Abstract
Vaccines that elicit a protective broadly neutralizing antibody (bNAb) response and monoclonal antibody therapies are critical for the treatment and prevention of viral infections. However, isolation of protective neutralizing antibodies has been challenging for some viruses, notably those with high antigenic diversity or those that do not elicit a bNAb response in the course of natural infection. Here, we discuss recent work that employs protein engineering strategies to design immunogens that elicit bNAbs or engineer novel bNAbs. We highlight the use of rational, computational, and combinatorial strategies and assess the potential of these approaches for the development of new vaccines and immunotherapeutics.
Keywords: Immunogen Design, Antiviral Immunotherapy, Antibody Engineering
Introduction
The introduction of viral vaccines during the 20th century has led to a significant decrease in viral disease burden worldwide [1]. Most viral vaccines are thought to work by inducing the production of antibodies that block infection or reduce viral load, thereby providing host protection or blunting infection such that cellular immunity can be effective [2, 3]. Antibodies can participate in host defense in several ways, including opsonization, the coating of viruses to enhance uptake by phagocytic cells, or activation of the complement family of proteins that can directly destroy pathogens or enhance phagocytic uptake. Here, we will focus on neutralizing antibodies, which bind the virus and prevent infection. Neutralizing antibodies are protective against many viruses in both animals and humans [4–11]; therefore there has been much interest in their identification and characterization for potential use as immunotherapeutic agents, or to serve as templates for immunogen design. Neutralizing antibodies have historically been identified by immunization of animals with viral components, or from B-cell repertoires of human vaccinees or survivors [11–17]. In recent years, an increasing amount of structural information about neutralizing antibodies – and their mechanisms of activity – has shifted focus toward structure-based design of both immunogens designed to elicit such antibodies and of the antibodies themselves [18–34].
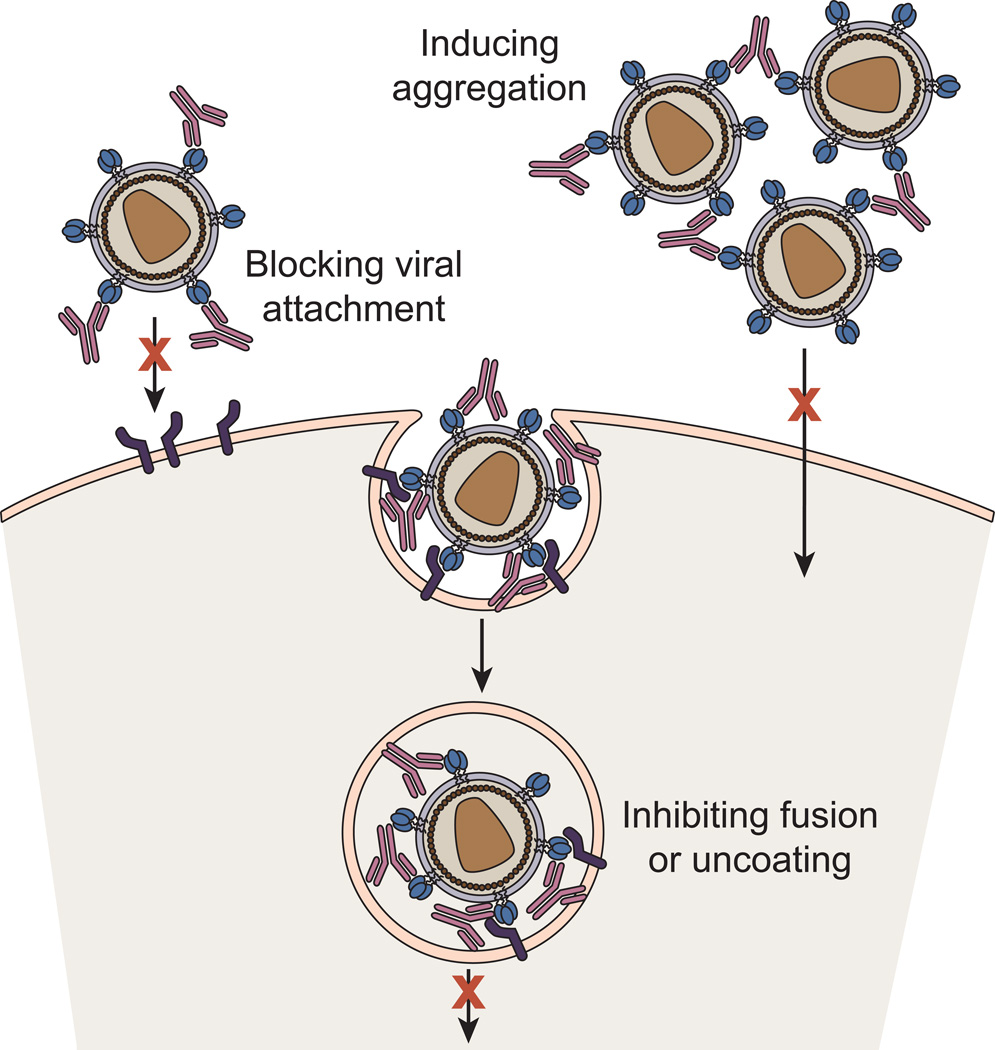
Neutralizing antibodies are thought to abrogate viral infectivity by three major mechanisms (Figure 1): (i) by blocking virus attachment to host cells; (ii) by inhibiting viral uncoating or conformational changes in viral envelope glycoproteins needed for cell entry; or (iii) by inducing the formation of noninfectious viral aggregates that cannot enter cells. In the case of enveloped viruses, those surrounded by a lipid bilayer, the primary neutralization targets are the virus envelope glycoproteins that are responsible for mediating membrane fusion between the viral and host cell membranes, a critical step for infection [35]. During the course of natural infection or vaccination, neutralizing antibodies against many viruses, such as polio, mumps, and measles, are elicited in both humans and animals. However, induction of effective neutralizing antibodies is rare or does not occur against some viruses, notably those with high antigenic diversity such as the human immunodeficiency virus-1 (HIV-1), hepatitis C virus, and influenza virus. Not surprisingly, this antigenic variation is reflected in the diverse sequences of the virus envelope glycoproteins among strains or clades, and thus antibodies that do not bind conserved epitopes have a narrow spectrum of activity.
Figure 1. Mechanism by which Neutralizing Antibodies Block Viral Infection.
Neutralizing antibodies are thought to abrogate viral infectivity by blocking virus attachment to host cells, inhibiting viral uncoating, blocking conformational changes in viral envelope glycoproteins needed for membrane fusion or prematurely triggering the fusion machinery, or by inducing the formation of noninfectious viral aggregates that cannot enter cells.
Various strategies have been employed to develop vaccines that elicit neutralizing antibodies for these high diversity viruses. In vaccination trials, the use of adjuvants to enhance the quality of antibody response to vaccination [36], nucleic-acid based methods for the delivery of antigen [37–40], and the administration of more than one type of vaccine to boost immunogenicity [41–43] have been attempted. However, effective vaccines for these viruses remain elusive. A major hurdle appears to be that the immunodominant antibody responses are directed against the most variable parts of the envelope glycoproteins, and therefore most neutralizing antibodies are narrowly strain-specific. An effective vaccine should be able to elicit “broadly neutralizing” antibodies (bNAbs) that engage conserved, less variable domains and can therefore protect across a spectrum of genetic isolates. Likewise, immunotherapeutics for these viruses should be directed at conserved viral epitopes or infection pathways. In this review, we highlight recent work that utilizes novel protein engineering strategies for the development of effective vaccines and immunotherapeutics against highly variable viruses and viruses for which a bNAb response does not arise during the course of natural infection.
1. Viral Antigen Design to Elicit Broadly Neutralizing Antibodies
One promising strategy for the generation of bNAbs by vaccintion is “reverse engineering,” where structural information gleaned from the binding of bNAbs raised in the course of natural infection is used to guide immunogen design [3, 44]. In theory, translation of this antibody binding information into an immunogen designed to display specific, critical epitopes should allow production of antibodies with similar broad neutralization capacity in vivo, provided that the immunological evolution pathway of the bNAb can be induced by vaccination. Thoughtful modification of the immunogen to reflect the specific, three-dimensional antibody-binding site is required (Figure 2). Since the goal of reverse engineering is to develop a peptide or protein scaffold that mimics the natural epitope, most strategies have utilized rational, combinatorial, or computational methods. Here we discuss several recent examples in which these methods were used to develop and evaluate immunogens.
Figure 2. Immunogen Design by Reverse Engineering.
Structural or biochemical studies are used to define the epitope of a bNAb. For both linear and discontinuous epitopes, residues involved in the antibody-antigen interaction can be grafted onto a stable scaffold in the appropriate conformation, or the original antigen can be modified to optimize recognition or other properties. Following confirmation of antibody binding, the engineered immunogen can then be used to elicit a broadly neutralizing antibody response in animal models. In this way, additional bNabs may be identified and the potential use of the immunogen in vaccine development may be assessed. Here, we have depicted the reverse engineering process using two HIV-1 bNAbs, b12, which recognizes a discontinuous epitope on gp120, and 2F5, which recognizes a continuous linear epitope on gp41. PDB files used in figure: b12: PDB ID 2NYZ, 3RPT, eRU8; 2F5: PDB ID ITJI, 3LEV; MAb 11f10: PDB ID 3LEX [19, 32].
1.1 Conformational Mimicry of Linear Epitopes from HIV-1 gp41 and gp120
HIV-1, a lentivirus, enters host cells by fusing its lipid bilayer with the host cell plasma membrane. This fusion is facilitated by the viral envelope glycoprotein, Env, which consists of a surface subunit, gp120, and a transmembrane subunit, gp41 [35]. Infection is initiated by gp120 binding to CD4 and a co-receptor on host cells, triggering large-scale conformational changes in gp41 that eventually lead to membrane fusion. Antibodies directed against Env have the potential to be neutralizing, but the generation of bNAbs has proven to be extremely challenging. This is likely because of the hypervariability encoded in the Env gene, the extensive glycoslyation of the surface of the Env protein, and structural heterogeneity associated with gp120 that is critical for its function as the triggering molecule for membrane fusion. During the course of chronic infection by HIV-1, ~10% of patients develop bNAbs, suggesting that a vaccine approach to prevent HIV-1 infection is possible [12, 45, 46]. A number of HIV-1 bNAbs target epitopes in the V3 region of gp120 or the membrane-proximal external region (MPER) of gp41. Structures of these bNAbs bound to peptide epitopes have demonstrated that these segments contain well-defined secondary structure when bound to the bNAbs. It is therefore hypothesized that immunogens designed to elicit antibodies that bind these segments in such conformations would be critical for a successful vaccination strategy.
Immunogens based on the V3 loop have been designed and have so far met with some limited success. Antibody 447-52D was isolated via hybridoma methods from a subtype B HIV-1-infected individual and found to bind to the tip of the V3 loop in a β-hairpin conformation [47]. Chakraborty et al. designed and synthesized a peptide immunogen to mimic the tip region by inserting the epitope of 447-52D into thioredoxin, a small and stable E. coli protein, and using newly introduced disulfide bonds to lock the epitope in the desired conformation [20]. This construct was able to generate a 447-52D-like response upon immunization in guinea-pigs. Although a fairly high antibody concentration was elicited by this immunization strategy (50–400 ug/mL serum), the serum was not able to effectively neutralize many primary viral isolates, perhaps because of the low accessibility of the V3 loop on many of these isolates [20].
Mor et al. synthesized a library of V3-based peptides in which they varied the position of disulfide bonds within the peptide [30]. The group found that V3-peptides containing a single disulfide bond, regardless of position, retained flexibility and did not form an ideal β-hairpin turn. However, installation of a second disulfide bond led to a significant improvement in peptide rigidity and many of these disulfide bond-containing peptides exhibited higher affinity to 447-52D than corresponding linear V3 peptides [29]. The constrained V3 peptides were linked to an 18-residue segment of the gp120 C4 region, known to induce a helper T-cell response, and were shown to elicit a 30-fold stronger HIV-1 neutralizing response in rabbits as compared to analogous linear V3 peptides or gp120 constructs displaying the V3 loop [31]. These studies suggest that carefully designed proteins that mimic natural HIV-1 bNAb binding sites have potential to elicit neutralizing responses.
Two of the most potent bNAbs known to target HIV-1, 2F5 and 4E10, bind linear epitopes on the MPER of gp41. The MPER is a highly conserved, tryptophan-rich region that is believed to play a crucial role in HIV-1 membrane fusion [48, 49]. The 2F5 and 4E10 epitopes neighbor one another and appear to require binding to only a few crucial residues within their respective epitopes [50]. Both antibodies have been shown to interact with the virion lipid membrane in addition to binding to gp41, suggesting that the structure of membrane-anchored MPER is crucial for binding by these mAbs [22]. Because of the breadth and potency of neutralization exhibited by these antibodies, strategies aimed at eliciting a 2F5- or 4E10-like response are the subject of many efforts for development of an effective anti-HIV vaccine. Both 2F5 and 4E10 were isolated well over a decade ago [48, 51, 52] and efforts to mimic their epitopes with designed immunogens have been ongoing since then. Recently, several novel bNAbs have been isolated against the MPER. One example is mAb 10E8, isolated from an HIV-infected donor by Huang et al. [53]. 10E8 is one of the most potent and broadly neutralizing anti-HIV antibodies yet identified. It was shown to bind the MPER in a conformation similar to 4E10, but has a novel binding epitope [53]. The presence of 10E8 and other MPER-binding antibodies in natural infection suggests that an appropriately designed immunogen would elicit similar antibodies.
In 2010, Ofek et al. used computational methods to construct an epitope scaffold using the 2F5 epitope [32]. The 2F5 epitope is conformationally flexible when not bound by the antibody, therefore posing a particular challenge for epitope design. Upon 2F5 binding, the MPER epitope adopts a kinked, extended structure and recognition of this specific structure is postulated to be a requirement for neutralizing activity. Ofek et al. therefore strove to mimic this structure in their computationally designed immunogen. The group first searched the protein data bank (PDB) for “acceptor proteins” that could be used as scaffolds, with segments that contained backbone structural similarity to the 2F5-bound gp41 epitope. The identified proteins were re-designed using RosettaDesign to introduce mutations such that the 2F5 MPER epitope side chains would be included in these scaffolds [32]. These constructs were used in vaccination trials using mice. Although some antibodies with similar binding modes to 2F5 were identified, the vaccine trials failed to produce neutralizing sera. However, crystal structures of the resulting antibodies in complex with the HIV MPER demonstrated that the segment corresponding to the 2F5 epitope adopted the desired kinked, extended structure [32]. Correia et al. performed a similar study using the linear epitope of 4E10 [21]. Appropriate scaffold proteins were again identified from the PDB and optimized using RosettaDesign. The resulting protein-4E10 epitope constructs were found to bind with higher affinity (in some cases 100-fold higher) to 4E10 than compared to the MPER peptide epitope alone [21]. These epitope-scaffolds were used in immunization trials with rabbits, and were shown to induce antibodies that were non-neutralizing but displayed high structural similarity to 4E10 [21]. As discussed above, it is known that both 2F5 and 4E10 require interaction with the virion lipid membrane for binding [22]. Therefore, this approach may require some modifications to incorporate membrane-like components to elicit 2F5- or 4E10-like antibodies. Nonetheless, these studies demonstrate that appropriate engineering of immunogens to contain segments that mimic conformational features of linear epitopes can be used to generate structure-specific antibodies against those epitopes.
Azoitei et al. have developed a computational and experimental methodology to incorporate both the backbone conformation and the side chains of functional motifs onto appropriate protein scaffolds [18, 19]. In the examples above, protein grafting involved transplantation of the protein side chains onto an “acceptor protein” scaffold that already contained native segments in which the backbone conformation matched that of the linear epitope in the bNAb-bound conformation. However, such an approach is limited in that an acceptor protein with a segment that matches the epitope conformation of the epitope region must be identified. In their work, Azoitei et al. developed a method to incorporate both side chains and backbones of an epitope into a scaffold and imposing the desired epitope conformation by protein design [18]. The authors found that epitope backbone grafting resulted in scaffolds that bound 2F5 with up to 30-fold higher affinity than the corresponding side-chain only grafting construct [18]. Therefore, backbone grafting may prove to be more successful for the generation of bNAbs than side chain only grafting, although no immunization trials have yet been performed with constructs designed using this methodology.
1.2 Computational and Combinatorial Redesign of HIV-1 gp120 Analogs
While some bNAbs, such as 2F5 and 4E10, target linear epitopes on viral glycoproteins, many known bNAbs target epitopes that consist of discontinuous protein segments. Therefore, neutralization requires recognition of a specific three-dimensional conformation of the viral antigen. This presents a challenge for immunogen design, which must accurately recapitulate the three-dimensional antibody binding epitope. An additional challenge is that many viral glycoproteins are structurally heterogeneous; therefore, it is crucial to design immunogens that mimic the structure of the epitope that is relevant for antibody neutralization. The case of HIV-1 gp120 highlights these problems facing effective viral immunogen design. In 2009, Chen et al. used modeling and binding experiments to explore the differences between the potent HIV-1 bNAb b12, which targets the CD4-binding site on gp120, and poorly neutralizing antibodies that also target the CD4-binding site [54]. They found that even slight differences in the binding site, on the order of a few angstroms, were sufficient to result in differing antibody neutralization capabilities. For example, antibody b13, whose angle of approach to the CD4-binding site differs from that of b12 only by a 17° rotation of the variable region, binds a substantially different conformation of gp120 than does b12 [54]. Therefore, one explanation for the failure of viral immunogens to be translated into effective vaccines thus far may be that immunogen design has not been sufficiently precise in the nature of the interactions with the antibodies they elicit.
Recent attempts to overcome these challenges have turned to advances in computational and combinatorial protein design methods. Azoitei et al. used a combined computational and experimental approach to graft the nonlinear, discontinous b12 epitope into an acceptor protein scaffold [19]. The resulting immunogen, 2bodx_43, was observed to bind tightly to b12 but not to other CD4 binding site-targeting or non-neutralizing antibodies such as b13 [19]. In 2010, Wu et al. used protein engineering to generate a variant of gp120 that preserved the CD4 binding site, but eliminated other antigenic regions by substituting surface exposed residues not included in the CD4 binding site with simian immunodeficiency virus homologs or other non-HIV residue identities (the “resurfaced gp120” core, Figure 3A). This design was intended to focus interactions on the CD4-binding site, since antibodies that interact with other segments of HIV-1 gp120 would not bind the resurfaced segments. The resurfaced gp120 core was used as bait for screening using broadly neutralizing serum from human patients, and a potent bNAb, VRC01, was identified [34]. As expected from the gp120 variant design, VRC01 binds the CD4 binding site [55]; other “VRC01-like” antibodies have been identified from HIV-infected individuals and have been shown to broadly neutralize across HIV genetic isolates [34, 56].
Figure 3. Computational and Combinatorial Redesign of HIV-1 gp120 Analogs.
A) Structure of the gp120 core showing the CD4 binding site in yellow and residues that were modified in the “resurfaced gp120” in red [34]. Residues for modification were chosen to eliminate other antigenic regions on gp120 and thereby focus the immune response to this molecule on the CD4 binding site. This modified gp120 molecule was used as bait for screening using broadly neutralizing serum from human patients, resulting in the identification of VRC01, a potent bNAb. (gp120 PDB ID: 2NXY). B) Schematic of gp120 engineering scheme by Jardine et al. in order to engage VRC01 germline precursors [27]. Inset shows native gp120 (green) and the engineered molecule, eOD-GT6 (pink) with binding to the VRC01 germline antibody Fab (cyan). (eOD-GT6 and the germline precursor were taken from PDB ID 4JPK; gp120 from PDB ID 2NXY). Potential steric clashes between gp120 glycans at positions N276 and N463 and the germline precursor were predicted; these glycans are drawn in on the inset figure in red sticks. Both Asn residues were mutated to Asp in eOD-GT6, thereby removing these clashes.
Interestingly, all of the “VRC01-like” antibodies derive from the IGVH1-2 germline segment, a promising observation for the induction of similar bNAbs since antibodies originating from this germline segment are estimated to be present in ~2% of the human antibody repertoire [27]. However, the predicted germline progenitors of VRC01 and VRC01-like antibodies do not show binding to wild-type gp120. Therefore, it is unlikely that gp120 itself could elicit VRC01-like antibodies since it is unable to engage the appropriate unmutated progenitors. To address this problem, Jardine et al. have recently used a combination of computationally-guided antigen design and in vitro screening to engineer an HIV-1 gp120 immunogen that binds to multiple VRC01-class bNAbs and their germline precursors (Figure 3B) [27]. Jardine et al. first built a homology model of a germline precursor to VRC01 bound to gp120; the model revealed areas of potential steric clashes between gp120 glycans and the germline antibody (Figure 3B, inset). The group generated a gp120 outer domain (OD) construct where these clashes were removed. Rosetta computational protein interface was then used to identify additional mutations at the CD4-binding site that were predicted to increase the affinity for the VRC01 germline progenitor, and libraries with these mutations were screened using yeast display. gp120 outer domain variants identified from this screen were subjected to further rounds of computational design and library screening, as well as selection for retention of binding to CD4. The final result of this iterative process was a construct termed eOD-GT6, which was shown to bind with nanomolar affinity to VRC01 and VRC01-like antibodies and with lower (micromolar) affinity to germline progenitors [27]. eOD-GT6 was fused to a self-assembling virus-like nanoparticles and was shown to activate germline and mature VRC01-class B cells [27]. The eOD-GT6 construct has not yet been evaluated in vaccination trials; rabbits, mice and macaques lack a germline segment that bears homology to IGVH1-2. Nonetheless, eOD-GT6 may be a promising immunogen candidate to elicit VRC01-like bNAbs. Furthermore, the concept of targeting of immunogens to engage both early and fully matured forms of antibodies along the immunological evolution pathway merits further evaluation.
1.3 Identification of Conserved Epitopes in Influenza Hemagglutinin
Influenza is an RNA virus that causes respiratory tract infection in mammals and some bird species [57]. Influenza is an enveloped virus containing two coat proteins: hemagglutinin (HA), which is responsible for host cell receptor binding and membrane fusion, and neuraminidase (NA), which cleaves sialic acid residues to release newly budding viral particles from the host cell [35, 58]. As is the case with HIV-1 Env, a high degree of antigenic diversity is tolerated in both HA and NA, thus explaining influenza’s ability to evade host immune responses and cause repeated infections in a single host. Highly infectious, pandemic influenza outbreaks occur approximately every 10–12 years, and are the result of large changes in or recombination of HA and/or NA, termed antigenic shift. These highly contagious and lethal outbreaks are cause for great concern worldwide. In particular, the H5N1 influenza resurgence in 2004 has led to concerns over its pandemic potential and highlighted the need for a vaccine that is effective against multiple H5N1 genetic isolates [57].
In 2011, Giles and Ross utilized a novel computational antigen design technique termed COBRA (computationally optimized broadly reactive antigen) to generate a synthetic HA protein capable of eliciting a broadly neutralizing antibody response against H5N1 avian influenza [26]. H5N1 viruses are divided among 10 distinct clades, which are geographically diverse and classified according to phylogentic distance among HA genes. One approach to address the vast sequence diversity in circulating H5N1 isolates has been to generate consensus–based viral proteins for use as immunogens, where a population of H5N1 sequences are aligned and the most common residue at each position within the viral protein is selected. This strategy has been met with some success, with consensus-based H5N1 HA immunogens eliciting broad antibody responses in mice, ferrets and macaques [59–61]. However, Giles and Ross raise concerns over the bias inherent in consensus-based methods, which rely on the available input sequences and are therefore subject to sampling bias. The authors point out that the majority of H5N1 HA sequences from human isolates arise from clade 2 and therefore there are concerns that these sequences do not accurately reflect the true diversity of vial sequences circulating in avian population. The COBRA method was used to overcome these concerns by using multiple rounds of consensus generation.
The COBRA method was performed as follows: 129 unique HA sequences representing clade 2 H5N1 viruses were grouped into phylogenetic subclades and further divided into individual outbreak groups based on the time and geographic location of isolation. Consensus HA sequences were generated for each outbreak group and then consensus sequences were generated for each subclade based on the results for the outbreak groups. These subclade consensus sequences were again aligned to generate a final consensus sequence, termed “clade 2 COBRA HA” [26]. Phylogenetic comparison of this final consensus sequence with all human isolates of H5N1 HA sequences demonstrated that clade 2 COBRA HA retained a clade 2-like sequence but did not fall specifically within any subclade classification. Indeed, clade 2 COBRA HA represents a unique HA sequence that has not been previously isolated [26]. VLPs displaying clade 2 COBRA HA on the surface were shown to bind to the HA receptor sialic acid; mice, ferrets, and cynomolgus macaques vaccinated with clade 2 COBRA HA VLPs demonstrated protective levels of anti-HA antibodies to a series of viral isolates representing each subclade of H5N1 clade 2 [25, 26]. Interestingly, the clade 2 COBRA HA VLPs were found to elicit higher-titer antibodies to a panel of H5N1 HA proteins than a mixture of VLP vaccines expressing representative HA molecules from clade 2.1, 2.2, and 2.3 [24]. This result suggests that the COBRA-derived sequence elicits a more efficient antibody response than an immunogen designed from a single genetic isolate.
2. Anti-Viral Monoclonal Antibodies
As an alternative to vaccination strategies, where immunogens are designed to elicit a neutralizing antibody response, monoclonal antibodies (mAbs) can be administered directly for therapeutic intervention. mAbs have been used clinically to treat a variety of diseases since the 1980s [62]. The first patient to be treated with a mAb in the United States was in 1980 for non-Hodgkins lymphoma [62]. Because of the ability of mAbs to precisely target specific cell proteins and receptors, mAb therapy for cancer therapeutics quickly accelerated. Despite the clear advantages that mAbs offered over traditional chemotherapeutic regimes in terms of specific cell targeting, early therapeutic trials were limited by the immunogenicity of murine antibody scaffolds upon repeated exposure, resulting in shorter mAb half-life, lack of antibody effector functions, and human anti-mouse antibody (HAMA) responses causing fever, chills, rash, nausea, headaches, and rarely anaphylaxis [62–64]. These limitations have been overcome by the development of human-mouse chimeras and fully humanized antibodies. Currently, there are 30 mAbs approved by the FDA for clinical use in the United States. Twenty-one of these are fully humanized, six are human-murine chimeras, and three are murine [65]. One of these antibodies, Palivizumab (Synagis), is a fully humanized mAb approved for prophylaxis against respiratory syncytial virus, demonstrating that mAb therapy can be effective against viral targets [16, 66]. Palivizumab remains the only therapy to significantly reduce RSV hospitalization rates in high risk infants (from 10.6% to 4.8%) and to reduce subsequent morbidity/mortality [66]. However, it is worth noting that the cost-effectiveness of Palivizumab has been questioned, as the antibody is administered in monthly injections for all five months of the RSV season. On average, seventeen children must be treated in this manner to prevent one RSV-related hospital admission, and fifty-nine must be treated to prevent one ICU admission [67]. Palivizumab is not covered by the national health services in Australia or the UK because of concerns over cost-effectiveness.
Traditionally, mAbs against viral targets have been isolated from immunized/infected animals or humans. The study of antibody-antigen complexes, mainly by X-ray crystallography, has allowed for antibody engineering (primarily within the CDRs) to enhance the potency of these naturally occurring antibodies. Computer-aided analysis has allowed for antibody engineering even in the absence of high-resolution structural data. Through engineering based on structural examination or computational methods, rational mutations can be introduced into mAbs in order to improve affinity or convey other desirable properties, such as cross-reactivity or solubility, thus facilitating the identification of neutralizing antibodies against therapeutically challenging viruses. Here we highlight some recent examples of the engineering of anti-viral mAbs. We end with a discussion of synthetic antibody engineering and the potential for this technique to be applied to viral therapeutics.
2.1 Rational Engineering of a Potent VRC01-like mAb
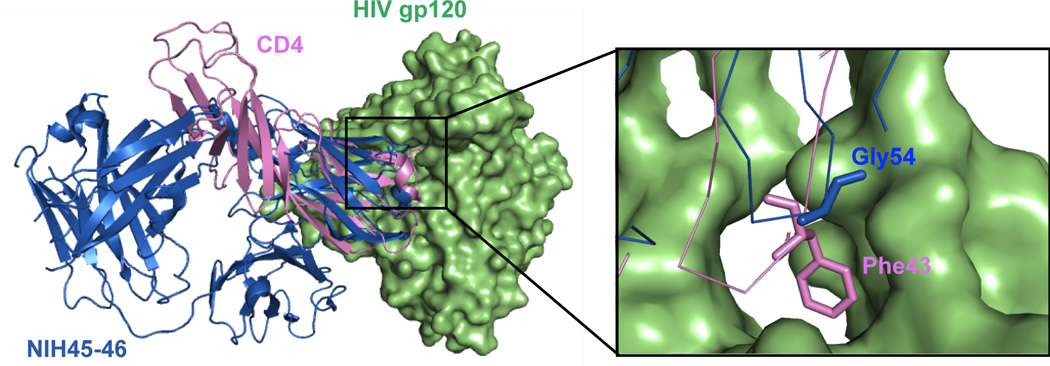
Diskin et al. used structural information to increase the potency of a VRC01-like mAb against HIV-1 [23]. The group started with NIH45–46, a clonal variant of VRC01 that was isolated from the same donor patient but has enhanced neutralization abilities. Structurally, the two mAbs are highly similar, containing 85% sequence identity in the heavy chain variable (VH) domains and 96% sequence identity in light chain variable (VL) domains. However, NIH45–46 includes a four-residue insertion within the third heavy chain complementarity determining region (CDR-H3) relative to VRC01; these residues were observed to contribute to binding of the antibody to the CD4 binding site on gp120. Diskin et al. carefully examined the crystal structures of both VRC01 and NIH45–46 bound to gp120. They noted that a key interaction between CD4 residue 43 (a phenylalanine) and a hydrophobic pocket on gp120 was not mimicked by either antibody, and hypothesized that mutating residues on the mAbs to interact with this hydrophobic pocket would increase mAb potency and breadth (Figure 4). A series of NIH45–46 mutants were therefore constructed containing hydrophobic amino acid substitutions at residue 54, a Gly in the native NIH45–46, which is in close proximity to the gp120 hydrophobic pocket. They saw that, as expected, NIH45–46 containing G54W or G54F mutations showed increased neutralization potency [23]. Remarkably, NIH45–46G54W showed a substantial increase in the breadth of HIV-1 strains that it neutralized compared to the parent NIH45–46, with up to 2000-fold higher neutralization abilities against some HIV-1 strains [23]. This work provides an elegant example of how structure based rational design can be used to construct potent anti-viral antibodies that have the potential to be used in passive immunization or treatment of viral infection.
Figure 4. Rational Engineering of a Potent VRC01-like mAb.
Diskin et al. noted that neither VRC01 nor mAb NIH45–46 (blue) recapitulated a key interaction between CD4 (pink) residue 43 (a Phe) and a hydrophobic pocket on gp120 (green) [23]. A series of NIH45–46 mutants were constructed to contain hydrophobic amino acid substitutions at residue 54, a Gly in NIH45–46 (shown in sticks in inset). NIH45–46 containing G54W or G54F mutations showed increased neutralization potency and breadth.
2.2 Computational Redesign of Broad Dengue Virus Antibodies
Recent work by Tharakaraman et al. utilizes a novel computation approach that is not reliant on crystal structure information to increase the potency of an antibody against Dengue Virus (DENV) [68]. DENV is a flavivirus responsible for 50–100 million human infections per year, ranging in severity from an acute febrile illness to a fatal hemorrhagic fever or shock syndrome [69]. DENV consists of four serotypes (DENV1–4), which vary from one another at the amino acid level by 25–40%. Importantly, DENV infection is characterized by a marked antibody-dependent enhancement of replication, whereby higher levels of viral replication and lethality are observed in DENV survivors during a second infection with the same DENV serotype or a primary infection by any of the other three serotypes [70]. Because of this phenomenon, previous vaccination trials against DENV have failed and current interest focuses on antibodies that can inhibit multiple serotypes.
In their work, Tharakaraman et al. computationally redesigned 4E11, an antibody directed against the DENV viral envelope glycoprotein (E) [68]. 4E11 was previously identified from mice and found have potent neutralizing ability for DENV1–3 but limited neutralization potential against DENV4. Therefore, the group strove to improve the affinity of 4E11 to DENV4, while maintaining the affinity of the antibody to DENV1–3 by relying on computational docking models. Recently, the structure of the 4E11 single chain variable fragment (scFv) in complex with E domain III from all four serotypes was reported [71], but when the Tharakaraman et al. study was initiated, these structures were not yet available [68]. The group first used a multivariate logistic regression analysis (MLR) to identify key physiochemical features that define natural antigen-antibody interfaces, which allowed them to more accurately predict the correct antibody-antigen complexes than standard docking protocols relying solely on energy minimizing functions. Using the MLR results, models of 4E11 bound to the E protein of DENV1–4 were generated. The models were used to design mutations that were predicted to increase antibody affinity for DENV4 while not detrimentally affecting contacts with DENV1–3. One mutant, 4E5A, contained five amino acid substitutions and was found to have the greatest increase in affinity towards DENV4. Compared to 4E11, the redesigned 4E5A displayed a 450-fold enhancement in affinity to DENV4 and a 15-fold enhancement in affinity to DENV2, while maintaining affinity to DENV1 and DENV3. In vitro neutralization assays showed that 4E5A neutralized DENV4 with >75-fold increased potency, while maintaining potency against DENV1–3. Additionally, 4E5A demonstrated potent antiviral activity against all four DENV strains in a mouse model of infection, causing a significant reduction in viremia at both 1 mg/kg and 5 mg/kg. [68].
Recent work from the Varani laboratory has also applied computational methods to improve the neutralization capabilities of an anti-DENV antibody [72]. Here, NMR epitope mapping was used to define the binding site of a broadly neutralizing human mAb, DV32.6, against all four DENV serotypes. This structural information was used to filter the results of computational docking of DV32.6 to DENV1–4. Analysis of the best docking models allowed the group to rationally engineer DV32.6 mutants that bound only to one serotype (thereby demonstrating an ability to increase antibody specificity) or that bound more tightly to the eptiopes on DENV1–4, resulting in a mutant that was up to 40 times more effective than the parent DV32.6 at neutralizing DENV [72]. Of note, this group has used computational modeling in the past to explore why an anti-DENV antibody failed to neutralize viral infection [73] and to examine the binding of two neutralizing mAbs to the Influenza HA protein [74]. The examples outlined here demonstrate how computational methods can be applied to the study of antibody-virus interactions, and how this information can be used to develop more effective anti-viral mAbs.
2.3 Synthetic Antibodies Targeting Intermediates of Ebola Virus Fusion
In contrast to computational modeling, where a starting antibody is required, synthetic antibody engineering shows promise for the development of novel antibodies against viral targets for which few antibodies are available. Synthetic antibodies contain antigen-binding sites that are constructed entirely from rationally designed, man-made diversity [75]. A key advantage compared to traditional methods is that synthetic antibody engineering does not rely on previous human or animal infection/immunization. Therefore, it is possible to select antibodies against traditionally non-immunogenic targets, or to target viral epitope conformations that may not arise during natural infection. Synthetic antibody libraries displayed on the surface of M13 filamentous bacteriophage (phage display) have been used to select antibodies that bind to targets such as ubiquitin [76], histones [77], and hemoglobins [78], and to select against precise antigenic conformations [79]. Antibodies with exquisite specificity have been obtained from synthetic antibody libraries, such as antibodies that can distinguish between chicken and quail lysozyme [80], which differ by only four amino acids, and those that can differentiate between two conformations of the same enzyme (caspase) [81]. An additional advantage is that the initial libraries are typically constructed on human scaffolds, therefore the resulting antibodies do not require extensive engineering to “humanize” prior to therapeutic use [75].
Despite the advantages offered by synthetic antibody phage display, this technique has only recently been applied to the discovery of antibodies targeting viral epitopes. Our laboratory used this technology to target fusion intermediates of the Ebola Virus (EBOV) envelope glycoprotein (GP) [82]. EBOV is a highly pathologic member of the Filoviridae family of viruses that causes severe hemorrhagic fever [83]. Viral entry is mediated by GP, which consists of three copies each of a surface subunit, GP1, and a transmembrane subunit, GP2 [35, 84]. GP1 binding to cell surface receptors initiates uptake of the virus into the endosome. Here, host cysteine proteases cleave GP removing most of GP1; this cleavage event has been shown to be necessary for EBOV entry [85, 86]. Cleavage is hypothesized to be important for viral entry for two reasons: i) cleavage is thought to unmask the receptor binding site for Neimann Pick C1, an endosomal cholesterol transporter which was recently shown to be a critical intracellular receptor for EBOV entry [87] and ii) cleavage appears to prime the GP2 subunit for large-scale conformational changes which ultimately lead to fusion of the viral and host endosomal membranes [35]. However, structural changes in GP associated with endosomal proteolytic cleavage are incompletely defined, and the precise timing of cleavage within the endosome is poorly understood. Additionally, it was unclear whether epitopes on the proteolytically cleaved GP were available for virus neutralization. Therefore, we strove to identify novel mAbs capable of distinguishing between the uncleaved (GPUNCL) and proteolytically cleaved (GPCL) forms of GP. However, there are limited sources of natural human Ebola virus antibodies since survivors generally have low serum antibody titers, and many responses are dominated by antibodies that bind preferentially with a secreted, dimeric form of GP (sGP) that is not relevant to membrane fusion [13, 88–91].
To identify mAbs with specific recognition profiles toward GPUNCL and GPCL, we used a synthetic antibody binding fragment (Fab) library based on a human anti-maltose binding protein Fab scaffold. This library, “Library F”, contains binomial tyrosine/serine randomization in non-structural positions of CDR-H1 and CDR-H2, and additional variation at CDR-H3 and CDR-L3 encoding the nine residues Tyr/Ser/Gly/Ala/Phe/Trp/His/Pro/Val in a 5/4/4/2/1/1/1/1/1 ratio [92]. This amino acid distribution mimics the observed distribution found in natural CDR segments [93]. We screened Library F against protein mimics of GPUNCL and GPCL. We identified antibodies with distinct recognition profiles: FabCL bound preferentially to GPCL (EC50 = 1.7 nM), whereas FabUNCL bound specificity to GPUNCL (EC50 = 75 nM) [82]. Neutralization assays with GP-containing pseudotyped viruses indicated that these antibodies inhibited GPCL or GPUNCL-mediated viral entry with specificity that matched their recognition profiles (IC50s: 87 nM for IgGCL; 1 µM for FabUNCL) [82]. This work demonstrates that epitopes on GPCL are available for neutralization by antibodies, and may lead to the development of new tools for dissecting intermediates of EBOV entry. Importantly, these results demonstrate the applicability of synthetic antibody engineering to the study of viral membrane fusion, paving the way for synthetic antibody libraries to be screened against other viral targets for which there are limited sources of natural, human antibodies.
Conclusions
Rational, computational, and combinatorial methods hold great promise for application of protein engineering principles to viral vaccine and immunotherapeutic development. Furthermore, the number of bNAb characterization and structural studies have been increasing steadily over recent years, providing additional information on which to base design. While much progress has been made in engineering new viral immunogens and antibodies, the challenge moving forward will be evaluation of these reagents in appropriate animal models. We expect this future work will provide new fundamental insight into requirements for protection from and neutralization of viruses in vivo.
Acknowledgments
Funding: J. R. L. acknowledges funding from the National Institutes of Health (R01-AI090249). J. F. K. was supported in part by NIH Medical Scientist Training Grant T32-GM007288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huang DB, Wu JJ, Tyring SK. A review of licensed viral vaccines, some of their safety concerns, and the advances in the development of investigational viral vaccines. The Journal of infection. 2004;49:179–209. doi: 10.1016/j.jinf.2004.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plotkin SA. Correlates of protection induced by vaccination. Clinical and vaccine immunology : CVI. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton DR. Antibodies, viruses and vaccines. Nature reviews Immunology. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 4.Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, Basler CF, Palese P. Neutralizing Antibodies Against Previously Encountered Influenza Virus Strains Increase over Time: A Longitudinal Analysis. Science translational medicine. 2013;5:198ra107. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khetsuriani N, Pallansch MA, Jabirov S, Saparova N, Oberste MS, Wannemuehler K, Ursu P, Wassilak S, Martin R. Population immunity to polioviruses in the context of a large-scale wild poliovirus type 1 outbreak in Tajikistan, 2010. Vaccine. 2013;31:4911–4916. doi: 10.1016/j.vaccine.2013.06.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takada A, Ebihara H, Jones S, Feldmann H, Kawaoka Y. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine. 2007;25:993–999. doi: 10.1016/j.vaccine.2006.09.076. [DOI] [PubMed] [Google Scholar]
- 7.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nature medicine. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 8.Hangartner L, Zinkernagel RM, Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nature reviews Immunology. 2006;6:231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
- 9.Smith TJ, Chase ES, Schmidt TJ, Olson NH, Baker TS. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung DT, Tam FC, Ma CH, Chan PK, Cheung JL, Niu H, Tam JS, Lim PL. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. The Journal of infectious diseases. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakurai H, Williamson RA, Crowe JE, Beeler JA, Poignard P, Bastidas RB, Chanock RM, Burton DR. Human antibody responses to mature and 25 immature forms of viral envelope in respiratory syncytial virus infection: significance for subunit vaccines. Journal of virology. 1999;73:2956–2962. doi: 10.1128/jvi.73.4.2956-2962.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. Journal of virology. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PW, Burton DR. Ebola virus can be effectively neutralized by antibody produced in natural human infection. Journal of virology. 1999;73:6024–3030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu X, Alimonti JB, Melito PL, Fernando L, Stroher U, Jones SM. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clinical immunology (Orlando, Fla) 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. The Journal of infectious diseases. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 17.Prabhu N, Prabakaran M, Ho HT, Velumani S, Qiang J, Goutama M, Kwang J. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. Journal of virology. 2009;83:2553–2562. doi: 10.1128/JVI.02165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azoitei ML, Ban YE, Julien JP, Bryson S, Schroeter A, Kalyuzhniy O, Porter JR, Adachi Y, Baker D, Pai EF, Schief WR. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. Journal of molecular biology. 2012;415:175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azoitei ML, Correia BE, Ban YE, Carrico C, Kalyuzhniy O, Chen L, Schroeter A, Huang PS, McLellan JS, Kwong PD, Baker D, Strong RK, Schief WR. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science (New York, NY) 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty K, Durani V, Miranda ER, Citron M, Liang X, Schleif W, Joyce JG, Varadarajan R. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. The Biochemical journal. 2006;399:483–491. doi: 10.1042/BJ20060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correia BE, Ban YE, Holmes MA, Xu H, Ellingson K, Kraft Z, Carrico C, Boni E, Sather DN, Zenobia C, Burke KY, Bradley-Hewitt T, Bruhn-Johannsen JF, Kalyuzhniy O, Baker D, Strong RK, Stamatatos L, Schief WR. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure (London, England : 1993) 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Dennison SM, Stewart SM, Stempel KC, Liao HX, Haynes BF, Alam SM. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. Journal of virology. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science (New York, NY) 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles BM, Bissel SJ, Dealmeida DR, Wiley CA, Ross TM. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutininbased H5N1 virus-like particle vaccines. Clinical and vaccine immunology : CVI. 2012;19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. The Journal of infectious diseases. 2012;205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine. 2011;29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science (New York, NY) 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGaughey GB, Citron M, Danzeisen RC, Freidinger RM, Garsky VM, Hurni WM, Joyce JG, Liang X, Miller M, Shiver J, Bogusky MJ. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry. 2003;42:3214–3223. doi: 10.1021/bi026952u. [DOI] [PubMed] [Google Scholar]
- 29.Mester B, Manor R, Mor A, Arshava B, Rosen O, Ding FX, Naider F, Anglister J. HIV-1 peptide vaccine candidates: selecting constrained V3 peptides with highest affinity to antibody 447-52D. Biochemistry. 2009;48:7867–7877. doi: 10.1021/bi900146g. [DOI] [PubMed] [Google Scholar]
- 30.Mor A, Segal E, Mester B, Arshava B, Rosen O, Ding FX, Russo J, Dafni A, Schvartzman F, Scherf T, Naider F, Anglister J. Mimicking the structure of the V3 epitope bound to HIV-1 neutralizing antibodies. Biochemistry. 2009;48:3288–3303. doi: 10.1021/bi802308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moseri A, Tantry S, Sagi Y, Arshava B, Naider F, Anglister J. An optimally constrained V3 peptide is a better immunogen than its linear homolog or HIV-1 gp120. Virology. 2010;401:293–304. doi: 10.1016/j.virol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD. Elicitation of structure-specific antibodies by epitope scaffolds. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaram R, Lynch MP, Rawale SV, Sun Y, Kazanji M, Kaumaya PT. De novo design of peptide immunogens that mimic the coiled coil region of human T-cell leukemia virus type-1 glycoprotein 21 transmembrane subunit for induction of native protein reactive neutralizing antibodies. The Journal of biological chemistry. 2004;279:24141–24151. doi: 10.1074/jbc.M313210200. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, NY) 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, Schlake T, Thess A, Kallen KJ, Stitz L, Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nature biotechnology. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 38.Luo M, Tao P, Li J, Zhou S, Guo D, Pan Z. Immunization with plasmid DNA encoding influenza A virus nucleoprotein fused to a tissue 28 plasminogen activator signal sequence elicits strong immune responses and protection against H5N1 challenge in mice. Journal of virological methods. 2008;154:121–127. doi: 10.1016/j.jviromet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Park KS, Seo YB, Lee JY, Im SJ, Seo SH, Song MS, Choi YK, Sung YC. Complete protection against a H5N2 avian influenza virus by a DNA vaccine expressing a fusion protein of H1N1 HA and M2e. Vaccine. 2011;29:5481–5487. doi: 10.1016/j.vaccine.2011.05.062. [DOI] [PubMed] [Google Scholar]
- 40.Ledgerwood JE, Graham BS. DNA vaccines: a safe and efficient platform technology for responding to emerging infectious diseases. Human vaccines. 2009;5:623–626. doi: 10.4161/hv.8627. [DOI] [PubMed] [Google Scholar]
- 41.Lu S. Heterologous prime-boost vaccination. Current opinion in Immunology. 2009;21:346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, Nabel GJ. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science (New York, NY) 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 43.Wei CJ, Yassine HM, McTamney PM, Gall JG, Whittle JR, Boyington JC, Nabel GJ. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Science translational medicine. 2012;4:147ra114. doi: 10.1126/scitranslmed.3004273. [DOI] [PubMed] [Google Scholar]
- 44.Burton DR. Scaffolding to build a rational vaccine design strategy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17859–17860. doi: 10.1073/pnas.1012923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. Journal of virology. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. Journal of virology. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conley AJ, Gorny MK, Kessler JA, 2nd, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. Journal of virology. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salzwedel K, West JT, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. Journal of virology. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. Journal of virology. 2005;79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, Nussenzweig MC. A method for identification of HIV gp140 binding memory B cells in human blood. Journal of immunological methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS research and human retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science (New York, NY) 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. Journal of virology. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural 30 convergence of broad and potent HIV antibodies that mimic CD4 binding. Science (New York, NY) 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. Avian influenza A (H5N1) infection in humans. The New England journal of medicine. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 58.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 59.Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laddy DJ, Yan J, Kutzler M, Kobasa D, Kobinger GP, Khan AS, Greenhouse J, Sardesai NY, Draghia-Akli R, Weiner DB. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PloS one. 2008;3:e2517. doi: 10.1371/journal.pone.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen MW, Cheng TJ, Huang Y, Jan JT, Ma SH, Yu AL, Wong CH, Ho DD. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13538–13543. doi: 10.1073/pnas.0806901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oldham RK, Dillman RO. Monoclonal antibodies in cancer therapy:25 years of progress. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:1774–1777. doi: 10.1200/JCO.2007.15.7438. [DOI] [PubMed] [Google Scholar]
- 63.Dillman Human antimouse and antiglobulin responses to monoclonal antibodies. Antibody Immunocon Radiopharm. 1990;3:1–15. [Google Scholar]
- 64.Legouffe E, Liautard J, Gaillard JP, Rossi JF, Wijdenes J, Bataille R, Klein B, Brochier J. Human anti-mouse antibody response to the injection of murine monoclonal antibodies against IL-6. Clinical and experimental Immunology. 1994;98:323–329. doi: 10.1111/j.1365-2249.1994.tb06145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reichert J. Therapeutic monoclonal antibodies approved or in review in the European Union or United States. The Antibody Society; Waban, Massachusetts: 2013. p. 02468. [Google Scholar]
- 66.Resch B, Resch E, Muller W. Should respiratory care in preterm infants include prophylaxis against respiratory syncytial virus infection? The case in favour. Paediatric respiratory reviews. 2013;14:130–136. doi: 10.1016/j.prrv.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Isaacs D. Should respiratory care in preterm infants include prophylaxis against respiratory syncytial virus? The case against. Paediatric respiratory reviews. 2013;14:128–129. doi: 10.1016/j.prrv.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 68.Tharakaraman K, Robinson LN, Hatas A, Chen YL, Siyue L, Raguram S, Sasisekharan V, Wogan GN, Sasisekharan R. Redesign of a cross-reactive antibody to dengue virus with broad-spectrum activity and increased in vivo potency. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1555–E1564. doi: 10.1073/pnas.1303645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Back AT, Lundkvist A. Dengue viruses - an overview. Infection ecology & epidemiology. 2013;3 doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annual review of Immunology. 2011;29:587–619. doi: 10.1146/annurev-immunol-031210-101315. [DOI] [PubMed] [Google Scholar]
- 71.Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure (London, England : 1993) 2012;20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Simonelli L, Pedotti M, Beltramello M, Livoti E, Calzolai L, Sallusto F, Lanzavecchia A, Varani L. Rational engineering of a human anti-dengue antibody through experimentally validated computational docking. PloS one. 2013;8:e55561. doi: 10.1371/journal.pone.0055561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simonelli L, Beltramello M, Yudina Z, Macagno A, Calzolai L, Varani L. Rapid structural characterization of human antibody-antigen complexes through experimentally validated computational docking. Journal of molecular biology. 2010;396:1491–1507. doi: 10.1016/j.jmb.2009.12.053. [DOI] [PubMed] [Google Scholar]
- 74.Pedotti M, Simonelli L, Livoti E, Varani L. Computational docking of antibody-antigen complexes, opportunities and pitfalls illustrated by influenza hemagglutinin. International journal of molecular sciences. 2011;12:226–251. doi: 10.3390/ijms12010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sidhu SS, Fellouse FA. Synthetic therapeutic antibodies. Nat Chem Biol. 2006;2:682–688. doi: 10.1038/nchembio843. [DOI] [PubMed] [Google Scholar]
- 76.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. Journal of molecular biology. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 77.Philibert P, Stoessel A, Wang W, Sibler AP, Bec N, Larroque C, Saven JG, Courtete J, Weiss E, Martineau P. A focused antibody library for selecting scFvs expressed at high levels in the cytoplasm. BMC biotechnology. 2007;7:81. doi: 10.1186/1472-6750-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parsons HL, Earnshaw JC, Wilton J, Johnson KS, Schueler PA, Mahoney W, McCafferty J. Directing phage selections towards specific epitopes. Protein engineering. 1996;9:1043–1049. doi: 10.1093/protein/9.11.1043. [DOI] [PubMed] [Google Scholar]
- 79.Geyer CR, McCafferty J, Dubel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods in molecular biology (Clifton, NJ) 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 80.Ayriss J, Woods T, Bradbury A, Pavlik P. High-throughput screening of single-chain antibodies using multiplexed flow cytometry. Journal of proteome research. 2007;6:1072–1082. doi: 10.1021/pr0604108. [DOI] [PubMed] [Google Scholar]
- 81.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koellhoffer JF, Chen G, Sandesara RG, Bale S, Saphire EO, Chandran K, Sidhu SS, Lai JR. Two synthetic antibodies that recognize and neutralize distinct proteolytic forms of the ebola virus envelope glycoprotein. Chembiochem : a European journal of chemical biology. 2012;13:2549–2557. doi: 10.1002/cbic.201200493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhn JH, Bao Y, Bavari S, Becker S, Bradfute S, Brister JR, Bukreyev AA, Cai Y, Chandran K, Davey RA, Dolnik O, Dye JM, Enterlein S, Gonzalez JP, Formenty P, Freiberg AN, Hensley LE, Honko AN, Ignatyev GM, Jahrling PB, Johnson KM, Klenk HD, Kobinger G, Lackemeyer MG, Leroy EM, Lever MS, Lofts LL, Muhlberger E, Netesov SV, Olinger GG, Palacios G, Patterson JL, Paweska JT, Pitt L, Radoshitzky SR, Ryabchikova EI, Saphire EO, Shestopalov AM, Smither SJ, Sullivan NJ, Swanepoel R, Takada A, Towner JS, van der Groen G, Volchkov VE, Wahl-Jensen V, Warren TK, Warfield KL, Weidmann M, Nichol ST. Virus nomenclature below the species level: a standardized nomenclature for laboratory animal-adapted strains and variants of viruses assigned to the family Filoviridae. Archives of virology. 2013;158:1425–1432. doi: 10.1007/s00705-012-1594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science (New York, NY) 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. Journal of virology. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee JE, Saphire EO. Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Current opinion in structural biology. 2009;19:408–417. doi: 10.1016/j.sbi.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohan GS, Li W, Ye L, Compans RW, Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS pathogens. 2012;8:e1003065. doi: 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Basler CF. A novel mechanism of immune evasion mediated by Ebola virus soluble glycoprotein. Expert review of anti-infective therapy. 2013;11:475–478. doi: 10.1586/eri.13.30. [DOI] [PubMed] [Google Scholar]
- 91.Dolnik O, Volchkova V, Garten W, Carbonnelle C, Becker S, Kahnt J, Stroher U, Klenk HD, Volchkov V. Ectodomain shedding of the glycoprotein GP of Ebola virus. The EMBO journal. 2004;23:2175–2184. doi: 10.1038/sj.emboj.7600219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Persson H, Ye W, Wernimont A, Adams JJ, Koide A, Koide S, Lam R, Sidhu SS. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. Journal of molecular biology. 2013;425:803–811. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koide S, Sidhu SS. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS chemical biology. 2009;4:325–334. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]