Abstract
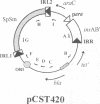
A system is described which permits visualization and analysis of a number of molecular species associated with transposition activity of the bacterial insertion sequence, IS1, in vivo. The technique involves induction of an IS1 transposase gene carried by a plasmid which also includes an IS1-based transposable element. It is, in principle, applicable to the identification of transposition intermediates as well as unstable transposition products and those which are not detectable by genetic means. Thirteen novel molecular species were detected after 4 h of induction. Five major species were characterized, based on their behaviour as a function of time, on their hybridization patterns and on the nucleotide sequences of the transposon-backbone junctions. All result from intramolecular IS1 transposition events. The two reciprocal partner products of IS1-mediated deletions, the intramolecular equivalent of co-integrates generated by intermolecular transposition, have been identified. Both carry a single copy of the transposable element and present complementary distributions of deletion endpoints. These results establish, by direct physical means, that adjacent IS1-mediated deletions are accompanied by duplication of the element. A second type of molecule identified was an excised circular copy of the transposon, raising the possibility that IS1 is capable of following an intermolecular transposition pathway, via excised transposon circles, leading to direct insertion.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S. W., Adelt G., Berg D. E. Transcriptional control of IS1 transposition in Escherichia coli. J Mol Biol. 1984 Apr 5;174(2):251–264. doi: 10.1016/0022-2836(84)90337-1. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J. Y., Woszczyk J., Repoila F., François V., Louarn J. M., Krisch H. M. Direct PCR sequencing of the ndd gene of bacteriophage T4: identification of a product involved in bacterial nucleoid disruption. Gene. 1994 Apr 8;141(1):9–16. doi: 10.1016/0378-1119(94)90121-x. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Chandler M., Galas D. J. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J Mol Biol. 1983 Oct 15;170(1):61–91. doi: 10.1016/s0022-2836(83)80227-7. [DOI] [PubMed] [Google Scholar]
- Cornelis G., Saedler H. Deletions and an inversion induced by a resident IS1 of the lactose transposon Tn951. Mol Gen Genet. 1980;178(2):367–374. doi: 10.1007/BF00270486. [DOI] [PubMed] [Google Scholar]
- Escoubas J. M., Lane D., Chandler M. Is the IS1 transposase, InsAB', the only IS1-encoded protein required for efficient transposition? J Bacteriol. 1994 Sep;176(18):5864–5867. doi: 10.1128/jb.176.18.5864-5867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas J. M., Prère M. F., Fayet O., Salvignol I., Galas D., Zerbib D., Chandler M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991 Mar;10(3):705–712. doi: 10.1002/j.1460-2075.1991.tb08000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982 Jan 15;154(2):245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Goosen N., van de Putte P. Role of ner protein in bacteriophage Mu transposition. J Bacteriol. 1986 Aug;167(2):503–507. doi: 10.1128/jb.167.2.503-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995 Jul;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R. Insertion element IS1 can generate a 10-base pair target duplication. Gene. 1986;45(2):233–235. doi: 10.1016/0378-1119(86)90260-x. [DOI] [PubMed] [Google Scholar]
- Iida S., Marcoli R., Bickle T. A. Variant insertion element IS1 generates 8-base pair duplications of the target sequence. Nature. 1981 Nov 26;294(5839):374–376. doi: 10.1038/294374a0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Regulation of transposition in bacteria. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- Lane D., Cavaillé J., Chandler M. Induction of the SOS response by IS1 transposase. J Mol Biol. 1994 Sep 30;242(4):339–350. doi: 10.1006/jmbi.1994.1585. [DOI] [PubMed] [Google Scholar]
- Lüthi K., Moser M., Ryser J., Weber H. Evidence for a role of translational frameshifting in the expression of transposition activity of the bacterial insertion element IS1. Gene. 1990 Mar 30;88(1):15–20. doi: 10.1016/0378-1119(90)90054-u. [DOI] [PubMed] [Google Scholar]
- Machida C., Machida Y. Base substitutions in transposable element IS1 cause DNA duplication of variable length at the target site for plasmid co-integration. EMBO J. 1987 Jun;6(6):1799–1803. doi: 10.1002/j.1460-2075.1987.tb02433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C., Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989 Aug 20;208(4):567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Prère M. F., Fayet O., Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992 Dec;11(13):5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Teter B., Chandler M., Galas D. J. Functional promoters created by the insertion of transposable element IS1. J Mol Biol. 1986 Oct 5;191(3):383–393. doi: 10.1016/0022-2836(86)90134-8. [DOI] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Lowe J. B., McDivitt L., Berg D. E. Control of transposon Tn5 transposition in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7450–7454. doi: 10.1073/pnas.79.23.7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Eisaki N., Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994 Feb 4;235(5):1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre M. C., Turlan C., Bortolin M., Chandler M. Mutagenesis of the IS1 transposase: importance of a His-Arg-Tyr triad for activity. J Bacteriol. 1995 Sep;177(17):5070–5077. doi: 10.1128/jb.177.17.5070-5077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Xu X., Chen J. M., Berg D. E., Berg C. M. Inversions and deletions generated by a mini-gamma delta (Tn1000) transposon. J Bacteriol. 1994 Mar;176(5):1332–1338. doi: 10.1128/jb.176.5.1332-1338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Schaus N. A., Grindley N. D. Insertion sequence duplication in transpositional recombination. Science. 1983 Nov 18;222(4625):755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Gamas P., Chandler M., Prentki P., Bass S., Galas D. Specificity of insertion of IS1. J Mol Biol. 1985 Oct 5;185(3):517–524. doi: 10.1016/0022-2836(85)90068-3. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Jakowec M., Prentki P., Galas D. J., Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987 Oct;6(10):3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D., Polard P., Escoubas J. M., Galas D., Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990 Mar;4(3):471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Zerbib D., Prentki P., Gamas P., Freund E., Galas D. J., Chandler M. Functional organization of the ends of IS1: specific binding site for an IS 1-encoded protein. Mol Microbiol. 1990 Sep;4(9):1477–1486. [PubMed] [Google Scholar]