Abstract
Circadian clocks coordinate physiology and behavior with the 24-hour solar day to provide temporal homeostasis with the external environment. The molecular clocks that drive these intrinsic rhythmic changes are based on interlocked transcription/translation feedback loops that integrate with diverse environmental and metabolic stimuli to generate internal 24-hour timing. In this review we highlight recent advances in our understanding of the core molecular clock and how it utilizes diverse transcriptional and post-transcriptional mechanisms to impart temporal control onto mammalian physiology. Understanding the way in which biological rhythms are generated throughout the body may provide avenues for temporally-directed therapeutics to improve health and prevent disease.
Keywords: circadian, transcription, post-transcription, peripheral clock
A clockwork physiology
Mammalian physiology and behavior are coordinated by an intrinsic molecular clock into rhythms that are synchronized with the 24-hour solar day. Circadian (Latin ‘circa diem’, meaning ‘about a day’) synchronization allows anticipation of regular environmental changes to influence molecular and behavioral decisions that impact fitness and survival, including food intake and metabolism, predator/prey interactions, and the evasion of DNA damage from environmental insults, amongst others [1]. Circadian rhythms therefore allow an animal to achieve temporal homeostasis (see Glossary) with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. For example, DNA damage induced by solar irradiation is preferentially repaired by the nucleotide excision repair pathway in the late afternoon and early evening, while the ability to repair such damage is at its lowest before dawn [2]. Temporal regulation of this pathway, which plays a critical role in maintaining genomic integrity, is conferred by the circadian clock through control of xeroderma pigmentosum A (XPA) protein expression, a rate-limiting factor in excision repair of UV-induced dipyrimidine photoproducts [3]. To better understand the powerful role of the circadian clock in coordinating physiology and behavior, we highlight recent advances in our understanding of the molecular mechanisms used to generate circadian rhythms of protein expression. Exciting new studies point towards an unanticipated integration of diverse transcriptional and post-transcriptional mechanisms to generate circadian rhythms of protein expression on a tissue-specific basis, demonstrating that the molecular clock utilizes many strategies to regulate circadian output and temporal homeostasis with the external environment.
Clocks throughout the body
A hierarchical timing system
Circadian rhythms are genetically encoded by a molecular clock located in nearly every cell that generates internal timing of approximately 24 hours in the absence of external cues (Box 1). Molecular clocks located throughout the body in peripheral tissues are organized into a coherent, hierarchical system by a ‘master’ clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus [4]. The SCN is comprised of approximately 20,000 neurons that form a highly unified circadian network [5]. This master clock is the only molecular clock to receive light input from the retina that synchronizes internal clock timing to the external solar day, which it passes on to peripheral clocks via endocrine and systemic cues [6, 7]. Molecular clocks located in neurons of the SCN and throughout peripheral tissues share the same molecular architecture and capacity to generate sustained circadian rhythms [8], although one key difference between master and peripheral clocks lies in the degree of their intercellular coupling [9]. A high degree of intercellular coupling among neurons of the SCN forms a neuronal network that is resistant to phase perturbations from internal cues [6], while the phase of peripheral clocks is susceptible to adjustment from the SCN clock via circulating hormones and other metabolic cues [10, 11], as well as systemic changes such as body temperature [12, 13]. This network logic ensures that the master clock faithfully keeps intrinsic ~24-hour timing to maintain temporal coordination with the external solar cycle, while peripheral clocks adapt to reflect the local metabolic status of the tissues in which they function [14].
Box 1. The molecular clock in mammals.
The cell-autonomous molecular clock in mammals is generated by two interlocking transcription/translation feedback loops (TTFL) that function together to produce robust 24-hour rhythms of gene expression. The core TTFL is driven by four integral clock proteins: two activators (CLOCK and BMAL1) and two repressors (PER and CRY), as well as kinases and phosphatases that regulate the localization and stability of these integral clock proteins (kinases: CKIα, CKIδ, and CKIε; phosphatases PP1, PP5). CLOCK and BMAL1 are subunits of the heterodimeric basic helix-loop-helix-PAS (PER-ARNT-SIM) transcription factor CLOCK:BMAL1 [59], which activates transcription of the repressor Per and Cry genes, as well as other clock-controlled output genes. PER and CRY proteins heterodimerize in the cytoplasm and translocate to the nucleus to interact with CLOCK:BMAL1, inhibiting further transcriptional activation. As PER and CRY proteins are degraded through ubiquitin-dependent pathways [72, 73, 109–111], repression on CLOCK:BMAL1 is relieved and the cycle begins again with ~24-hour periodicity. The casein kinases CKIδ and CKIε play an important role in determining the intrinsic period of the clock by controlling the rate at which the PER:CRY complexes are either degraded or enter the nucleus, and their activity is either counteracted or regulated by the phosphatases PP1 and PP5, respectively [48, 112]. Notably, familial mutations resulting in the loss of a single phospho-acceptor site on PER2 (S662G) [113] or a loss-of-function mutation in CKIδ (T44A) [114] shorten the intrinsic period of the clock in mice and give rise to sleep phase disorders in humans. A key role for the casein kinases in establishing period length has also been demonstrated pharmacologically via modulation of the kinases with small molecule inhibitors, which dramatically lengthen the period by modulating PER localization and stability [104, 107]. A second TTFL is generated through transcriptional activation by the retinoid-related orphan receptors (RORa, b, c) [115] and repression by REV-ERBα/REVERBβ [32]. TTFL drives rhythmic changes in Bmal1 transcription and introduces a delay in Cry1 mRNA expression that offsets it from genes regulated strictly by CLOCK:BMAL1 [55]. While rhythmic changes in BMAL1 abundance are not required to drive the core TTFL loop [17], the ROR/REV TTFL-induced delay in Cry1 expression is critical for proper circadian timing [55]. The presence of cooperative, interlocking feedback loops provides robustness against noise and environmental perturbations to help keep accurate circadian timing, and also helps to generate phase delays in circadian transcriptional output that optimally time gene expression for local physiology [44].
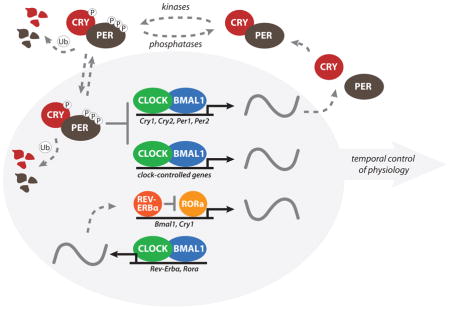
→ box footnotes: Abbreviations: BMAL1, brain and muscle ARNT-like 1; CLOCK, circadian locomotor output cycles kaput; CKI, casein kinase I; CRY, cryptochrome; PER, period; PP, protein phosphatase
Circadian regulation of physiology is locally controlled
Recent studies utilizing genetic tools have explored how master and peripheral clocks are integrated with circadian control of physiology. Single knockouts of most integral clock genes (Box 1) do not completely disrupt behavioral rhythms due to apparent functional redundancy of paralogs or compensation by the coupled SCN network [9, 15–17], both of which provide resiliency to maintain clock function. For example, both Period genes (Per 1 and Per2) are required for cycling [18], perhaps due to their low abundance as the stoichiometrically limiting factor in forming key clock protein complexes [19].
Deletion of Bmal1 is the only single knockout that eliminates clock function in both the SCN and peripheral tissues; Bmal1−/− mice lack all molecular and behavioral circadian phenotypes, and present additional phenotypes such as decreased activity and body weight, as well as a shortened lifespan [20–22]. Transgenic expression of Bmal1 in the brain of Bmal1−/− knockout mice restores circadian behavioral rhythms, but does not rescue other comorbid phenotypes; however, rescue of Bmal1 expression solely in muscle tissue improves activity levels, body weight, and increases lifespan, while the animals remain behaviorally arrhythmic [21]. These surprising data suggest that local control of peripheral clock output is sufficient to drive tissue-specific physiology. As predicted from this observation, tissue-specific deletions of Bmal1 in the context of an otherwise rhythmic animal consistently have profound effects on local tissue function that resonate throughout the physiology of the animal [23–26]. Disruption of the peripheral clock in the liver by targeted deletion of Bmal1 eliminates circadian regulation of glucose regulatory genes, which interferes with the export of hepatic blood glucose in a time-of-day-dependent manner and leads to a systemic disruption of blood glucose buffering over the course of a 24-hour behavioral cycle [24]. Nearly all clock-controlled genes in the liver require the local oscillator for proper temporal expression, although a small number of genes expressed with circadian timing are systemically controlled by the master clock in the SCN [27]. Therefore, circadian timing of tissue function stems predominantly from local control of clock-controlled proteins with some modulation by systemic pathways. Learning how peripheral clocks integrate signals to generate tissue-specific, rhythmic output will be important to understand how the circadian clock provides temporal homeostasis in normal and pathophysiological settings.
Transcriptional control of circadian output
Peripheral clocks regulate tissue-specific expression patterns
Early microarray studies revealed that peripheral clocks regulate vast transcriptional programs to induce a single peak of expression once per day for each of the clock-controlled genes, representing approximately 10% of all expressed genes. These oscillating messenger RNA transcripts are specifically coordinated with tissue function, resulting in only ~1–3% convergence in oscillating transcripts between liver, heart, muscle and the SCN [28–30]. Comparing clock-driven transcripts in the liver and heart, commonly regulated transcripts include integral clock genes and others that play a key role in maintenance of basic oscillator function and output, such as Rev-Erbα and the PAR bZIP transcription factors Dbp and Tef [31, 32], while genes regulated in a tissue-specific manner reveal functional ties to amino acid and glucose metabolism (liver only) or G-protein-coupled receptor pathways (heart only) [30]. These studies established the paradigm that circadian output is primarily controlled at the transcriptional level (Box 1), but raised the question of how molecular clocks built upon the same molecular architecture can regulate such divergent gene expression in a tissue-dependent manner. Certainly, tissue-restricted expression of transcriptional co-regulators and/or downstream transcription factors can contribute to unique circadian transcriptional responses in different tissues [31, 33]. However, a proteomic analysis of rhythmically expressed hepatic proteins provided the first glimpse that a purely transcription-driven model could not fully explain circadian rhythms of protein expression in the liver. At least 50% of the soluble proteins examined had robust changes in abundance throughout the 24-hour day with no detectable mRNA oscillation [34]. This is consistent with new findings that circadian control of poly(A) tail length can regulate translation independent of the levels of steady-state mRNA for some genes [35]. Furthermore, the peak expression of some alternatively spliced protein isoforms can also vary significantly [36], suggesting important roles for post-transcriptional, translational and/or post-translational regulation in generating circadian output.
How does the interplay of transcriptional and post-transcriptional regulatory mechanisms determine circadian output? Recently, many groups have begun to address this question by performing genome-wide studies examining the temporal recruitment of integral clock proteins, transcriptional machinery and epigenetic modifications to chromatin to determine how the clock confers temporal control over transcriptional output [15, 37–43]. By quantifying nascent transcripts relative to mature mRNAs, surprisingly, it was found that only ~22–30% of cycling mRNAs have robust, underlying circadian rhythms of transcription [39, 41], indicating that the vast majority of circadian changes in mRNA levels may result from post-transcriptional regulation. However is it currently unknown what factors determine how a specific gene is regulated (i.e. transcriptionally and/or post-transcriptionally) and how these regulatory processes are integrated between different tissues to exert circadian control over physiology. Future studies focusing on the peripheral clock in the liver, which is an ideal model for how the molecular clock integrates internal timing with systemic and metabolic cues, will provide significant advances towards answering these questions.
Dynamics of circadian transcriptional regulation
Insight into the molecular clock mechanism
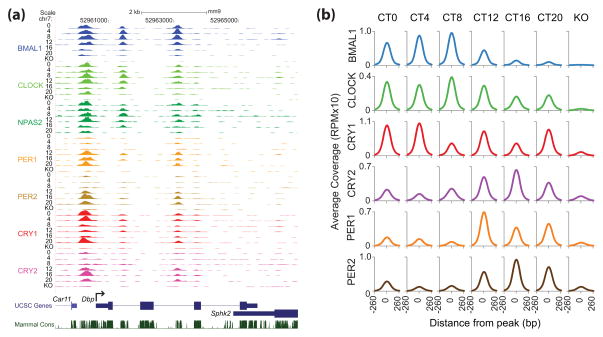
The heterodimeric CLOCK:BMAL1 complex is the essential positive regulator of circadian transcription in mammals, binding to several thousand sites throughout the genome in the liver with peak occupancy mid-day at circadian time (CT) CT5–8 (approximately 11 a.m. to 2 p.m.) [39, 42]. CLOCK:BMAL1 binding occurs primarily at consensus E-box DNA motifs and recruits complexes of the integral clock protein transcriptional repressors PER1, PER2, CRY1 and CRY2 with peak occupancy in the evening at CT15–18 [39]. In total, over 1400 distinct sites in the liver genome are bound by all six integral clock proteins with this temporal profile (Figure 1). The current model of the molecular clock (Box 1) posits that cyclical repression of CLOCK:BMAL1 activity by PER:CRY complexes establishes rhythms of transcriptional output occuring with ~24-hour periodicity, which are expanded beyond direct CLOCK:BMAL1 target genes through circadian regulation of additional transcription factors and co-regulators [44]. Outside of their role in establishing the negative arm of the core circadian transcription/translation feedback loop, PER and CRY proteins are also recruited to a staggering number of other sites throughout the genome (>5000 sites independent of CLOCK:BMAL1), that are enriched for nuclear hormone receptor binding sites [39]. This enrichment is in line with their ability to bind and regulate the transcriptional activity of a diverse array of nuclear hormone receptors [45, 46]. These data provide a glimpse into the widespread temporal regulation of transcription factors outside of the core molecular clock loop by PER and CRY, and demonstrate how clock-driven oscillations in their abundance over the 24-hour day may convey timing cues to downstream transcription factors through rhythmic transcriptional regulation.
Figure 1.
Temporal resolution of clock protein recruitment to genes that are transcriptionally regulated by the molecular circadian clock. (a) UCSC genome browser view of BMAL1 (blue), CLOCK (light green), NPAS2 (green), PER1 (orange), PER2 (gold), CRY1 (red) and CRY2 (purple) occupancy at the Dbp locus. Each track represents the normalized ChIP-seq read coverage (wiggle plot) at a single time point. For each transcription factor, six time points every 4 hr over a circadian cycle are shown beginning at CT0 and ending at CT20. Knockout (KO) mice were used as a negative control for each factor except NPAS2. The conservation track shows 30-Way Multiz Alignment & Conservation scores (PhastCons) provided by the UCSC genome browser. (b) Binding coverage profiles from ChIP-Seq experiments illustrate the orchestrated recruitment of core circadian proteins to over 1400 shared genomic sites and how the core circadian transcriptional regulatory complex remodels over the 24-hour day. The binding of individual clock proteins BMAL1 (blue), CLOCK (green), CRY1 (red), CRY2 (purple), PER1 (orange), and PER2 (brown) are shown from −260 to +260 base pairs surrounding the shared binding sites at different Circadian Times (CT) throughout the day in wild-type mice or in mice with a deletion of the indicated clock gene (KO). Reprinted with permission from [39].
Significant insight into the core transcription/translation feedback loop model of the molecular clock is gained by examining, at high genomic resolution, how formation of the core circadian transcriptional machinery is controlled as a function of time. PER proteins transport cryptochromes into the nucleus with appropriate circadian timing [19, 47, 48], but may also contribute directly to repression of clock-controlled target genes through interaction with several classes of RNA-binding proteins, helicases, and other transcriptional repressors [49–52]. Remarkably, PER appears to regulate circadian control of transcription by at least two different modes. First, PER interacts directly with the CLOCK:BMAL1 at the tail end of the nascent transcript peak to recruit complexes containing SIN3-HDAC that remodel chromatin to repress transcription [50]. Second, PER complexes are also recruited to the Per1 3′ termination site through interactions with RNA-binding proteins and helicases that may play a role in transcription termination to regulate transcription independently of CLOCK:BMAL1 interactions [52].
Cryptochromes are potent repressors of CLOCK:BMAL1 transcriptional activation, although their mechanism of action has not yet been determined [53]. Of the two mammalian cryptochromes, only CRY1 can sustain circadian cycling in the absence of compensation from SCN network properties [9, 15], and it requires a significant delay in expression relative to CRY2 that is encoded by multiple clock-regulated promoter elements [54, 55]. Expressing Cry2 from a Cry1 promoter provides the delayed phase of expression but does not restore cycling [56]; instead, the ability of CRY1 to sustain circadian cycling may be encoded biochemically [57], perhaps by mediating a unique repressive function or perhaps through increased affinity for the CLOCK:BMAL1 complex. CRY1 can interact with CLOCK:BMAL1 independently of PER proteins [58] and is found at CLOCK:BMAL1-bound sites in the early morning at CT0 (Figure 1) [39, 54], suggesting that CRY1 may act as a molecular gatekeeper to maintain CLOCK:BMAL1 in a poised and repressed state until the proper time for transcriptional activation.
Towards atomic resolution: structural studies provide new insight
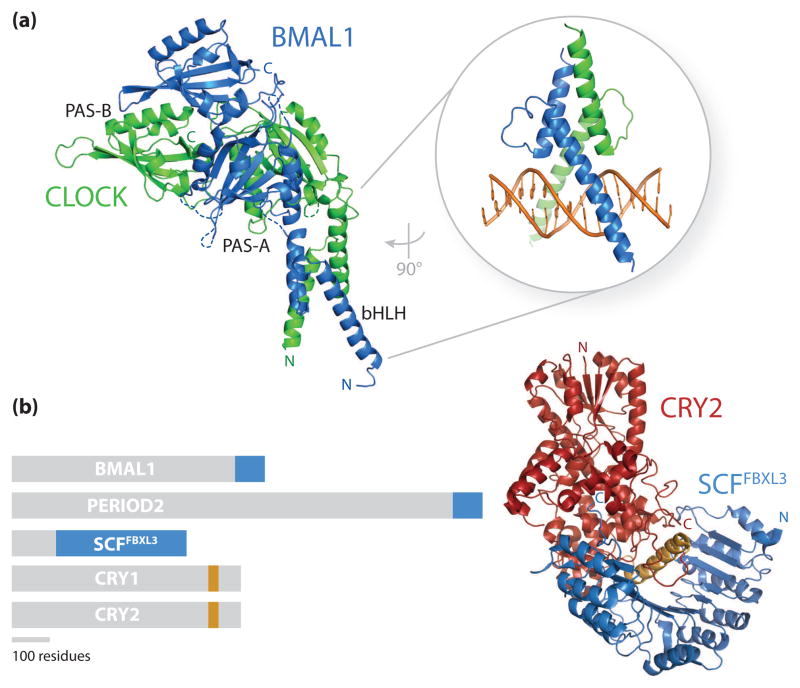
Recent high-resolution structures of clock proteins are beginning to provide fresh insight into the architecture of the circadian transcriptional machinery. Structures of the DNA binding and PAS domains of CLOCK:BMAL1 [59, 60] illustrate how the heterodimeric bHLH-PAS (basic helix-loop-helix PER-ARNT-SIM) complex interacts with E-box sites on DNA and uses modular PAS domains to provide selectivity for each other (Figure 2a) within the bHLH-PAS family of environmentally sensitive transcription factors [61]. Within this family, both the PAS domains and the intrinsically disordered regions that follow regulate dynamic binding of coactivators and repressors to control transcriptional activation [62]. The PAS domains of CLOCK and BMAL1 have been implicated in PER binding [47, 63], potentially through direct interaction with the tandem PAS homodimers of PER [64, 65]. Moreover, an overlapping region on CLOCK PAS-B has been implicated in direct binding of CRY1 [66], as has a region at the C-terminus of BMAL1 [63, 67], suggesting that the architecture of the core clock protein complex may remodel throughout the day to control transcriptional output. The unstructured C-terminal regions of CLOCK and BMAL1 exhibit structural plasticity, allowing them to interact with several transcriptional regulators that control genomic targeting through chromatin remodeling and the recruitment of transcriptional machinery [66, 68, 69]. The concept that clock function may be partially driven by competition for binding to a few critical signaling ‘hot spots’ on the core clock proteins is underscored by the observation that BMAL1, PER [70, 71] and the E3 ubiquitin ligase FBXL3 [72, 73] all compete for interaction with cryptochromes at a conserved, surface accessible helix (Figure 2b) [74, 75]. Future mechanistic work examining how interactions between these proteins regulate CRY function may provide insight into the timely progression of repressed and activated states of the CLOCK:BMAL1 complex.
Figure 2.
Structural determinants of clock protein complex assembly and competition. (a) Ribbon diagram of the mouse CLOCK:BMAL1 heterodimer bHLH-PAS structure (Protein Data Bank (PDB) code 4F3L, taken from [59]) illustrating how the DNA-binding bHLH domain and tandem PAS domains each contribute to complex formation between CLOCK (green) and BMAL1 (blue). This structure lacks the C-terminal regions of each protein (471 of 855 residues in CLOCK and 179 of 626 residues in BMAL1) that lack ordered structure but are required for CLOCK:BMAL1 activity and clock function. Inset, a closer view of the CLOCK:BMAL1 bHLH domain bound to a canonical E-box element in DNA (PDB code 4H10 [60]). (b) A schematic representation of clock protein domain organization highlights regions on BMAL1, PER, and the E3 ubiquitin ligase SCFFBXL3 (blue) that compete for interaction with the conserved CC helix of cryptochromes (orange), as illustrated in the mouse CRY2: SCFFBXL3 complex structure (PDB code 4I6J [75]). The CC helix (orange) of CRY2 (red) serves as the primary docking site for the SCFFBXL3 E3 ubiquitin ligase (blue), which competes with PER2 for binding at this site [74, 75] to regulate the stability of cryptochromes.
The clock links daily epigenetic changes and RNAPII occupancy to transcriptional activation
Given the complexity of the circadian transcriptional machinery, how is temporal control of transcriptional output regulated so robustly over the 24-hour day? The clock appears to exert temporal control over RNA polymerase II (RNAPII) recruitment, rather than a rhythmic transition from paused to productive elongation, to execute circadian changes in transcription [39, 40]. Phosphorylation of the RNAPII C-terminal domain at serine 5 (RNAPII-Ser5P) correlates with promoter escape and transcriptional initiation [76]. Genomic analysis of RNAPII recruitment to sites bound by CLOCK:BMAL1 over the 24-hour day shows that transcriptionally poised RNAPII-Ser5P exhibits a circadian peak of binding to CLOCK:BMAL1 sites in the early morning at CT0.6 [39]. Transcriptional initiation likely requires removal of CRY1 from CLOCK:BMAL1 in favor of the histone acetyltransferase coactivators p300 and CBP [63, 77], which begins at CT4 and continues for the next four hours with a simultaneous increase in histone acetylation [39]. Transcriptional initiation occurs mid-day with a peak in the formation of nascent transcripts throughout the afternoon and early evening, abruptly decreasing upon recruitment of the repressive PER:CRY complexes to sites of CLOCK:BMAL1-regulated genes [39, 41]. Rhythms of histone modifications at promoters, enhancers, and throughout gene bodies over the 24-hour day correlate with predicted accessibility to transcriptional machinery [39, 40, 43], consistent with the requirement of diverse histone modifying enzymes for robust circadian cycling [68, 77–82]. Temporal changes in chromatin remodeling therefore play an important role in priming the epigenetic landscape for circadian regulation of transcription and serve as an additional point of integration with metabolic cues [83].
Different modes of circadian transcriptional regulation
The integral clock genes and well-characterized clock-controlled output genes that have been studied for over a decade typically exhibit high amplitude rhythms of transcription that confer high amplitude rhythms of mRNA abundance. What distinguishes these ‘model’ clock-controlled genes from others that exhibit either low amplitude transcriptional rhythms or lack them altogether? The significant overlap between genes common to molecular clocks and genes highly transcriptionally regulated and required for the generation of molecular rhythms suggests that peripheral clocks are hard-wired to encode clock function at the level of transcription [41, 42]. Furthermore, a subset of these core clock genes is defined by the presence of a tandem E-box motif (E1–E2) with precisely defined spacing of two E-boxes that engages CLOCK:BMAL1 cooperatively to increase recruitment of circadian transcriptional machinery at the promoters [42]. Computational studies suggest these regulatory motifs may help cluster specific transcription factors to regulate circadian dependent transcription. [44]. Notably, a comparison of the degree of circadian transcription factor occupancy and the surrounding chromatin environment correlates with the degree of cycling [43], suggesting that temporal regulation of transcription is achieved through synergistic remodeling of the epigenetic landscape and recruitment of the clock transcriptional regulatory complex.
One surprise from genome-wide surveys is the finding that both chromatin remodeling and RNAPII recruitment exhibit a circadian-dependent variation at nearly all actively expressed genes, even at genes that do not exhibit robust circadian changes in transcription [39, 40, 43]. Widespread changes in epigenetic modifications, such as H3K4 trimethylation and H3K9/H3K27 acetylation, create favorable environments on chromatin for RNAPII recruitment, transcriptional initiation and elongation. These changes could contribute to an overall increase in transcription over a specific period within the 24-hour day, and may suggest that gene expression itself correlates with internal timing provided by the circadian clock. Nearly 70% of genes that have a rhythmic change in mRNA abundance lack a detectable circadian rhythm of transcription, although most of these transcriptionally arrhythmic genes have elevated levels of nascent transcription that precede their cycling mRNA peaks [39, 41]. Moreover, increased variability in transcription rates over a 24-hour day correlates with rhythmic mRNA expression even in the absence of a defined 24-hour transcriptional rhythm [41], demonstrating that transcriptional discontinuity may play a significant role in the emergence of rhythmic properties in the mammalian clock [84].
Cis-acting regulatory elements may play a role in shaping the kinetics of transcriptional bursts that contribute to this variability, or noise [85, 86], likely through recruitment and retention of general transcription machinery and co-regulators by factors that remodel the local environment [87], such as poly-ADP ribose polymerase (PARP) and the histone deacetylase SIRT1. Notably, the activity of both PARP and SIRT1 is regulated by circadian changes in the abundance of required coenzyme nicotinamide adenine dinucleotide (NAD+) [88, 89], which in turn allows the enzymes to influence timing of the molecular clock [82, 90]. NAD+ levels vary with metabolic state, potentially serving as an additional point of integration between peripheral clock function and metabolism at the level of transcriptional regulation [91]. Whether by targeted regulation of specific genes or by influencing chromatin structure and RNAPII occupancy genome-wide, molecular clocks utilize many strategies to regulate transcriptional output. These strategies may contribute to the flexibility necessary to confer temporal regulation of physiology on a tissue-specific basis.
Post-transcriptional/translational control of circadian output
Generation of the 24-hour molecular clock that drives circadian changes in physiology and behavior is rooted in transcriptional regulation of integral clock genes, although it is surprisingly resilient to a reduction in overall transcriptional rates [92]. Outside of these integral clock genes, post-transcriptional/translational regulation likely helps to provide additional flexibility by generating rhythms in mRNA transcripts or proteins in a tissue- or stimulus-dependent manner. For example, genes with arrhythmic transcription and rhythmic mRNA are enriched for metabolic functions and sensory detection [41], suggesting that one purpose of post-transcriptional regulation of clock targets may be to integrate metabolic or environmental cues more rapidly than transcription alone could allow. Studies are just beginning to shed light on the multitude of post-transcriptional/translational mechanisms that provide this regulation, although our understanding of how these mechanisms collaborate to regulate circadian biology is far from complete. Future work should be focused on identifying key proteins that confer post-transcriptional regulation to clock target genes, elucidating their mechanism of action, and demonstrating their sufficiency for generating rhythms at their respective molecular levels.
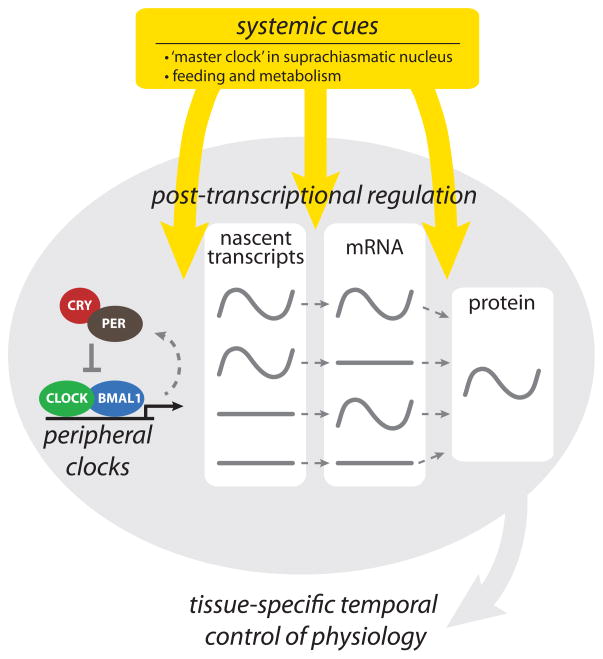
Notably, among the small set of systemically controlled genes in the liver (those that remained rhythmic when the local clock in the liver was specifically ablated) are several that encode proteins involved in post-transcriptional control, such as FUS, CIRP and Nocturnin [27]. CIRP (Cold-inducible RNA-binding protein) plays an important role in the core clock mechanism through its interaction with Clock mRNA, which results in stabilization of Clock mRNA in the cytoplasm. A reduction in CIRP levels causes low levels of CLOCK protein as well as other circadian components, leading to significant blunting of molecular circadian rhythms [93]. By contrast, Nocturnin is not part of the core clock mechanism, but instead is a deadenylase that regulates the stability and/or translatability of circadian output mRNAs through control of poly(A) tail length [94, 95]. The importance of this post-transcriptional circadian control mechanism is clear since loss of Nocturnin results in strong metabolic phenotypes in mice, including resistance to diet-induced obesity [96, 97]. Importantly, the interaction of systemically controlled genes with the molecular machinery of peripheral clocks helps to give rise to tissue-specific patterns and 12-hour harmonics of circadian gene expression, driven by the integration of transcriptional and post-transcriptional mechanisms (Figure 3) [27, 98, 99]. Understanding the interplay between transcriptional and post-transcriptional mechanisms, as well as how locally controlled clock genes are additionally regulated by systemic cues will be important for a complete understanding of the circadian gene expression landscape in each tissue and cell type.
Figure 3.
Schematic outline of the relationship between molecular mechanisms that contribute to the generation of circadian rhythms. Local control of circadian transcriptional and post-transcriptional mechanisms is integrated with systemic cues from the master clock in the SCN, as well as feeding and metabolism, to generate tissue-specific changes in transcription, mRNA accumulation and protein production. Different classes of clock-controlled genes are illustrated with either flat lines, to demonstrate constitutive expression at a given step (e.g. nascent transcript or mRNA), or sinusoidal lines, to demonstrate circadian regulation giving a once-per-day peak. Recent studies support the existence of these discrete classes [35, 39, 41], although the mechanisms used to discriminate between regulation of specific target genes are not well understood. The flexibility afforded by this complex, integrative approach likely allows for the maintenance of a core molecular clock in each cell (primarily driven by transcription) while allowing the tissue-specific control of clock-controlled genes necessary for temporal regulation of physiology by the clock.
Concluding remarks
Our lives are intimately tied to the external environment through the interaction of our intrinsic molecular clocks with the solar cycle. Knowledge of how internal clocks drive 24-hour timing to temporally coordinate physiology has improved in recent years, but we still lack fundamental insight into the molecular basis of circadian timing and tissue-dependent control of physiology. Although built upon transcription-based feedback loops, it now appears that the majority of clock-controlled processes arise from some type of post-transcriptional regulation [39, 41], possibly to provide flexibility to peripheral clocks to afford local, tissue-specific control over physiology. Looking ahead to translational studies that aim to capitalize on circadian control of temporal homeostasis and to control physiological integration at the organism level, it will be important to identify the specific molecular mechanisms that regulate each peripheral clock, alongside the required systemic and local metabolic cues. Furthermore, while structural and mechanistic studies [58, 59, 64, 65, 67, 74, 75, 100–102] are beginning to provide insight via much-needed atomic resolution of the core clock proteins that drive circadian TTFLs, more studies are needed to explore how 24-hour timing arises and how interindividual differences affect this timing through inherited mutations and polymorphisms [103]. Such studies would also help identify molecular targets and/or mechanisms for pharmacological modulators of circadian timing identified from high throughout screens [104–106]. The recent identification of “clock enhancing molecules” that increase the robustness of circadian cycling, even in the presence of mutant clock proteins [107], suggests that the dysfunctional clocks can be targeted with novel therapeutics that reinforce internal timing for personalized medical treatment regimes in disease and throughout the decline of circadian function in aging [108].
Highlights.
The circadian clock generates molecular rhythms with 24-hour periodicity.
Circadian control of physiology is distributed to peripheral tissues.
24-hour timing arises from the ordered recruitment of clock proteins to promoters.
Many mechanisms are used to generate rhythmic output from the core molecular clock.
Clocks integrate with systemic cues to give flexibility to circadian physiology.
Acknowledgments
We would like to thank the anonymous reviewers for their insightful comments and helpful suggestions. This work was supported by the National Institutes of Health (NIH P50 MH074924 and R01 MH078024 to J.S.T. and NIH P50 MH074924, R01 GM090247 and R21 NS079986 to C.B.G.) and by start-up funds from the University of California Santa Cruz (C.L.P.). J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Glossary
- Circadian time (CT)
a standard of time based on the internal free-running period of a circadian clock. By convention, the onset of activity in diurnal organisms defines circadian time zero (CT 0; usually 6 a.m.), while the onset of activity in nocturnal organisms defines circadian time twelve (CT 12)
- Entrainment
synchronization of an internal circadian oscillator to an environmental stimulus that occurs at regular intervals (usually with ~24-hour periodicity)
- Free-running
the state of a self-sustaining molecular rhythm (oscillation) in the absence of any external cues that may affect the period of the oscillator
- Homeostasis
the tendency to maintain internal equilibrium by adjusting physiological processes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lowrey P, Takahashi J. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annual review of genomics and human genetics. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang TH, et al. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang TH, et al. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko C, Takahashi J. Molecular components of the mammalian circadian clock. Human molecular genetics. 2006;15(Spec No 2):7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 5.Mohawk J, Takahashi J. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends in neurosciences. 2011 doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buhr E, et al. Temperature as a universal resetting cue for mammalian circadian oscillators. Science (New York, NY) 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dibner C, et al. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 8.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu A, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralph M, et al. Transplanted suprachiasmatic nucleus determines circadian period. Science (New York, NY) 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, et al. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 12.Brown S, et al. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 13.Saini C, et al. Simulated body temperature rhythms reveal the phase- shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26:567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 15.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBruyne J, et al. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nature neuroscience. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu A, et al. Redundant function of REV-ERBalpha and beta and non- essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, et al. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondratov R, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes & development. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDearmon E, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science (New York, NY) 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, et al. The mortality of MOP3 deficient mice with a systemic functional failure. Journal of biomedical science. 2006;13:845–851. doi: 10.1007/s11373-006-9108-4. [DOI] [PubMed] [Google Scholar]
- 23.Cheng B, et al. Tissue-intrinsic dysfunction of circadian clock confers transplant arteriosclerosis. Proc Natl Acad Sci U S A. 2011;108:17147–17152. doi: 10.1073/pnas.1112998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamia K, et al. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storch KF, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007:5. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller B, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 30.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 31.Gachon F, et al. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell metabolism. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 33.Hamaguchi H, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. The Biochemical journal. 2004;382:43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy A, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Kojima S, et al. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes & development. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlincy N, et al. Regulation of alternative splicing by the circadian clock and food related cues. Genome biology. 2012:13. doi: 10.1186/gb-2012-13-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science (New York, NY) 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatanaka F, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol. 2010;30:5636–5648. doi: 10.1128/MCB.00781-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (New York, NY) 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Martelot G, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012:10. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menet J, et al. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012:1. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011:9. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell metabolism. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown S, et al. (Re)inventing the circadian feedback loop. Developmental cell. 2012;22:477–487. doi: 10.1016/j.devcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Lamia K, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmutz I, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H-m, et al. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A. 2011;108:16451–16456. doi: 10.1073/pnas.1107178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown S, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science (New York, NY) 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 50.Duong H, et al. A molecular mechanism for circadian clock negative feedback. Science (New York, NY) 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalska E, et al. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol. 2012;32:4585–4594. doi: 10.1128/MCB.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Padmanabhan K, et al. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science (New York, NY) 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 53.Tamanini F, et al. Structure function analysis of mammalian cryptochromes. Cold Spring Harb Symp Quant Biol. 2007;72:133–139. doi: 10.1101/sqb.2007.72.066. [DOI] [PubMed] [Google Scholar]
- 54.Stratmann M, et al. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev. 2010;24:1317–1328. doi: 10.1101/gad.578810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Khan S, et al. Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J Biol Chem. 2012;287:25917–25926. doi: 10.1074/jbc.M112.368001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarthy E, et al. Generation of a novel allelic series of cryptochrome mutants via mutagenesis reveals residues involved in protein-protein interaction and CRY2-specific repression. Mol Cell Biol. 2009;29:5465–5476. doi: 10.1128/MCB.00641-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye R, et al. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang N, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science (New York, NY) 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, et al. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Research. 2012;2012:1–12. doi: 10.1038/cr.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McIntosh B, et al. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annual review of physiology. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 62.Partch C, Gardner K. Coactivator recruitment: a new role for PAS domains in transcriptional regulation by the bHLH-PAS family. Journal of cellular physiology. 2010;223:553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiyohara Y, et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc Natl Acad Sci U S A. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hennig S, et al. Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol. 2009:7. doi: 10.1371/journal.pbio.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kucera N, et al. Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc Natl Acad Sci U S A. 2012;109:3311–3316. doi: 10.1073/pnas.1113280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao WN, et al. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nature cell biology. 2007;9:268–275. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
- 67.Czarna A, et al. Quantitative analyses of cryptochrome-mBMAL1 interactions: mechanistic insights into the transcriptional regulation of the mammalian circadian clock. The Journal of biological chemistry. 2011;286:22414–22425. doi: 10.1074/jbc.M111.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature structural & molecular biology. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahata S, et al. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes to cells: devoted to molecular & cellular mechanisms. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- 70.Chaves I, et al. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Molecular and cellular biology. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozber N, et al. Identification of two amino acids in the C-terminal domain of mouse CRY2 essential for PER2 interaction. BMC molecular biology. 2010;11:69. doi: 10.1186/1471-2199-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science (New York, NY) 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 73.Siepka S, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Czarna A, et al. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013;153:1394–1405. doi: 10.1016/j.cell.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 75.Xing W, et al. SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuda N, et al. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Etchegaray JP, et al. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 78.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science (New York, NY) 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doi M, et al. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 80.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 81.Jones M, et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masri S, et al. The circadian clock transcriptional complex: metabolic feedback intersects with epigenetic control. Annals of the New York Academy of Sciences. 2012;1264:103–109. doi: 10.1111/j.1749-6632.2012.06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suter D, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science (New York, NY) 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 86.Suter D, et al. Origins and consequences of transcriptional discontinuity. Current opinion in cell biology. 2011;23:657–662. doi: 10.1016/j.ceb.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 87.Motta-Mena L, et al. The three Rs of transcription: recruit, retain, and recycle. Mol Cell. 2010;40:855–858. doi: 10.1016/j.molcel.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science (New York, NY) 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramsey K, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science (New York, NY) 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Asher G, et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 91.Peek C, et al. Nutrient sensing and the circadian clock. Trends in endocrinology and metabolism: TEM. 2012;23:312–318. doi: 10.1016/j.tem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dibner C, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. The EMBO journal. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morf J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science (New York, NY) 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 94.Baggs J, Green C. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Current biology: CB. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 95.Garbarino-Pico E, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA (New York, NY) 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Current biology: CB. 2011;21:1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Green C, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hughes M, et al. Harmonics of circadian gene transcription in mammals. PLoS genetics. 2009:5. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hughes M, et al. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS genetics. 2012:8. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pardee K, et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta. PLoS Biol. 2009:7. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Phelan C, et al. Structure of Rev-erbalpha bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nature structural & molecular biology. 2010;17:808–814. doi: 10.1038/nsmb.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z, et al. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell research. 2012 doi: 10.1038/cr.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ptácek L, et al. Novel insights from genetic and molecular characterization of the human clock. Cold Spring Harb Symp Quant Biol. 2007;72:273–277. doi: 10.1101/sqb.2007.72.017. [DOI] [PubMed] [Google Scholar]
- 104.Hirota T, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Isojima Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Solt L, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen Z, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pagani L, et al. Serum factors in older individuals change cellular clock properties. Proc Natl Acad Sci U S A. 2011;108:7218–7223. doi: 10.1073/pnas.1008882108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Godinho S, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science (New York, NY) 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 110.Reischl S, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 111.Shirogane T, et al. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 112.Partch C, et al. Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci U S A. 2006;103:10467–10472. doi: 10.1073/pnas.0604138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Toh K, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science (New York, NY) 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 114.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 115.Sato T, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]