Abstract
The apicomplexan protozoan Toxoplasma gondii is a significant human and veterinary pathogen. As an obligate intracellular parasite, Toxoplasma depends on nutrients provided by the host cell and needs to adapt to limitations in available resources. In mammalian cells, translational regulation via GCN2 phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α) is a key mechanism for adapting to nutrient stress. Toxoplasma encodes two GCN2-like protein kinases, TgIF2K-C and TgIF2K-D. We previously showed that TgIF2K-D phosphorylates T. gondii eIF2α (TgIF2α) upon egress from the host cell, which enables the parasite to overcome exposure to the extracellular environment. However, the function of TgIF2K-C remained unresolved. To determine the functions of TgIF2K-C in the parasite, we cloned the cDNA encoding TgIF2K-C and generated knockout parasites of this TgIF2α kinase to study its function during the lytic cycle. The TgIF2K-C knockout did not exhibit a fitness defect compared with parental parasites. However, upon infection of human fibroblasts that were subsequently cultured in glutamine-free medium, the intracellular TgIF2K-C knockout parasites were impeded for induced phosphorylation of TgIF2α and showed a 50% reduction in the number of plaques formed compared with parental parasites. Furthermore, we found that this growth defect in glutamine-free media was phenocopied in parasites expressing only a non-phosphorylatable TgIF2α (TgIF2α-S71A), but not in a TgIF2K-D knockout. These studies suggest that Toxoplasma GCN2-like kinases TgIF2K-C and TgIF2K-D evolved to have distinct roles in adapting to changes in the parasite’s environment.
Keywords: eIF2 kinase, Translational control, Stress, Parasite, Apicomplexa, Glutamine
1. Introduction
Cells adjust to diverse stresses ranging from oxidative and endoplasmic reticulum (ER) stress to fluctuations in the availability of nutrients (Sonenberg and Hinnebusch, 2009; Walter and Ron, 2011; Baird and Wek, 2012). Central to this stress response is translational control by phosphorylation of the alpha subunit of eukaryotic initiation factor-2 (eIF2α) (Sonenberg and Hinnebusch, 2009; Baird and Wek, 2012; Donnelly et al., 2013). Following phosphorylation of a regulatory serine residue, eIF2α becomes an inhibitor of its own guanine nucleotide exchange factor (eIF2B), leading to diminished levels of global protein production but preferential translation of key transcription factors such as mammalian ATF4 and yeast GCN4, which reprogram gene expression for an adaptive response (Sonenberg and Hinnebusch, 2009; Baird and Wek, 2012). Mammals encode four eIF2α kinases that harbor a signature protein kinase domain flanked by unique regulatory domains that sense different cellular stresses. Whereas HRI (EIF2AK1) is induced upon heme deprivation in erythroid tissues, PKR (EIF2AK2) is activated upon viral infection. GCN2 (EIF2AK4) and PERK (EIF2AK3) serve as sensors for the depletion of amino acids and accumulation of misfolded proteins in the ER, respectively (Chen, 2007; Raven and Koromilas, 2008; Baird and Wek, 2012; Donnelly et al., 2013).
Toxoplasma gondii is an obligate intracellular protozoan parasite in the phylum Apicomplexa and the causative agent of toxoplasmosis, an important opportunistic disease of immunocompromised patients (Montoya and Liesenfeld, 2004). During acute Toxoplasma infection, tachyzoites progress through the lytic cycle, which consists of host cell infection, replication within the parasitophorous vacuole and egress into the extracellular environment to invade a new host cell (Montoya and Liesenfeld, 2004). In infected hosts, a chronic infection is established by tachyzoites that convert into latent bradyzoites residing within protective tissue cysts. Bradyzoites are not eliminated by the host immune response and may resume acute infection by reconverting into tachyzoites when the immune system becomes compromised (Montoya and Liesenfeld, 2004).
While it has been well-established that cellular stresses such as alkaline pH, oxidative stress and nutrient limitation induce bradyzoite formation in vitro, the molecular mechanisms involved in this developmental process are largely unknown (Sullivan and Jeffers, 2012). In previous studies we showed that inducers of bradyzoite conversion also trigger phosphorylation of Toxoplasma gondii eIF2α (TgIF2α), coincident with a reduction in global protein synthesis, suggesting that translational control mechanisms are involved in the formation of bradyzoites (Narasimhan et al., 2008; Konrad et al., 2013). Supporting this idea, we also found that TgIF2α phosphorylation is maintained in mature bradyzoites (Narasimhan et al., 2008). Most recently, we showed that inhibitors of TgIF2α dephosphorylation impede tachyzoite replication in vitro and in vivo, and block the reactivation of bradyzoites into tachyzoites in vitro (Konrad et al., 2013).
Phosphorylation of TgIF2α also plays a key role in promoting survival of extracellular tachyzoites during the lytic cycle. Tachyzoites genetically modified to express TgIF2α in which the phosphorylated serine is substituted for alanine (TgIF2αS71A) fail to phosphorylate TgIF2α while in the extracellular environment and exhibit a recovery defect after being deprived of host cells (Joyce et al., 2010). In a follow up study, we also identified that intracellular TgIF2αS71A parasites displayed a recovery defect following nutrient deprivation (Konrad et al., 2011).
Toxoplasma possesses four eIF2α kinases (TgIF2Ks). Similar to PERK, TgIF2K-A localizes to the ER and is suggested to be involved in sensing accumulation of malfolded proteins in the ER (Narasimhan et al., 2008). TgIF2K-B has no clear homologue in other species but likely senses stresses that disrupt cytosolic homeostasis, such as oxidative stress (Narasimhan et al., 2008). TgIF2K-C and TgIF2K-D share the highest sequence identity with GCN2, the mammalian eIF2α kinase that facilitates adaptation to nutrient limitation (Sonenberg and Hinnebusch, 2009; Baird and Wek, 2012). Previous studies have determined that TgIF2K-D is involved in the adaptation of tachyzoites to the extracellular environment (Konrad et al., 2011). Similar to TgIF2α-S71A tachyzoites, parasites deficient in TgIF2K-D (Δtgif2k-d) fail to phosphorylate TgIF2α when extracellular and exhibit a growth recovery defect following exposure to extracellular stress (Konrad et al., 2011). In contrast to TgIF2α-S71A tachyzoites, replicating Δtgif2k-d parasites continue to phosphorylate TgIF2α when deprived of nutrients, suggesting that an independent TgIF2K is activated when intracellular parasites experience limiting nutrients (Konrad et al., 2011).
Here we describe a second cytoplasmic GCN2-like eIF2α kinase in Toxoplasma (TgIF2K-C) that is activated upon amino acid deprivation of intracellular tachyzoites. Viable TgIF2K-C knockout parasites were generated which, in contrast to the TgIF2K-D knockout, phenocopy the inability of the TgIF2α-S71A mutant to adapt to glutamine starvation. These findings establish that the two GCN2-like eIF2α kinases in Toxoplasma function independently to alleviate distinct stresses.
2. Materials and methods
2.1. Parasite culture
Unless indicated otherwise, Toxoplasma tachyzoites were maintained in human foreskin fibroblasts (HFF) in DMEM containing 25 mM glucose and 4 mM glutamine (Invitrogen, USA) supplemented with 1% heat inactivated FBS (Gibco, USA) and 100 U/ml of penicillin/100 μg/ml of streptomycin (Fisher Scientific, USA) at 37°C and 5% CO2.
2.2. Cloning of tgif2k-c cDNA
Cloning of the tgif2k-c cDNA, as well as 5′- and 3′-rapid amplification of cDNA ends (RACE), were carried out as described previously for the tgif2k-d cDNA (Konrad et al., 2011). The oligonucleotides employed in the PCRs were designed according the annotation of the tgif2k-c gene in the Toxoplasma database (www.toxodb.org version 8.1, TGME49_204110).
2.3. Generation of TgIF2K-C knockout parasites
TgIF2K-C knockout parasites (Δtgif2k-c) were generated following a strategy employed previously to make TgIF2K-D knockout parasites (Δtgif2k-d) (Konrad et al., 2011). Approximately 1.5 kb DNA fragments upstream and downstream of the open reading frame (ORF) for tgif2k-c (TGME49_204110) were amplified and inserted into the pDHFR*-TSc3 vector, flanking the ends of a modified dihydrofolate reductase-thymidylate synthase (DHFR*-TS) minigene that confers resistance to pyrimethamine (Roos et al., 1994). The 5′ flanking sequence was amplified with oligonucleotides #85 and #86 while the 3′ flanking sequence was amplified with oligonucleotides #87 and #88 (oligonucleotide sequences used in this study are listed in Supplementary Table S1). Fifty μg of the resulting knockout vector, Δtgif2k-c::DHFR*, were linearized with NotI and transfected into RH strain tachyzoites lacking Ku80 (Fox et al., 2009; Huynh and Carruthers, 2009), as previously described (Roos et al., 1994). To select for the presence of the DHFR*-TS minigene, transfected parasites were cultured in HFFs using DMEM supplemented with 1 μM pyrimethamine. Following cloning by limiting dilution, individual clones were screened by PCR for the correct insertion of the DHFR*-TS minigene and ablation of the tgif2k-c genomic locus. Genomic DNA from candidate clones was isolated and analyzed by PCR using oligonucleotides complementary to the 3′-untranslated region (UTR) of the DHFR* minigene (#99) and upstream of the insertion site (#165). Loss of tgif2k-c mRNA was verified by reverse transcriptase (RT)-PCR using oligonucleotides #134 and #129, which amplify the sequence encoding the kinase domain, and oligonucleotides #143 and #144, which amplify a ~1000 bp DNA fragment immediately upstream of the kinase coding domain As a control for RNA quality, a portion of Toxoplasma actin (TgME49_009030) mRNA was amplified by RT-PCR using primers #13 and #14.
2.4. Generation of parasites expressing endogenously tagged TgIF2K-C
To generate Toxoplasma parasites expressing TgIF2K-C endogenously tagged with three tandem hemagglutinin (HA) epitopes at the C-terminus, a ~1.4 kb DNA fragment spanning from intron X to exon 12 from Toxoplasma genomic DNA using oligonucleotides #214 and #216 was amplified. As described for TgIF2K-D (Konrad et al., 2011), this fragment was cloned into the vector 3xHA-LIC-DHFR-TS (Huynh and Carruthers, 2009) using the ligation-independent cloning method (Lucy Stolsa et al., 2002). Fifty μg of the TgIF2K-C-HA3x tagging plasmid were linearized with SbfI and subsequently transfected into RHΔku80 parasites (Huynh and Carruthers, 2009). Both the LIC vector and RHΔku80 parasites were kindly provided by Dr. Vern Carruthers (University of Michigan, USA). Transfected parasites were selected in media with 1 μM pyrimethamine and cloned by limiting dilution. Resistant clones were first screened by PCR using genomic DNA and oligonucleotides #219 and #215, which are complementary to DNA sequences upstream of the SbfI restriction site and to the HA tag-encoding fragment, respectively. In a second step, positive clones from the PCR-based screen were then examined by western blot using a monoclonal antibody specific for the HA tag (Roche, USA #11867423001, 1:2000 dilution).
2.5. Competitive fitness assay
The competitive parasite fitness assay was carried out largely as described by Joyce et al. (2010). Briefly, intracellular parental RHΔku80 and Δif2k-c parasites were mechanically released from host cells via syringe passage (25 gauge). Following filter purification, 5×105 parasites of each strain were mixed in 10 ml of Toxoplasma culture medium and transferred onto a confluent HFF monolayer in a T-25 cm2 cell culture flask. After 2 days, 5×105 parasites of the mixed population were transferred onto a new confluent HFF monolayer in a T-25 cm2 cell culture flask. The remaining mixed parasites were harvested and the genomic DNA was isolated using a DNeasy kit (Qiagen, USA). After 5 days, the mixed population was harvested again and the genomic DNA isolated. The relative presence of each parasite line was analyzed employing a SYBR-green based PCR assay. Oligonucleotides #173 and #172 recognize both parasite lines, #171 and #182 recognize only parental RHΔku80 parasites, and #174 and #185 recognize only Δif2k-c parasites. All PCRs were performed in triplicate using the 7500 Real-Time PCR System and analyzed with relative quantification software (SDS software, version 1.2.1; Applied Biosystems, USA).
2.6. In vivo virulence of Δtgif2k-c
The in vivo virulence of Δtgif2k-c KO1 was analyzed similarly as previously described for TgIF2α-S71A parasite mutants and this study protocol was approved by the Institutional Care and Use Committee at the Indiana University, USA (Joyce et al., 2010). In brief, 10–11 female BALB/c mice were infected via i.p. injection with 100 parental RHΔku80, Δtgif2k-c or TgIF2α-S71A tachyzoites resuspended in pre-warmed 1xPBS, which had been released via syringe passage and filter purified from host cell material. Mice were monitored at least twice a day and the time to moribund state was recorded. The statistical analyses were carried out using a two-tailed student’s t-test with equal variance.
2.7. Western blot and IFAs
Western blot analyses of total Toxoplasma protein lysate using an anti-HA antibody (1:2000 dilution, Roche #11867423001) and antibodies specific for TgIF2α (1:5000 dilution) and TgIF2α~P (1:1000 dilution) were carried out as described (Konrad et al., 2011). For immunofluorescence analysis of TgIF2K-C3xHA, intracellular tachyzoites were fixed with 3% paraformaldehyde and further analyzed using an anti-HA antibody as described (Konrad et al., 2011).
2.8. Glutamine starvation assay
Intracellular parental RHΔku80, TgIF2α-S71A, Δif2k-c and Δif2k-d tachyzoites were mechanically released from host cells through syringe passage as described in Section 2.5. A confluent HFF monolayer grown in 12-well plates was infected with 200 filter-purified tachyzoites resuspended in Toxoplasma culture medium. After 4 h, extracellular tachyzoites were removed and the monolayer was washed twice with 1xHank’s Balanced Salt Solution (HBSS) (Gibco) pre-warmed to 37°C. The infected host cells were incubated for 5 days in DMEM lacking glutamine (DMEM ΔQ; US Biological, USA) supplemented with 1% dialyzed FBS (Gibco) and 100 U/ml of penicillin/ 100 μg/ml of streptomycin (Fisher Scientific) at 37°C and 5% CO2. As a control, some cultures contained DMEM ΔQ supplemented with 4 mM glutamine. After 5 days, the HFF monolayer was fixed with ice-cold methanol and stained with crystal violet to visualize parasite plaques.
For the analysis of TgIF2α phosphorylation, HFFs grown in T-75 cm2 flasks were infected with 4×106 freshly purified tachyzoites for 24 h. At this time point, infected HFF monolayers were washed twice with pre-warmed 1xHBSS as described above. To induce glutamine starvation, host cells were incubated for up to 12 h in DMEM ΔQ or DMEM ΔQ supplemented with 4 mM glutamine. At the indicated time point, the medium was removed and the intracellular tachyzoites were mechanically released and filter-purified. TgIF2α phosphorylation was assessed by western blotting and Image J software (Version 1.46r) was used for densitometric analysis.
2.9. Bioinformatic analysis
The TgIF2K-C amino acid sequence was analyzed for homologous proteins and protein domains using BLASTp (Altschul et al., 1997), PFAM (Bateman et al., 2002), MOTIF (http://www.genome.jp/tools/motif/) and SMART (Letunic et al., 2012). A phylogenetic tree was generated using sequences of the protein kinase domain of TgIF2K-C and those from the listed protein kinases and Clustal Omega (Goujon et al., 2010; Sievers et al., 2011).
3. Results
3.1. Characterization of the GCN2-like kinase, TgIF2K-C
We previously showed that the GCN2-like eIF2α kinase called TgIF2K-D is critical for promoting survival of extracellular tachyzoites following egress from host cells (Konrad et al., 2011). A second GCN2-like eIF2α kinase, termed TgIF2K-C, was predicted to be encoded in the Toxoplasma genome (Narasimhan et al., 2008), but its function had yet to be defined. To verify the annotation of TgIF2K-C (TGME49_204100) in ToxoDB (toxodb.org), we cloned the complete cDNA and carried out 5′- and 3′-RACE. The tgif2k-c cDNA contains an ORF of 9810 bp in length, encoding for a 3269 amino acid protein with a predicted molecular weight of 349 kDa (GenBank accession number KF241722) (Supplementary Fig. S1). This ORF is slightly shorter than the predicted 9816 bp due to a misannotation at exon 2. RACE reactions indicate that the 5′-UTR is 837 nucleotides (nt) and the 3′-UTR extends to 1110 nt. Our analysis indicates that the TgIF2K-C locus consists of 12 exons and 11 introns.
An alignment between the protein kinase domains of TgIF2K-C, TgIF2K-D and eIF2α kinases from other species revealed that TgIF2K-C (residues 898 to 1558) contains hallmark features of eIF2α kinases, including an insert between subdomains IV and V (Fig. 1, Supplementary Figs. S1, S2). According to a BLASTp analysis, the kinase domain of TgIF2K-C is most closely related to putative GCN2-related protein kinases from Dictyostelium discoideum (IFKC_DICDI, Q75JN1.1, 5e-18; IFKB_DICDI, Q550L8.1, 5e-15; IFKA_DICDI, Q558U1.1, 1e-14), followed by GCN2 orthologues in Mus musculus (Q9QZ05.2, 9e-11) (Berlanga et al., 1999; Sood et al., 2000a), Schizosaccharomyces pombe (Q9HGN1.2, 3e-9) (Zhan et al., 2004), Saccharomyces cerevisiae (P15442.3, 5e-9) (Wek et al., 1989) and Arabidopsis thaliana (Q9lX30.2, 4e-8) (Zhang et al., 2003). A phylogenetic analysis suggests that TgIF2K-C is most closely related to GCN2-related sequences from apicomplexan parasites, followed by GCN2 in plants, yeast and metazoic animals (Supplementary Fig. S3).
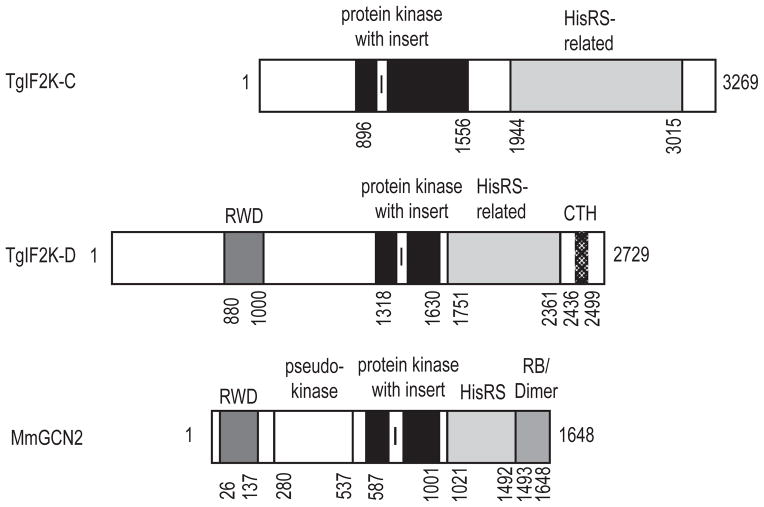
Fig. 1.
Diagram of Toxoplasma gondii (Tg)IF2K-C protein with catalytic and regulatory domains. The TgIF2K-C primary sequence contains the eIF2α kinase domain with characteristic kinase insert, as well as a histidyl-tRNA synthetase (HisRS)-related domain. Additional domain structures are shown for the other Toxoplasma GCN2-like eIF2α kinase, TgIF2K-D, and mouse GCN2. TgIF2K-D contains the RWD and the C-terminal homology domains (CTH). The domain structure of mouse GCN2 (MmGCN2) contains the RWD and ribosomal binding/dimerization (RB/Dimer) domains.
In addition to the kinase domain, GCN2-like kinases contain signature regulatory domains (Fig. 1). A central feature of GCN2 eIF2α kinases is the juxtaposition of the protein kinase domain to carboxy-terminal regulatory sequence with homology to histidyl tRNA synthetase (HisRS) enzymes (Fig. 1) (Wek et al., 1989). The HisRS-related domain is suggested to stimulate its eIF2α kinase activity by directly binding to uncharged tRNAs that accumulate during nutrient deprivation (Wek et al., 1995; Garcia-Barrio et al., 2000; Hinnebusch, 2005; Baird and Wek, 2012). TgIF2K-C contains a HisRS-like domain (amino acid residues 1944–3015), which is approximately twice as large as the HisRS-domain in mammalian GCN2 (Sood et al., 2000b). A central feature of the TgIF2K-C HisRS is the sequence ASGGRYD2741, which matches the histidine B motif that is a hallmark feature of HisRS domains (Fig. 1, Supplementary Fig. S1) (Berlanga et al., 1999; Sood et al., 2000a).
The N-terminus of yeast and mammalian GCN2 includes the RWD domain that was reported to bind directly to the coactivator GCN1 in S. cerevisiae upon amino acid starvation (Fig. 1) (Hinnebusch, 2005). The RWD domain was named after three proteins in which this domain had been identified: RING finger containing proteins, WD40 containing proteins and DEXD-like helicases (Nameki et al., 2004). Sequence comparison identified the YPXXXP motif to be characteristic for RWD domains (Nameki et al., 2004; Hinnebusch, 2005). In contrast to TgIF2K-D (Konrad et al., 2011), we did not detect this motif in TgIF2K-C, indicating that a RWD domain is absent. Interestingly, the RWD domain is also absent in GCN2-related protein kinase Plasmodium PfeIK1 and Dictyostelium IFKA and IFKB (Rai et al., 2006). In addition, alternative splice variants of mammalian GCN2 have been identified, which lead to the deletion of the RWD domain (Sood et al., 2000b); however, consistent with results on the ToxoDB, we detected no alternatively spliced products during cloning of TgIF2K-C derived from multiple different cDNA library preparations. Additionally, no RWD domain was detected in predicted amino acid sequences that would be encoded in TgIF2K-C intron regions or 5′ intragenic regions. These findings indicate that the absence of a RWD domain in a GCN2-like kinase can be found in diverse eukaryotic organisms, including apicomplexan parasites Toxoplasma and Plasmodium.
In common with TgIF2K-D and other protozoan GCN2-like kinases (Fennell et al., 2009; Konrad et al., 2011), TgIF2K-C lacks the pseudokinase domain, which is believed to contribute to eIF2α kinase activity in mammalian and yeast GCN2 (Fig. 1) (Qiu et al., 1998). The pseudokinase region of these GCN2 orthologues has sequence similarity to many of the subdomains of eukaryotic protein kinases, although the key residues critical for ATP binding and catalysis appear to be absent (Qiu et al., 1998; Boudeau et al., 2006). The degenerate kinase domain of yeast GCN2 can interact with the bona fide protein kinase domain and may participate in the repression of eIF2α kinase activity that is released upon GCN2 binding to uncharged tRNA. In conclusion, while lacking a clear RWD domain, TgIF2K-C contains a signature eIF2α kinase domain and a well-conserved histidine B motif. Therefore, we propose that TgIF2K-C is a GCN2-related protein kinase that may play a role in nutrient deprivation responses.
3.2. TgIF2K-C is expressed in the cytosol of Toxoplasma tachyzoites
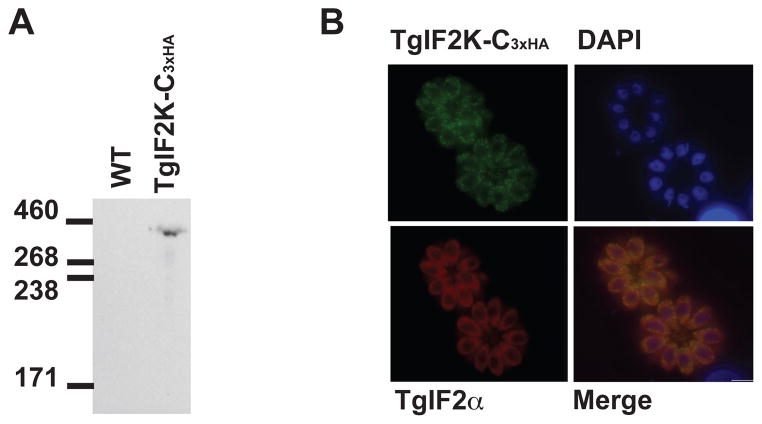
To determine whether TgIF2K-C is expressed in proliferating Toxoplasma tachyzoites, TgIF2K-C was endogenously tagged with 3xHA (TgIF2K-C3xHA) at the C-terminus in the RHΔku80 strain (Fox et al., 2009; Huynh and Carruthers, 2009). A western blot using parasite lysates probed with a monoclonal HA-antibody detected a single protein with a size similar to the deduced molecular weight of 349 kD (Fig. 2A). IFAs of these parasites revealed that TgIF2K-C is distributed throughout the parasite cytosol, with no signal detected in the nucleus (Fig. 2B).
Fig. 2.
TgIF2K-C is expressed in the cytosol ofToxoplasma gondii tachyzoites. (A) TgIF2K-C was endogenously tagged with three tandem hemagglutinin (HA) tags (TgIF2K-C3xHA) and expression was analyzed by western blot of parasite lysate using anti-HA antibody. Protein lysate prepared from the parental line (wild-type; WT) was loaded as a control. Sizes are shown in kilodaltons. (B) IFA using HA-antibody reveals a cytoplasmic distribution for TgIF2K-C3xHA (green). DAPI (blue) was used as a co-stain to highlight the parasite nucleus and total TgIF2α highlights the parasite cytosol (red). Scale bar = 5 um.
3.3. TgIF2K-C is dispensable for in vitro tachyzoite replication in complete medium
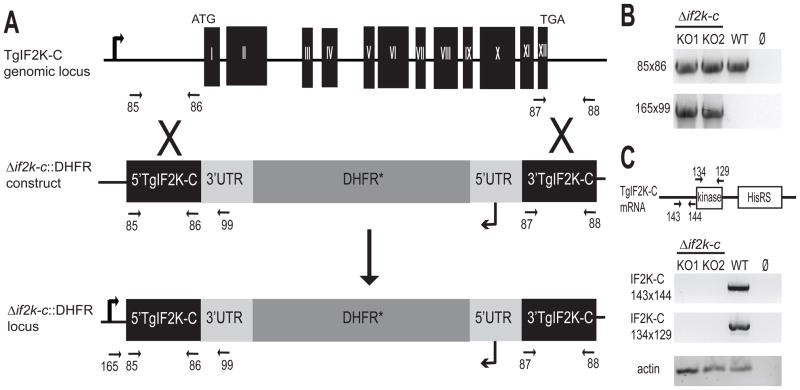
To address the function of TgIF2K-C in tachyzoites, TgIF2K-C knockout parasites were generated using homologous recombination in the RHΔku80 background, which is referred to as parental wild-type (WT) in the subsequent studies. As shown in Fig. 3A, the complete TgIF2K-C ORF was replaced with a modified DHFR-TS minigene that confers resistance to pyrimethamine (Donald and Roos, 1993). Candidate TgIF2K-C knockout clones were analyzed by PCR using primers designed to amplify a fragment of the DHFR-TS minigene when inserted in the TgIF2K-C genomic locus (Fig. 3B). Additionally, RT-PCR was performed to verify the absence of TgIF2K-C mRNA. While we could amplify mRNA fragments coding for a portion of the N-terminus (oligonucleotides #143 and #144) and the protein kinase domain (oligonucleotides #134 and #129) of TgIF2K-C from the parental WT parasites, these gene sequences were no longer detected in TgIF2K-C knockout parasites, Δif2k-c (Fig. 3C). Two independent knockout clones were identified and shown to have similar mutant phenotypes that are described below.
Fig. 3.
Generation of Toxoplasma gondii (Tg)IF2K-C knockout (KO) parasites. (A) Genomic fragments upstream and downstream of the open reading frame were amplified by PCR using genomic DNA and oligonucleotide primers #85 and #86, and #87 and #88 (indicated by arrows), respectively. The resulting DNA fragments flank a dihydrofolate reductase (DHFR*) selection marker to comprise the knockout plasmid Δif2k-c::DHFR. The knockout construct was used to replace the complete TgIF2K-C genomic locus via homologous recombination in RHΔku80 strain (referred to as wild-type or WT). (B) The replacement of the TgIF2K-C locus in two independent clones (KO1, KO2) was analyzed by PCR using oligonucleotides complementary to DNA sequences upstream of the replacement site (oligonucleotide #165) and in the 3′-untranslated region (UTR) of the DHFR* minigene (oligonucleotide #99). WT genomic DNA was included as a positive control for amplification. (C) Reverse transcriptase (RT)-PCR using oligonucleotides that amplify a ~1.0 kb fragment at the 5′ end (oligonucleotides #143 and #144) and the fragment encoding the protein kinase domain (oligonucleotides #132 and #129) were carried out to verify the absence of TgIF2K-C mRNA in the knockout clones. As a control for the mRNA preparation, a portion of Toxoplasma actin mRNA was amplified by RT-PCR. To exclude contaminating genomic DNA, the template was omitted from one PCR sample (Ø).
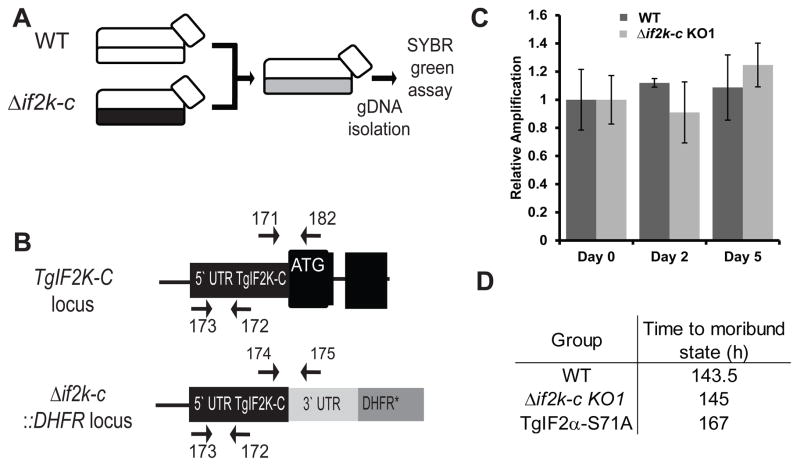
To determine whether loss of TgIF2K-C compromised tachyzoite viability, parental WT and Δif2k-c parasites were compared in a head-to-head fitness assay (Joyce et al., 2010; Konrad et al., 2011). A confluent HFF host cell monolayer was co-infected with an equal number of WT and Δif2k-c parasites (Fig. 4A) and the relative amount of each parasite line was determined with SYBR-green based PCR using strain-specific oligonucleotides (Fig. 4B). After up to 5 days of co-cultivation, Δif2k-c and WT parasites were still present in approximately equal numbers, indicating that the absence of TgIF2K-C does not cause a general fitness defect in tachyzoites progressing through the lytic cycle (Fig. 4C). This finding was further supported when Δif2k-c parasites showed similar virulence in vivo as WT parasites. Control TgIF2α-S71A mutant parasites, as expected, displayed a statistically significant defect in virulence compared with WT parasites (P < 0.001) (Fig. 4D).
Fig. 4.
Toxoplasma gondii (Tg)IF2K-C is dispensable for normal progression through the lytic cycle. (A) Schematic diagram of the experimental set up for the competitive fitness assay. Equal numbers of parental wild-type (WT) and Δif2k-c parasites were co-cultured in the same flask. Genomic (g)DNA from the mixed population was isolated for SYBR-green PCR analysis at days 0, 2 and 5. (B) Diagram of genomic loci and oligonucleotides that were used to distinguish WT and Δif2k-c parasites. (C) Relative levels of parasite genomic DNA in the co-cultured flasks were determined using a SYBR-green assay and oligonucleotide primers #171 and #182 or oligonucleotides #174 and #175, which are specific for WT or Δif2k-c parasites, respectively. A fragment of the 5′-untranslated region (UTR) conserved between WT and Δif2k-c parasites was amplified with the oligonucleotides #173 and #172 to normalize the samples. The results of Δif2k-c clone KO1 are shown. Error bars indicate the S.D. from a triplicate experiment. Analysis of the Δif2k-c clone KO2 showed a similar result. (D) BALB/c mice were infected i.p. with 100 WT, Δif2k-c or TgIF2α-S71A parasites. The average time to a moribund state was determined for each group of infected mice.
3.4. TgIF2K-C is required for adaptation to glutamine starvation
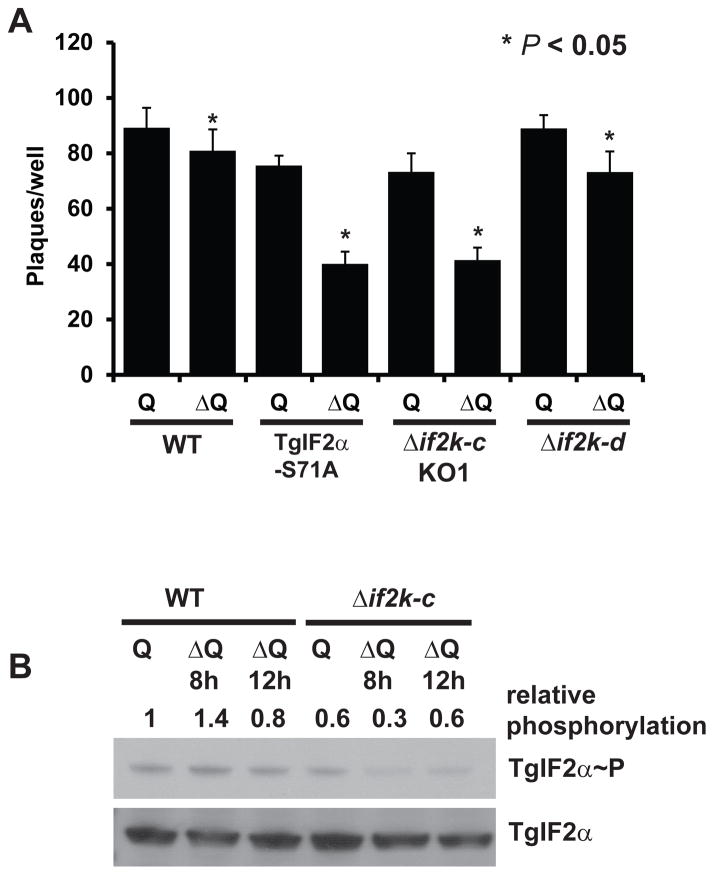
We previously established that the other GCN2-like kinase, TgIF2K-D, is critical in tachyzoites experiencing extracellular stress following departure from its host cell (Konrad et al., 2011). As TgIF2K-C was unable to compensate for this defect, we tested whether TgIF2K-C responded to nutrient stress experienced when tachyzoites were intracellular. Recent reports have implicated a critical role for individual amino acids during the proliferation of Toxoplasma tachyzoites (Fox et al., 2004; Ghosh et al., 2011; Macrae et al., 2012), including the non-essential amino acid glutamine (Chaudhary and Roos, 2005), which is a key metabolite in eukaryotic cells (Curthoys and Watford, 1995). Plaque assays were performed to analyze the contribution of TgIF2K-C in the adaptation to the absence of this amino acid. Following infection, parental WT, TgIF2α-S71A, Δif2k-c and Δif2k-d parasites were cultured in DMEM lacking glutamine (DMEM ΔQ) or DMEM ΔQ supplemented with 4 mM glutamine. After 5 days in DMEM ΔQ, WT parasites formed a modestly reduced number of plaques compared with the rich medium control (Fig. 5A), indicating that the absence of glutamine has minimal deleterious effects on parasite viability. However, TgIF2α-S71A parasites exhibited a significant defect adapting to glutamine starvation as indicated by the 50% reduction in the number of plaques (Fig. 5A). These results are consistent with the idea that TgIF2α phosphorylation promotes survival of intracellular parasites experiencing nutrient depletion. While Δif2k-d parasites exhibited no defect under glutamine deprivation, Δif2k-c parasites exhibited an adaptation defect similar to TgIF2α-S71A (Fig. 5A). These results suggest that the two TgGCN2-like kinases respond to distinct stress signals to promote parasite adaptation to different stresses.
Fig. 5.
Toxoplasma gondii (Tg)IF2K-C promotes parasite viability during glutamine starvation. (A) Human foreskin fibroblast monolayers were infected with parental wild-type (WT), TgIF2α-S71A, Δif2k-c clone KO1 or Δif2k-d parasites and cultured in medium lacking glutamine (ΔQ) or supplemented with glutamine (Q). After 5 days, infected monolayers were fixed and the number of plaques was determined. Graph shows the data compiled from three independent experiments performed in triplicate. Error bars indicate the S.D. The asterisk indicates a statistically significant difference between samples (unpaired two-tailed student’s t-test, P < 0.05). Analysis of the Δif2k-c clone KO2 showed a similar result (Supplementary Fig. S3). (B) Intracellular WT or Δif2k-c parasites were incubated for 8 or 12 h in Q or ΔQ medium. Phosphorylation of TgIF2α was analyzed by western blot of parasite lysate using an antibody specific for phosphorylated TgIF2α (TgIF2α~P) or total TgIF2α. Densitometry analysis was carried out to determine relative TgIF2α phosphorylation normalized to total TgIF2α. The experiment was repeated with similar results.
We next addressed whether TgIF2K-C is activated by glutamine starvation to phosphorylate TgIF2α. Intracellular WT and Δif2k-c tachyzoites were cultured up to 12 h in DMEM ΔQ. By 8 h of glutamine starvation, TgIF2α~P increased approximately 40% in WT parasites compared with non-stressed parasites, indicating that the lack of glutamine induces a stress response in Toxoplasma. At later time points, the amount of TgIF2α~P returned to basal levels, suggesting that the parasites adapted to the absence of glutamine by this time (Fig. 5B). In contrast, Δif2k-c parasites not only fail to induce TgIF2α phosphorylation in response to glutamine depletion, but they show a decrease in TgIF2α~P levels relative to unstressed samples, which could also be indicative of a feedback regulatory loop (Fig. 5B). Feedback mechanisms have been described for controlling phosphorylated eIF2α in mammalian cells (Marciniak and Ron, 2006; Baird and Wek, 2012). Altogether, these findings suggest that phosphorylation of TgIF2α by TgIF2K-C is required for proliferating tachyzoites to adapt to the absence of glutamine.
4. Discussion
Regulation of translation via phosphorylation of eIF2α is a key stress response pathway in eukaryotic cells. Toxoplasma expresses two GCN2-like eIF2α kinases, one of which (TgIF2K-D) was previously linked to promoting survival of tachyzoites following egress from host cells (Konrad et al., 2011). Here, we characterize the second GCN2-like kinase, TgIF2K-C. In contrast to parasites lacking TgIF2K-D, the TgIF2K-C knockout does not exhibit a fitness defect under standard in vitro culture conditions nor does it show reduced virulence in an in vivo mouse model maintained on a standard chow diet. However, our findings show that TgIF2K-C is required to promote survival of intracellular tachyzoites cultured under glutamine starvation conditions.
We addressed whether TgIF2K-C phosphorylates TgIF2α upon nutrient starvation by analyzing the phosphorylation status of TgIF2α in parental WT and Δif2k-c parasites in response to depleted glutamine, which is an important metabolite in Toxoplasma (Fox and Bzik, 2002; Macrae et al., 2012) and mammalian cells (Curthoys and Watford, 1995). Following 8 h of glutamine deprivation, TgIF2α is phosphorylated at its regulatory serine residue (Ser71). TgIF2α~P returns to basal levels by 12 h, suggesting that adaptation to this stress has occurred. Consistent with the defect in TgIF2α phosphorylation, Δif2k-c parasites form approximately 50% fewer plaques when cultured in glutamine deprived medium. Since the non-phosphorylatable TgIF2α-S71A mutant exhibited a similar defect, but Δtgif2k-d parasites did not, we conclude that TgIF2K-C is the primary TgIF2α kinase managing the intracellular response to glutamine starvation. Given the large size of the TgIF2K-C gene and the data showing that Δif2k-c phenocopies the TgIF2α-S71A mutant, attempts were not made to complement Δif2k-c parasites.
TgIF2K-C is an unusual GCN2-like homologue. While TgIF2K-C possesses a signature HisRS-like domain with the well-conserved histidine B motif, it appears to lack an RWD domain that interacts with GCN1 to enhance eIF2α kinase activity. A predicted GCN1 homologue is present in the Toxoplasma genome (TGME49_231480) and TgIF2K-D harbors the RWD domain, but an interaction has yet to be examined (Konrad et al., 2011). In yeast and mammalian GCN2, the HisRS-related domain serves as an activator of kinase activity upon binding to uncharged tRNAs that accumulate during amino acid starvation (Hinnebusch, 2005). A HisRS-like domain with the histidine B motif is also conserved in the GCN2-like PfeIK1, the Plasmodium homologue of TgIF2K-C that is activated by amino acid starvation in Plasmodium falciparum (Fennell et al., 2009). Interestingly, PfeIK1 also lacks the RWD domain. This suggests that an RWD domain is not required for enhanced TgIF2K-C and PfeIK1 phosphorylation of eIF2α, at least in response to nutrient stresses such as glutamine deprivation. In this case, these GCN2-like protein kinases may be activated independently of GCN1, or by engagement with GCN1 through an alternative sequence motif. We performed an alignment of the N-terminal extension sequences of P. falciparum and Toxoplasma homologues but no regions of significant homology were detected at the primary amino acid sequence level.
Multiple GCN2-like protein kinases have also been identified in Dictyostelium (IFKA, B and C) and recent studies have linked nutrient starvation to the activation of IFKs, the phosphorylation of Dictyostelium eIF2α and to the regulation of developmental processes (Rai et al., 2006; Bowman et al., 2011; Singleton et al., 2012). While IFKC possesses both RWD and HisRS-related domains, the RWD is absent in IFKA and IFKB, suggesting different mechanisms of regulation for these eIF2α kinases (Rai et al., 2006; Bowman et al., 2011). IFKA and IFKB were reported to have redundant roles in Dictyostelium morphogenesis, cell-cell and cell-substrate adhesion, and in spatial patterning (Rai et al., 2006). Although mammals encode only one GCN2 gene, alternative splicing of GCN2 mRNA has been identified. Alternative splicing occurs within the 5′-portion of mouse GCN2 mRNA in a tissue-specific manner, which leads to the elimination of the N-terminal RWD domain (Sood et al., 2000b). Further studies are needed to analyze these different variants of GCN2 and their functions in translation control.
In addition to its use in protein synthesis, glutamine is an amino group donor in numerous biosynthetic pathways. Glutamine has been suggested to be important for de novo pyrimidine synthesis pathway of the parasite (Fox and Bzik, 2002, 2010) and can fuel the TCA cycle via a newly identified γ-aminobutyric acid (GABA) shunt (Macrae et al., 2012). Parasites defective in the former step show reduced virulence and become auxotrophic for uracil, which is used in a salvage pathway for pyrimidine synthesis and can be rescued by supplementing the medium with uracil (Fox and Bzik, 2002, 2010). However, neither supplementing the medium with 0.4 mM uracil or up to 4 mM GABA rescued the growth defect of Δif2k-c and TgIF2α-S71A parasites in glutamine-free medium (data not shown). This could indicate that glutamine starvation affects a different metabolic pathway in Toxoplasma or has an indirect effect via starvation of the host cells. Further studies are needed to discriminate between these two possibilities.
In conclusion, this study characterizes the second of two GCN2-like eIF2α kinases present in Toxoplasma, which have now been shown to possess distinct, non-overlapping functions during the tachyzoite stage. Both TgIF2K-C and –D are largely dispensable for parasite viability, but are important regulators of adaptive responses to stresses associated with nutrient deprivation, consistent with their GCN2-like attributes. TgIF2K-A has previously been shown to localize to the parasite ER and is a likely candidate regulating the recently characterized unfolded protein response (UPR) in Toxoplasma (Narasimhan et al., 2008; Joyce et al., 2013). TgIF2K-B is a cytoplasmic eIF2α kinase that has yet to be studied (Sullivan Jr et al., 2004; Narasimhan et al., 2008; Joyce et al., 2010; Konrad et al., 2011). As thwarting adaptive responses is deleterious to parasite replication as well as the conversion into latent bradyzoite forms, the continued study of translational control through eIF2α phosphorylation promises to expose novel opportunities for therapeutic intervention.
Supplementary Material
Supplementary Fig. S1. Sequence of Toxoplasma gondii (Tg)IF2K-C mRNA. Start and stop codons are indicated in bold. Lower case indicates the 5′- and 3′-untranslated regions (UTRs). The deduced amino acid sequence is also given. The kinase domain and the histidyl tRNA synthetase (HisRS)-like domain are highlighted in red and green, respectively. The invariant lysine residue in the kinase domain’s active site and the histidine B motif in the HisRS-like domain are underlined.
Supplementary Fig. S2. Alignment of Toxoplasma gondii (Tg)IF2K-C protein kinase domain with other eIF2α kinases. The kinase domain of TgIF2K-C is comprised of 12 regions indicated by solid bars and roman numerals (I-XII) based on amino acid sequence alignments with known eIF2α kinases. The TgIF2K-C kinase domain contains an insert between regions IV and V that is characteristic for this class of protein kinases. For better visualization of the alignment, this insert and other identified inserts were replaced by the number of omitted residues. Identical residues between the eIF2α kinases are highlighted in black, and similar residues are shaded in gray. The asterisk indicates the invariant lysine residue in the active site. The numbers on the right indicate the respective amino acid residue positions in these eIF2α kinases: Tg: Toxoplasma gondii, Nc: Neospora caninum, Pf: Plasmodium falciparum, Pb: Plasmodium berghei, Tb: Trypanosoma brucei, At: Arabidopsis thaliana, Dm: Drosophila melanogaster, Sc: Saccharomyces cerevisiae, Mm: Mus musculus, Hs: Homo sapiens.
Supplementary Fig. S3. Phylogenetic tree showing relatedness between Toxoplasma gondii (Tg)IF2K-C and different GCN2 orthologues and other protein kinases. A phylogenetic tree was generated by using sequences of the protein kinase domains of TgIF2K-C and the indicated protein kinases derived from their respective species. The distances represent the degrees of difference between each protein kinase sequence, and the colors indicate GCN2 orthologues (blue), other eIF2α kinases (green), and an unrelated protein kinase (red). The apicomplexan parasite sequences cluster together and are highlighted in the gray box. Protein kinase sequences included in the phylogenetic tree and their accession numbers (in the order presented in the tree) are as follows: Homo sapiens CDK1 (NP_203698.1), Homo sapiens PERK/PEK (AF193339.1), Drosophila melanogaster PERK/PEK (AF193340.1), Schizosaccharomyces pombe HRI1 (AF536223.1), Toxoplasma gondii IF2K-D (AED01979.1), Neospora caninum IF2K-D (XP_003880619.1), T. gondii IF2K-C (KF241722), N. caninum IF2K-C (XP_003882346.1 and deduced from genomic sequence FR823388.1), Cryptosporidium muris GCN2 (XP_002141492.1), Plasmodium falciparum GCN2 (XP_001348597.1), Brachypodium distachyon GCN2 (XP_003580041.1), Cicer arietinum GCN2 (XP_004507335.1), Arabidopsis thaliana GCN2 (NP_191500.2), Saccharomyces cerevisiae GCN2 (NP_010569.3), Schizosaccharomyces pombe GCN2 (AAU11313.1), Drosophila melanogaster GCN2 (NP_477230.1), Strongylocentrotus purpuratus GCN2 (XP_785160.3), Danio rerio GCN2 (XP_002667149.3), Xenopus tropicalis GCN2 (NP_001243855.1), Gallus gallus GCN2 (XP_004941777.1), Mus musculus GCN2 (NP_001171277.1), and H. sapiens GCN2 (Q9P2K8.3).
Highlights.
A second cytosolic GCN2-like eIF2α kinase was identified in Toxoplasma gondii
TgIF2K-C is dispensable for normal tachyzoite replication and virulence
eIF2α is phosphorylated by TgIF2K-C in tachyzoite cultures deprived of glutamine
Adaptation to glutamine starvation is facilitated by TgIF2K-C
Two GCN2-like kinases have distinct functions in the Toxoplasma gondii stress responses
Acknowledgments
We thank Vern Carruthers (University of Michigan Medical School, USA) for providing RHΔku80 parasites and the LIC vector. This study was supported by grants from the National Institutes of Health (NIH), USA including AI084031 (WJS and RCW) and GM049164 (RCW) and a postdoctoral fellowship from the American Heart Association (11POST7730010 to CK). The authors also thank members of the Sullivan and Wek laboratories for helpful discussions.
Footnotes
Note: Supplementary data associated with this article
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homologu of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Bowman RL, Xiong Y, Kirsten JH, Singleton CK. eIF2{alpha} Kinases Control Chalone Production in Dictyostelium discoideum. Eukaryotic Cell. 2011;10:494–501. doi: 10.1128/EC.00270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary K, Roos DS. Protozoan genomics for drug discovery. Nat Biotechnol. 2005;23:3. doi: 10.1038/nbt0905-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Ann Rev Nutrition. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- Donald RG, Roos DS. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc Natl Acad Sci U S A. 1993;90:11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Molec Life Sci. 2013;70(19):3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell C, Babbitt S, Russo I, Wilkes J, Ranford-Cartwright L, Goldberg DE, Doerig C. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar J. 2009;8:99. doi: 10.1186/1475-2875-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Bzik DJ. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature. 2002;415:926–929. doi: 10.1038/415926a. [DOI] [PubMed] [Google Scholar]
- Fox BA, Bzik DJ. Avirulent uracil auxotrophs based on disruption of orotidine-5′-monophosphate decarboxylase elicit protective immunity to Toxoplasma gondii. Infect Immun. 2010;78:3744–3752. doi: 10.1128/IAI.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA, Gigley JP, Bzik DJ. Toxoplasma gondii lacks the enzymes required for de novo arginine biosynthesis and arginine starvation triggers cyst formation. Int J Parasitol. 2004;34:323–331. doi: 10.1016/j.ijpara.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio M, Dong J, Ulfano S, Hinnebusch AG. Association of GCN1-GCN20 regulatory complex with the N-terminus of eIF2alpha kinase GCN2 is required for GCN2 activation. EMBO J. 2000;19:1887–1899. doi: 10.1093/emboj/19.8.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Walton JL, Roepe PD, Sinai AP. Autophagy is a cell death mechanism in Toxoplasma gondii. Cell Microbiol. 2012;14(4):589–607. doi: 10.1111/j.1462-5822.2011.01745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Ann Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce BR, Queener SF, Wek RC, Sullivan WJ., Jr Phosphorylation of eukaryotic initiation factor-2{alpha} promotes the extracellular survival of obligate intracellular parasite Toxoplasma gondii. Proc Natl Acad Sci U S A. 2010;107:17200–17205. doi: 10.1073/pnas.1007610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce BR, Tampaki Z, Kim K, Wek RC, Sullivan WJ. The Unfolded Protein Response in the protozoan parasite Toxoplasma gondii features translational and transcriptional control. Eukaryotic Cell. 2013;12(7):979–89. doi: 10.1128/EC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C, Queener SF, Wek RC, Sullivan WJ., Jr Inhibitors of eIF2alpha dephosphorylation slow replication and stabilize latency in Toxoplasma gondii. Antimicrob Agents Chemother. 2013;57(4):1815–22. doi: 10.1128/AAC.01899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C, Wek RC, Sullivan WJ. A GCN2-like eukaryotic initiation factor 2 kinase increases the viability of extracellular Toxoplasma gondii parasites. Eukaryotic Cell. 2011;10:1403–1412. doi: 10.1128/EC.05117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolsa Lucy, Gub Minyi, Dieckmanb Lynda, Raffenb Rosemarie, Collartb Frank R, Donnellya MI. A New Vector for High-Throughput, Ligation-Independent Cloning Encoding a Tobacco Etch Virus Protease Cleavage Site. Protein Expr Purif. 2002;25:8. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- Macrae JI, Sheiner L, Nahid A, Tonkin C, Striepen B, McConville MJ. Mitochondrial Metabolism of Glucose and Glutamine Is Required for Intracellular Growth of Toxoplasma gondii. Cell Host Microbe. 2012;12:682–692. doi: 10.1016/j.chom.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- Nameki N, Yoneyama M, Koshiba S, Tochio N, Inoue M, Seki E, Matsuda T, Tomo Y, Harada T, Saito K, Kobayashi N, Yabuki T, Aoki M, Nunokawa E, Matsuda N, Sakagami N, Terada T, Shirouzu M, Yoshida M, Hirota H, Osanai T, Tanaka A, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kinoshita K, Guntert P, Kigawa T, Yokoyama S. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004;13:2089–2100. doi: 10.1110/ps.04751804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, Coppens I, Wek RC, Sullivan WJ., Jr Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem. 2008;283:16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Garcia-Barrio MT, Hinnebusch AG. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol Cell Biol. 1998;18:2697–2711. doi: 10.1128/mcb.18.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M, Xiong Y, Singleton CK. Disruption of the ifkA and ifkB genes results in altered cell adhesion, morphological defects and a propensity to form pre-stalk O cells during development of Dictyostelium. Differentiation. 2006;74:583–595. doi: 10.1111/j.1432-0436.2006.00085.x. [DOI] [PubMed] [Google Scholar]
- Raven JF, Koromilas AE. PERK and PKR: old kinases learn new tricks. Cell Cycle. 2008;7:1146–1150. doi: 10.4161/cc.7.9.5811. [DOI] [PubMed] [Google Scholar]
- Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Sys Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton CK, Xiong Y, Kirsten JH, Pendleton KP. eIF2alpha kinases regulate development through the BzpR transcription factor in Dictyostelium discoideum. PLoS One. 2012;7:e32500. doi: 10.1371/journal.pone.0032500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC. Pancreatic eukaryotic initation factor-2a kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to ER stress. Biochem J. 2000a;346:281–293. [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000b;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan WJ, Jr, Narasimhan J, Bhatti MM, Wek RC. Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem J. 2004;380:523–531. doi: 10.1042/BJ20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan WJ, Jeffers V. Mechanisms of Toxoplasma gondii persistence and latency. FEMS Microbiol Rev. 2012;36:717–733. doi: 10.1111/j.1574-6976.2011.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Molec Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan K, Narasimhan J, Wek RC. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics. 2004;168:1867–1875. doi: 10.1534/genetics.104.031443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dickinson JR, Paul MJ, Halford NG. Molecular cloning of an Arabidopsis homologue of GCN2, a protein kinase involved in co-ordinated response to amino acid starvation. Planta. 2003;217:668–675. doi: 10.1007/s00425-003-1025-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Sequence of Toxoplasma gondii (Tg)IF2K-C mRNA. Start and stop codons are indicated in bold. Lower case indicates the 5′- and 3′-untranslated regions (UTRs). The deduced amino acid sequence is also given. The kinase domain and the histidyl tRNA synthetase (HisRS)-like domain are highlighted in red and green, respectively. The invariant lysine residue in the kinase domain’s active site and the histidine B motif in the HisRS-like domain are underlined.
Supplementary Fig. S2. Alignment of Toxoplasma gondii (Tg)IF2K-C protein kinase domain with other eIF2α kinases. The kinase domain of TgIF2K-C is comprised of 12 regions indicated by solid bars and roman numerals (I-XII) based on amino acid sequence alignments with known eIF2α kinases. The TgIF2K-C kinase domain contains an insert between regions IV and V that is characteristic for this class of protein kinases. For better visualization of the alignment, this insert and other identified inserts were replaced by the number of omitted residues. Identical residues between the eIF2α kinases are highlighted in black, and similar residues are shaded in gray. The asterisk indicates the invariant lysine residue in the active site. The numbers on the right indicate the respective amino acid residue positions in these eIF2α kinases: Tg: Toxoplasma gondii, Nc: Neospora caninum, Pf: Plasmodium falciparum, Pb: Plasmodium berghei, Tb: Trypanosoma brucei, At: Arabidopsis thaliana, Dm: Drosophila melanogaster, Sc: Saccharomyces cerevisiae, Mm: Mus musculus, Hs: Homo sapiens.
Supplementary Fig. S3. Phylogenetic tree showing relatedness between Toxoplasma gondii (Tg)IF2K-C and different GCN2 orthologues and other protein kinases. A phylogenetic tree was generated by using sequences of the protein kinase domains of TgIF2K-C and the indicated protein kinases derived from their respective species. The distances represent the degrees of difference between each protein kinase sequence, and the colors indicate GCN2 orthologues (blue), other eIF2α kinases (green), and an unrelated protein kinase (red). The apicomplexan parasite sequences cluster together and are highlighted in the gray box. Protein kinase sequences included in the phylogenetic tree and their accession numbers (in the order presented in the tree) are as follows: Homo sapiens CDK1 (NP_203698.1), Homo sapiens PERK/PEK (AF193339.1), Drosophila melanogaster PERK/PEK (AF193340.1), Schizosaccharomyces pombe HRI1 (AF536223.1), Toxoplasma gondii IF2K-D (AED01979.1), Neospora caninum IF2K-D (XP_003880619.1), T. gondii IF2K-C (KF241722), N. caninum IF2K-C (XP_003882346.1 and deduced from genomic sequence FR823388.1), Cryptosporidium muris GCN2 (XP_002141492.1), Plasmodium falciparum GCN2 (XP_001348597.1), Brachypodium distachyon GCN2 (XP_003580041.1), Cicer arietinum GCN2 (XP_004507335.1), Arabidopsis thaliana GCN2 (NP_191500.2), Saccharomyces cerevisiae GCN2 (NP_010569.3), Schizosaccharomyces pombe GCN2 (AAU11313.1), Drosophila melanogaster GCN2 (NP_477230.1), Strongylocentrotus purpuratus GCN2 (XP_785160.3), Danio rerio GCN2 (XP_002667149.3), Xenopus tropicalis GCN2 (NP_001243855.1), Gallus gallus GCN2 (XP_004941777.1), Mus musculus GCN2 (NP_001171277.1), and H. sapiens GCN2 (Q9P2K8.3).