymatic degradation. MeGO-GelMA hydrogels were treated with type II collagenase, and the weight of the remaining hydrogel at various time points were measured over time. Degradation of GO-GelMA hydrogels was also evaluated as a control.
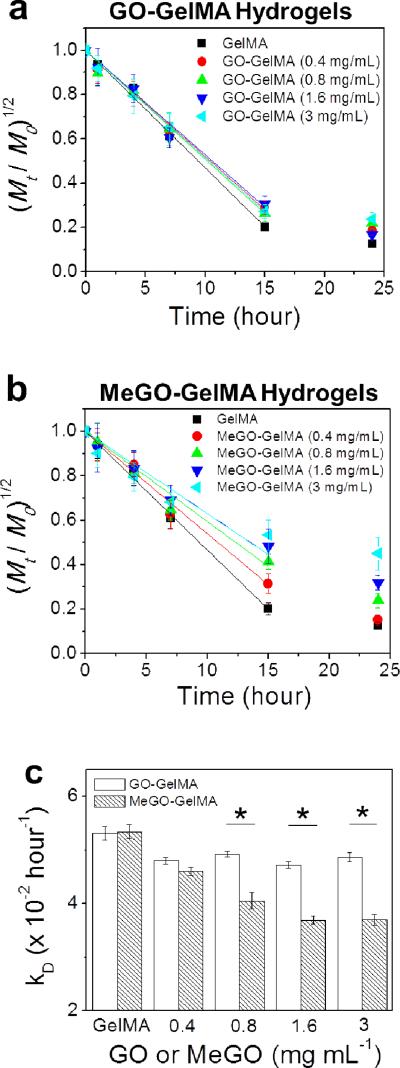
Figure 6a & 6b show the plots of (Mt / M0)1/2 vs. t, where Mt/M0 represents the fractional weight of the hydrogel at time, t, for GO-GelMA hydrogels and MeGO-GelMA hydrogels, respectively. The plots were then fitted with eq. (1) to obtain the degradation rates (kD) of the hydrogels. kD values for GO-GelMA hydrogels did not change regardless of the amount of GO, which indicates physical association of GO with the GelMA network had little effect on the enzymatic cleavage of the gelatin backbone (Figure 6a & 6c). However, there was a significant decrease in kD values with increasing amount of MeGO in the MeGO-GelMA hydrogels (Figure 6b & 6c). GO sheets covalently linked to GelMA molecules were likely able to bridge the cleaved GelMA chains, and delayed the hydrogel decomposition. This result further confirms that MeGO was able to covalently incorporate into the hydrogel network.
Figure 6.
Biodegradation of (a) GO-GelMA hydrogels and (b) MeGO-GelMA hydrogels, induced by treating the hydrogels with collagenase (1 U mL−1). The concentration of GO or MeGO was varied from 0 to 3 mg mL−1 (c) The degradation rates (kD) of the hydrogels were obtained by fitting the linear region (first 15 hours) of the plots in (a) and (b) with Equation 1. (*p<0.05)
2.6. Cell Encapsulation in MeGO-GelMA Hydrogels
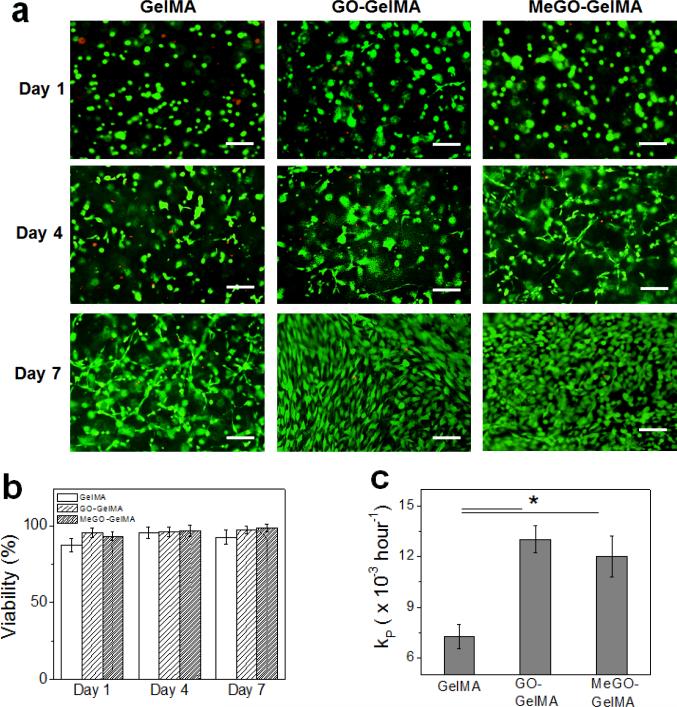
To assess the biocompatibility of MeGO-linked hydrogel, NIH-3T3 fibroblasts were encapsulated within MeGO-GelMA hydrogels (0.8 mg mL−1 MeGO) and their viability and proliferation were evaluated. As a control, cells encapsulated in pure GelMA hydrogels and GO-GelMA hydrogels (0.8 mg mL−1 GO) were evaluated. The initial viability of encapsulated cells, measured one hour after encapsulation, showed that the cell viability in MeGO-GelMA hydrogels (92 ± 2 %) and GO-GelMA hydrogel (94 ± 5 %) was higher than that in GelMA hydrogel (84 ± 4 %) (Figure S3 in Supporting Information). This suggests that the presence of GO, regardless of mode of incorporation within GelMA hydrogels, protected the cells from harmful environment during the crosslinking reaction. Shin et al. have recently reported a similar finding in which cells cultured on CNT-reinforced scaffold were protected against induced oxidative stress.[19] The decrease in the initial viability of cells encapsulated within radically polymerized hydrogels is often attributed to the free radicals and reactive oxidative species affecting the cells. It is suggested that the GO within the hydrogel may have acted as a scavenger that removes unreacted free radicals and prevented cell death, since GO is well known to readily react with free radicals due to its electron-rich surface.[26]
The viability of encapsulated cells was continuously monitored over the period of 7 days (Figure 7a, Figure S4 in Supporting Information). In all conditions, the cell viability remained high throughout the experiment and the cells were able to spread and proliferate over time, demonstrating that the presence of GO or MeGO in the hydrogels did not have any adverse effect on the long term viability of the encapsulated cells (Figure 7a & 7b). Interestingly, however, the proliferation rate was significantly higher in GO-GelMA hydrogels and MeGO-GelMA hydrogels as compared with pure GelMA hydrogels (Figure 7c). The cells became more elongated in GO-GelMA hydrogels as compared to those in MeGO-GelMA hydrogels, likely due to the increased crosslinking density by covalent incorporation of MeGO more constrained the cells in MeGO hydrogels. However, no significant difference in proliferation rate between GO-GelMA hydrogels and MeGO-GelMA hydrogels was observed, indicating the presence of GO layers within the hydrogels, not the mode of linkage to the hydrogel, was responsible for the effect on the cells. Several previous studies have also reported the enhanced cell behavior on graphene-based materials.[27] Khang et al. proposed that the presence of carbon nanomaterials within the polymeric matrix increased protein adsorption due to increased surface roughness in the nano-scale, which was also shown in MeGO-GelMA (Fig. 5).[28] Although the exact mechanism of biological responses have not been fully elucidated to date, these results further demonstrate the feasibility of utilizing GO-incorporated hydrogels demonstrating high mechanical strength for tissue engineering applications.
Figure 7.
(a) Fluorescent images of fibroblasts encapsulated in GelMA, GO-GelMA and MeGO-GelMA hydrogels over time. The cells were stained with calcein-AM and ethidium homodimer-1 to visualize live (green) and dead (red) cells.(Scale bar: 100 μm) (b) Viability of the encapsulated cells at various time points. (c) Proliferation rate (kP) determined from Equation 2.(*p<0.05)
3. Conclusion
Taken together, we have chemically modified GO to introduce methacrylate functional groups onto GO and generated methacrylic graphene oxide (MeGO) in order to covalently conjugate GO into hydrogel systems via radical copolymerization. Photocrosslinkable gelatin (GelMA) hydrogels with varying amounts of MeGO were fabricated, and the resulting hydrogels displayed improved mechanical toughness with increased concentrations of MeGO, whereas hydrogels incorporated with GO showed mechanical failure at lower GO concentration than MeGO. Morphological study of the hydrogels showed that covalently incorporating GO by using MeGO allowed stable dispersion and interfacial bonding between GO and polymeric network. Interestingly, the effect of MeGO on hydrogel mechanics was more pronounced on toughness than rigidity, which could be attributed to the conformational flexibility of GO layer effectively dissipated the energy accumulated within the polymeric network, but had smaller effect on the rigidity. Thus, incorporating GO into hydrogel can be used to enhance the fracture strength while minimizing the change in rigidity which is known to influence cell behavior. Furthermore, the biocompatibility of MeGO-GelMA hydrogels was confirmed by evaluating the viability and proliferation of encapsulated fibroblasts. Therefore, we believe that the strategy of covalently incorporating GO presented in this study can be successfully utilized to significantly improve the structural integrity and resistance to fracture in a wide range of cell-encapsulating hydrogels without inadvertently affecting their rigidity.
4. Experimental Section
Synthesis of MeGO
Graphene oxide (GO) was first prepared using modified Hummer's method.[29] Dried GO flakes were suspended in ethanol (1 mg mL−1) and sonicated for 20 minutes, which resulted in stable homogeneous dispersion. 3-(trimethoxysilyl)propyl methacrylate (Sigma Aldrich) was slowly added to GO suspension (50 μL per each mg of GO) with sonication, and continuously stirred for 12 hours at 50 °C. The mixture was dialyzed against ethanol, and then dried under vacuum to obtain the product. MeGO was dispersed in deionized (DI) water at 4 mg mL−1 as a stock solution.
Synthesis of GelMA
5 g of gelatin and 0.5 g of 4-(dimethylamino)pyridine (Sigma Aldrich) were dissolved in 50 mL of dimethyl sulfoxide at 50 °C. Then, 2 mL of glycidyl methacrylate (Sigma Aldrich) was slowly added to the mixture. The mixture was continuously stirred for 48 hours at 50 °C under dry N2 gas, and then dialyzed against DI water to remove byproducts. The powdered product, GelMA, was obtained by lyophilization.
Spectroscopic analyses of MeGO
For Fourier transform infrared (FT-IR) spectroscopic analysis, dried GO or MeGO sample was first mechanically ground and pressed into a pellet with KBr powder. FT-IR transmittance spectra in a wavelength region between 500 and 4000 cm−1 were acquired using a FT-IR spectrometer (Spectrum BX, Perkin Elmer).
For atomic force microscopic (AFM) analysis, GO or MeGO dispersed in ethanol (0.05 mg mL−1) was spin coated onto a circular silicon substrate (8 mm diameter). Then, AFM images were taken in tapping mode using a silicon-SPM tip (POINTPROBE®, NanoWorld), with a scan rate of 1.5 Hz (Digital Instruments Dimension 3000).
UV-vis spectroscopy was used to analyze the dispersion of GO or MeGO within GelMA solution. Varying concentrations of GO or MeGO was dissolved in 8 wt% GelMA solution in phosphate buffered saline (PBS, pH 7.4), and sonicated for 30 minutes. Then, absorbance between 200 and 600 nm was measured using a spectrophotometer (ND-1000, Thermo Fisher).
Hydrogel fabrication
Pre-gel solution was prepared by mixing 8 wt% GelMA with varying concentrations of GO or MeGO in PBS. 0.2 wt% of Irgacure® 2959 (Ciba) was also added to each solution as a photoinitiator. Each pre-gel solution was then placed in a custom-made cylinderical mold, and then irradiated with UV for 2 minutes (output power of 850 mW, OmniCure® S2000) to form a hydrogel disk (8 mm diameter, 2 mm thickness). The hydrogels were then incubated in PBS at 37 °C for 24 hours before characterization.
SEM was used to analyze the morphological features of hydrogels. Hydrogels were first washed with DI water and lyophilized. Then, the dried hydrogel samples were sputter-coated with gold (2 nm thickness, IBS/TM200S, VCR Group, Inc.), then visualized with SEM (Quanta 200 FEG, FEI™) under high vacuum.
Evaluation of hydrogel mechanical properties
The hydrogel disks were compressed at 1 mm min−1 until they fractured using a mechanical testing system (Model 5943, Instron®) equipped with a computer-based control/analysis system (Bluehill® 3).[30] Elastic modulus was calculated from the slope of a stress-strain curve at the first 10 % strain where the curve was linear. Ultimate stress was determined as the maximum stress before the hydrogel fractured.
Evaluation of hydrogel degradation rate
The hydrogel disks were incubated in 1 U mL−1 of collagenase (type II, Worthington Biochemical Co.) at 37 °C. At various time points, a hydrogel sample was taken out and its dried weight was measured. The results were reported as (Mt / M0)1/2 vs. t, where M0 is the original dry weight of the hydrogel and Mt is the dried weight at time, t. The degradation rates (kD) were obtained by fitting the linear region of the plots (first 15 hours) with the following equation,[31]
| (1) |
Cell studies
NIH-3T3 fibroblasts were suspended in a pre-gel solution (1 × 106 cells mL−1), and then crosslinked to fabricate hydrogels via photocrosslinking, as mentioned above. The cell-encapsulated hydrogels were incubated in the culture media (Dulbecco's Modified Eagle Medium, supplemented with 10 % fetal bovine serum and penicillin/streptomycin, all purchased from Invitrogen) at 37 °C with 5 % CO2. To measure the viability of encapsulated cells, the cells were fluorescently labeled with calcein-AM (green, live) and ethidium homodimer-1 (red, dead) using LIVE/DEAD® Viability/Cytotoxicity Assay kit (Invitrogen), and then visualized with a fluorescence microscope (Eclipse Ti, Nikon). The viability was quantified as the percentage of live cells from total encapsulated cells. Proliferation rate (kP) was obtained from the following equation,[32]
| (2) |
where N0 is the initial number of live cells and Nt is the number of live cells at time, t.
Statistical Analysis
All numerical data obtained in this work were averaged from four independent experiments. The statistical difference between two values were determined from one-way ANOVA (Tukey's post-hoc method), and p values below 0.05 was considered statistically significant and reported here.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (HL092836, EB02597, AR057837, HL099073 (A.K.)), the National Science Foundation (DMR0847287 (A.K)), the Office of Naval Research Young Investigator Award (A.K.), the Presidential Early Career Award for Scientists and Engineers (A.K.), and the Natural Science and Engineering Research Council of Canada (Discovery grant (X.G. & X. T.)).
Footnotes
Supporting Information
Supporting Information is available online from the Wiley Online Library or from the author.
Contributor Information
Dr. Chaenyung Cha, Harvard-MIT Division of Health Sciences and Technology Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital Harvard Medical School 65 Landsdowne St, Cambridge, MA 02139, USA
Dr. Su Ryon Shin, Harvard-MIT Division of Health Sciences and Technology Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital Harvard Medical School 65 Landsdowne St, Cambridge, MA 02139, USA
Xiguang Gao, Department of Chemistry, University of Waterloo 200 University Avenue West, Waterloo, Ontario N2L 3G1, Canada.
Dr. Nasim Annabi, Harvard-MIT Division of Health Sciences and Technology Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital Harvard Medical School 65 Landsdowne St, Cambridge, MA 02139, USA
Dr. Mehmet R. Dokmeci, Harvard-MIT Division of Health Sciences and Technology Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital Harvard Medical School 65 Landsdowne St, Cambridge, MA 02139, USA
Xiaowu (Shirley) Tang, Department of Chemistry, University of Waterloo 200 University Avenue West, Waterloo, Ontario N2L 3G1, Canada
Ali Khademhosseini, Harvard-MIT Division of Health Sciences and Technology Center for Biomedical Engineering, Department of Medicine, Brigham and Women's Hospital Harvard Medical School 65 Landsdowne St, Cambridge, MA 02139, USA.
References
- 1.a Drury JL, Mooney DJ. Biomaterials. 2003;24:4337. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]; b Lutolf MP, Hubbell JA. Nat. Biotech. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]; c Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F. Tissue Eng. Pt. B: Rev. 2010;16:371. doi: 10.1089/ten.teb.2009.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv. Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandl F, Sommer F, Goepferich A. Biomaterials. 2007;28:134. doi: 10.1016/j.biomaterials.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 3.a Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]; b Kim S, Healy KE. Biomacromolecules. 2003;4:1214. doi: 10.1021/bm0340467. [DOI] [PubMed] [Google Scholar]; c Cha C, Liechty WB, Khademhosseini A, Peppas NA. ACS Nano. 2012;6:9353. doi: 10.1021/nn304773b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Yang S, Leong K-F, Du Z, Chua C-K. Tissue Eng. 2001;7:679. doi: 10.1089/107632701753337645. [DOI] [PubMed] [Google Scholar]; b Wang N, Tytell JD, Ingber DE. Nat. Rev. Mol. Cell. Biol. 2009;10:75. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]; c Chen CS, Tan J, Tien J. Ann. Rev. Biomed. Eng. 2004;6:275. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 5.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Adv. Mater. 2010;22:3484. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Nielsen LE, Landel RF. Mechanical properties of polymers and composites. Marcel Dekker; New York: 1994. [Google Scholar]; b Kong HJ, Wong E, Mooney DJ. Macromolecules. 2003;36:4582. [Google Scholar]
- 7.a Xiang Y, Peng Z, Chen D. Eur. Polym. J. 2006;42:2125. [Google Scholar]; b Huang T, Xu HG, Jiao KX, Zhu LP, Brown HR, Wang HL. Adv. Mater. 2007;19:1622. [Google Scholar]; c Schexnailder P, Schmidt G. Colloid Polym. Sci. 2009;287:1. [Google Scholar]; d Coleman JN, Cadek M, Ryan KP, Fonseca A, Nagy JB, Blau WJ, Ferreira MS. Polymer. 2006;47:8556. [Google Scholar]
- 8.Knauert ST, Douglas JF, Starr FW. J. Polym. Sci. Pol. Phys. 2007;45:1882. [Google Scholar]
- 9.Zhu Y, Murali S, Cai W, Li X, Suk JW, Potts JR, Ruoff RS. Adv. Mater. 2010;22:3906. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
- 10.Cha C, Shin SR, Annabi N, Dokmeci MR, Khademhosseini A. ACS Nano. 2013;7:2891. doi: 10.1021/nn401196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyer DR, Park S, Bielawski CW, Ruoff RS. Chem. Soc. Rev. 2010;39:228. doi: 10.1039/b917103g. [DOI] [PubMed] [Google Scholar]
- 12.a Zhang L, Wang Z, Xu C, Li Y, Gao J, Wang W, Liu Y. J. Mater. Chem. 2011;21:10399. [Google Scholar]; b Zhang N, Li R, Zhang L, Chen H, Wang W, Liu Y, Wu T, Wang X, Wang W, Li Y, Zhao Y, Gao J. Soft Matter. 2011;7:7231. [Google Scholar]; c Adhikari B, Banerjee A. Soft Matter. 2011;7:9259. [Google Scholar]; d Qi YY, Tai ZX, Sun DF, Chen JT, Ma HB, Yan XB, Liu B, Xue QJ. J. Appl. Polym. Sci. 2013;127:1885. [Google Scholar]
- 13.Balazs AC, Emrick T, Russell TP. Science. 2006;314:1107. doi: 10.1126/science.1130557. [DOI] [PubMed] [Google Scholar]
- 14.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a Shin SR, Bae H, Cha JM, Mun JY, Chen Y-C, Tekin H, Shin H, Farshchi S, Dokmeci MR, Tang S, Khademhosseini A. ACS Nano. 2011;6:362. doi: 10.1021/nn203711s. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen Y-C, Lin R-Z, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Adv. Funct. Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a Bai H, Li C, Shi G. Adv. Mater. 2011;23:1089. doi: 10.1002/adma.201003753. [DOI] [PubMed] [Google Scholar]; b Bai H, Li C, Wang X, Shi G. Chem. Comm. 2010;46:2376. doi: 10.1039/c000051e. [DOI] [PubMed] [Google Scholar]
- 17.Paredes JI, Villar-Rodil S, Martínez-Alonso A, Tascón JMD. Langmuir. 2008;24:10560. doi: 10.1021/la801744a. [DOI] [PubMed] [Google Scholar]
- 18.Bao Q, Zhang H, Yang J.-x., Wang S, Tang DY, Jose R, Ramakrishna S, Lim CT, Loh KP. Adv. Funct. Mater. 2010;20:782. [Google Scholar]
- 19.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim S. b., Nikkhah M, Khabiry M, Azize M, Kong J, Wan K.-t., Palacios T, Dokmeci MR, Bae H, Tang X, Khademhosseini A. ACS Nano. 2013;7:2369. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yildirim ED, Yin X, Nair K, Sun W. J. Biomed. Mater. Res. B. 2008;87B:406. doi: 10.1002/jbm.b.31118. [DOI] [PubMed] [Google Scholar]
- 21.Behler KD, Stravato A, Mochalin V, Korneva G, Yushin G, Gogotsi Y. ACS Nano. 2009;3:363. doi: 10.1021/nn800445z. [DOI] [PubMed] [Google Scholar]
- 22.Wan C, Frydrych M, Chen B. Soft Matter. 2011;7:6159. [Google Scholar]
- 23.Gupta S, Zhang Q, Emrick T, Balazs AC, Russell TP. Nat. Mater. 2006;5:229. [Google Scholar]
- 24.Bowick M, Cacciuto A, Thorleifsson G, Travesset A. Phys. Rev. Lett. 2001;87:148103. doi: 10.1103/PhysRevLett.87.148103. [DOI] [PubMed] [Google Scholar]
- 25.Van den Steen PE, Dubois B, Nelissen I, Rudd PM, Dwek RA, Opdenakker G. Crit. Rev. Biochem. Mol. Biol. 2002;37:375. doi: 10.1080/10409230290771546. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Liu Y, Zhu D. J. Mater. Chem. 2011;21:3335. [Google Scholar]
- 27.a Ku SH, Park CB. Biomaterials. 2013;34:2017. doi: 10.1016/j.biomaterials.2012.11.052. [DOI] [PubMed] [Google Scholar]; b Shi X, Chang H, Chen S, Lai C, Khademhosseini A, Wu H. Adv. Funct. Mater. 2012;22:751. [Google Scholar]; c Sebaa M, Nguyen TY, Paul RK, Mulchandani A, Liu H. Mater. Lett. 2013;92:122. [Google Scholar]
- 28.Khang D, Kim SY, Liu-Snyder P, Palmore GTR, Durbin SM, Webster TJ. Biomaterials. 2007;28:4756. doi: 10.1016/j.biomaterials.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Hummers WS, Offeman RE. J. Am. Chem. Soc. 1958;80:1339. [Google Scholar]
- 30.Cha C, Jeong JH, Shim J, Kong H. Acta Biomater. 2011;7:3719. doi: 10.1016/j.actbio.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Okada T, Hayashi T, Ikada Y. Biomaterials. 1992;13:448. doi: 10.1016/0142-9612(92)90165-k. [DOI] [PubMed] [Google Scholar]
- 32.Chu C, Schmidt JJ, Carnes K, Zhang Z, Kong HJ, Hofmann M-C. Tissue Eng. Pt. A. 2009;15:255. doi: 10.1089/ten.tea.2008.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.