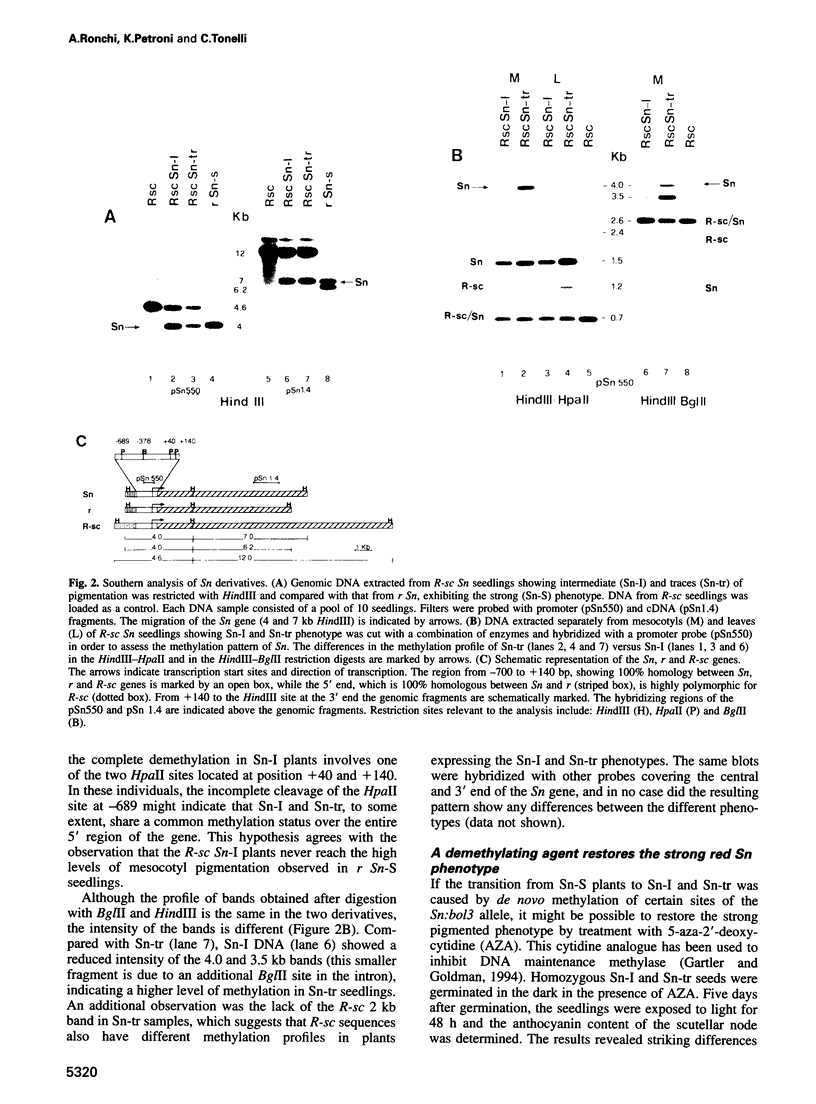
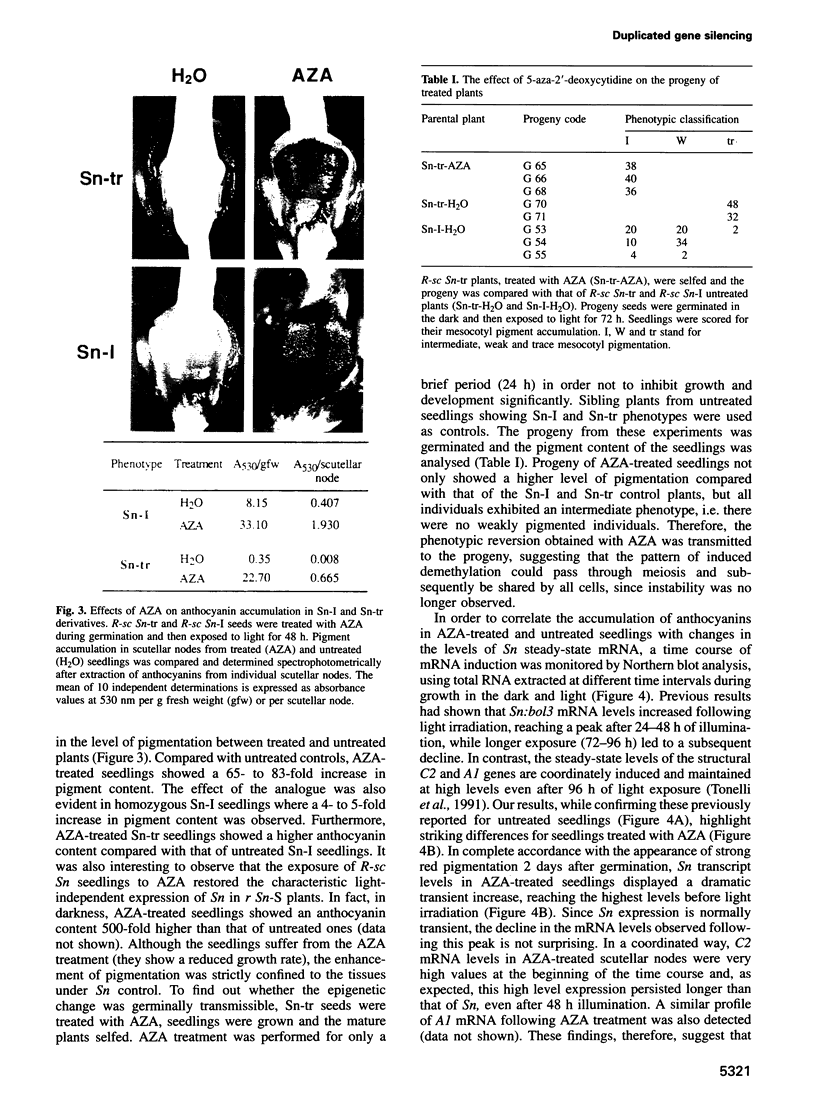
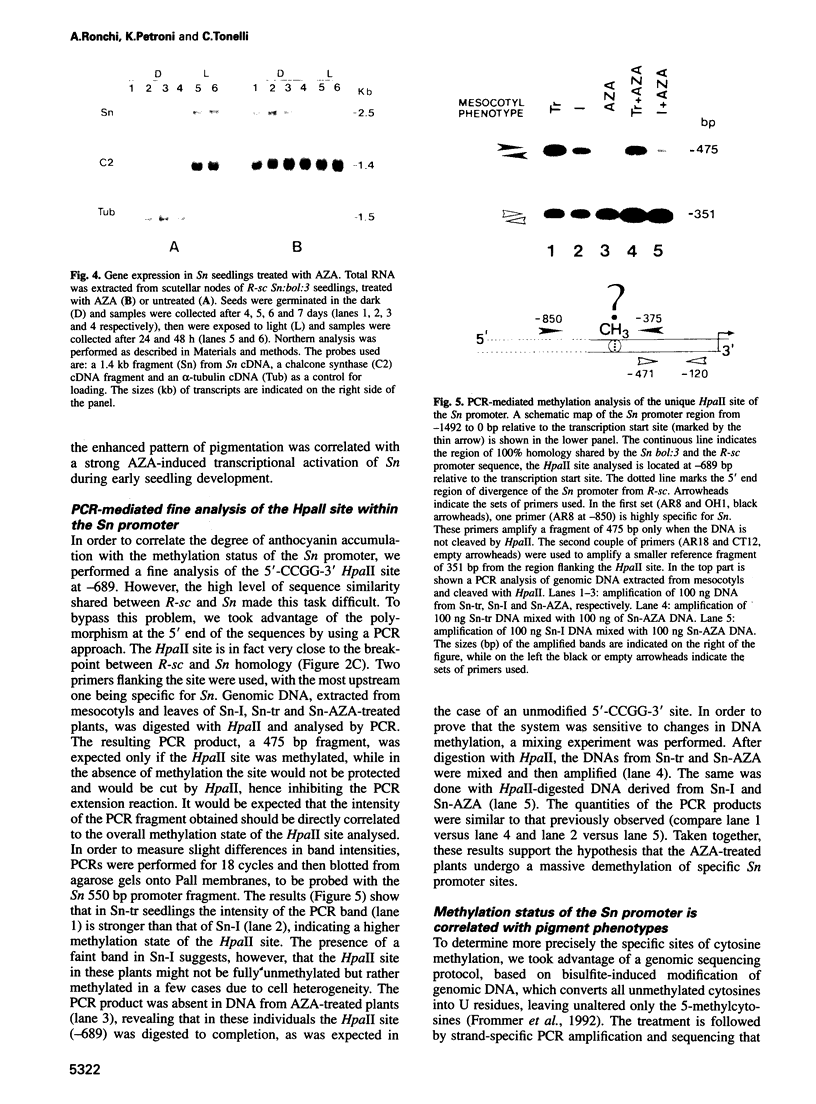
Abstract
The duplicated R and Sn genes regulate the maize anthocyanin biosynthetic pathway and encode tissue-specific products that are homologous to helix-loop-helix transcriptional activators. As a consequence of their coupling in the genome, Sn is partially silenced. Genomic restriction analysis failed to reveal gross structural DNA alterations between the strong original phenotype and the weak derivatives. However, the differences in pigmentation were inversely correlated with differences in the methylation of the Sn promoter. Accordingly, treatment with 5-azacytidine (AZA), a demethylating agent, restored a strong pigmentation pattern that was transmitted to the progeny and that was correlated with differential expression of the Sn transcript. Genomic sequencing confirmed that methylation of the Sn promoter was more apparent in the less pigmented seedlings and was greatly reduced in the AZA revertants. In addition, some methylcytosines were located in non-symmetrical C sequences. These findings provide an insight into Sn and R interaction, a process that we have termed Reduced Expression of Endogenous Duplications (REED). We propose that increasing the copy number of regulatory genes by endogenous duplication leads to such epigenetic mechanisms of silencing. Further understanding of the REED process may have broader implications for gene regulation and may identify new levels of regulation within eukaryotic genomes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aparicio O. M., Billington B. L., Gottschling D. E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991 Sep 20;66(6):1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Assaad F. F., Tucker K. L., Signer E. R. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993 Sep;22(6):1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer U., Sippel A. E., Klempnauer K. H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988 Oct 27;335(6193):835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors and the control of Drosophila development. Trends Genet. 1989 Nov;5(11):377–383. doi: 10.1016/0168-9525(89)90173-x. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri T., Ariel M., Chaillet J. R., McCarrey J., Razin A., Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993 Sep;12(9):3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R. A. Paramutation. Annu Rev Genet. 1973;7:129–152. doi: 10.1146/annurev.ge.07.120173.001021. [DOI] [PubMed] [Google Scholar]
- Brutnell T. P., Dellaporta S. L. Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics. 1994 Sep;138(1):213–225. doi: 10.1093/genetics/138.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Razin A. DNA methylation and development. Biochim Biophys Acta. 1990 May 24;1049(1):1–8. doi: 10.1016/0167-4781(90)90076-e. [DOI] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Chomet P. S., Wessler S., Dellaporta S. L. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 1987 Feb;6(2):295–302. doi: 10.1002/j.1460-2075.1987.tb04753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. J., Harrison J., Paul C. L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994 Aug 11;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Carpenter R. A semi-dominant allele, niv-525, acts in trans to inhibit expression of its wild-type homologue in Antirrhinum majus. EMBO J. 1988 Apr;7(4):877–883. doi: 10.1002/j.1460-2075.1988.tb02891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni G., Geuna F., Gavazzi G., Tonelli C. Molecular homology among members of the R gene family in maize. Plant J. 1993 Feb;3(2):335–346. doi: 10.1111/j.1365-313x.1993.tb00185.x. [DOI] [PubMed] [Google Scholar]
- Das O. P., Messing J. Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics. 1994 Mar;136(3):1121–1141. doi: 10.1093/genetics/136.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini S., Consonni G., Mereghetti M., Tonelli C. Antiparallel expression of the sense and antisense transcripts of maize alpha-tubulin genes. Mol Gen Genet. 1993 Oct;241(1-2):161–169. doi: 10.1007/BF00280213. [DOI] [PubMed] [Google Scholar]
- Dooner H. K., Kermicle J. L. Reconstitution of the R compound allele in maize. Genetics. 1974 Oct;78(2):691–701. doi: 10.1093/genetics/78.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faugeron G., Rhounim L., Rossignol J. L. How does the cell count the number of ectopic copies of a gene in the premeiotic inactivation process acting in Ascobolus immersus? Genetics. 1990 Mar;124(3):585–591. doi: 10.1093/genetics/124.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M., Sprenger M., Dinkelbach M., Yazaki K., Harter K., Weisshaar B. Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell. 1994 Nov;6(11):1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J. S., Verhave M., Kasper S., Tsukada T., Mandel G., Goodman R. H. The CGTCA sequence motif is essential for biological activity of the vasoactive intestinal peptide gene cAMP-regulated enhancer. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6662–6666. doi: 10.1073/pnas.85.18.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler S. M., Goldman M. A. Reactivation of inactive X-linked genes. Dev Genet. 1994;15(6):504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- Gavazzi G., Mereghetti M., Consonni G., Tonelli C. Sn, a light-dependent and tissue-specific gene of maize: the genetic basis of its instability. Genetics. 1990 May;125(1):193–199. doi: 10.1093/genetics/125.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin P. M., Sarokin L., Memelink J., Chua N. H. Molecular light switches for plant genes. Plant Cell. 1990 May;2(5):369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992 Dec;2(6):907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Jacobsen K., Laursen N. B., Jensen E. O., Marcker A., Poulsen C., Marcker K. A. HMG I-like proteins from leaf and nodule nuclei interact with different AT motifs in soybean nodulin promoters. Plant Cell. 1990 Jan;2(1):85–94. doi: 10.1105/tpc.2.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990 Dec;8(12):340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- Katagiri F., Lam E., Chua N. H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989 Aug 31;340(6236):727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kermicle J. L. Recombination between Components of a Mutable Gene System in Maize. Genetics. 1984 Jul;107(3):489–500. doi: 10.1093/genetics/107.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kricker M. C., Drake J. W., Radman M. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1075–1079. doi: 10.1073/pnas.89.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Interaction of a common cellular transcription factor, ATF, with regulatory elements in both E1a- and cyclic AMP-inducible promoters. Proc Natl Acad Sci U S A. 1988 May;85(10):3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn F., Heidmann I., Saedler H., Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet. 1990 Jul;222(2-3):329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- Martienssen R., Barkan A., Taylor W. C., Freeling M. Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 1990 Mar;4(3):331–343. doi: 10.1101/gad.4.3.331. [DOI] [PubMed] [Google Scholar]
- McGowan R., Campbell R., Peterson A., Sapienza C. Cellular mosaicism in the methylation and expression of hemizygous loci in the mouse. Genes Dev. 1989 Nov;3(11):1669–1676. doi: 10.1101/gad.3.11.1669. [DOI] [PubMed] [Google Scholar]
- Meyer P., Heidmann I., Niedenhof I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993 Jul;4(1):89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- Meyer P., Niedenhof I., ten Lohuis M. Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J. 1994 May 1;13(9):2084–2088. doi: 10.1002/j.1460-2075.1994.tb06483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. I., Thorpe C. J., Chandler V. L. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics. 1993 Nov;135(3):881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P. A., Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987 Dec 1;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot G. H., Cone K. C. Nucleotide sequence of the maize R-S gene. Nucleic Acids Res. 1989 Oct 11;17(19):8003–8003. doi: 10.1093/nar/17.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K., Sapienza C. Imprinting the genome: imprinted genes, imprinting genes, and a hypothesis for their interaction. Annu Rev Genet. 1993;27:7–31. doi: 10.1146/annurev.ge.27.120193.000255. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Riggs A. D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991 Jun;5(6):1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. A new bind for Myc. Trends Genet. 1992 Mar;8(3):91–96. doi: 10.1016/0168-9525(92)90196-b. [DOI] [PubMed] [Google Scholar]
- Rhounim L., Rossignol J. L., Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992 Dec;11(12):4451–4457. doi: 10.1002/j.1460-2075.1992.tb05546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Schütte J., Viallet J., Nau M., Segal S., Fedorko J., Minna J. jun-B inhibits and c-fos stimulates the transforming and trans-activating activities of c-jun. Cell. 1989 Dec 22;59(6):987–997. doi: 10.1016/0092-8674(89)90755-1. [DOI] [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Tonelli C., Consonni G., Dolfini S. F., Dellaporta S. L., Viotti A., Gavazzi G. Genetic and molecular analysis of Sn, a light-inducible, tissue specific regulatory gene in maize. Mol Gen Genet. 1991 Mar;225(3):401–410. doi: 10.1007/BF00261680. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992 Apr;1(1):45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- van Tunen A. J., Koes R. E., Spelt C. E., van der Krol A. R., Stuitje A. R., Mol J. N. Cloning of the two chalcone flavanone isomerase genes from Petunia hybrida: coordinate, light-regulated and differential expression of flavonoid genes. EMBO J. 1988 May;7(5):1257–1263. doi: 10.1002/j.1460-2075.1988.tb02939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]