Abstract
Striatal-enriched tyrosine phosphatase (STEP) is an important regulator of neuronal synaptic plasticity, and its abnormal level or activity contributes to cognitive disorders. One crucial downstream effector and direct substrate of STEP is extracellular signal-regulated protein kinase (ERK), which has important functions in spine stabilisation and action potential transmission. The inhibition of STEP activity toward phospho-ERK has the potential to treat neuronal diseases, but the detailed mechanism underlying the dephosphorylation of phospho-ERK by STEP is not known. Therefore, we examined STEP activity toward pNPP, phospho-tyrosine-containing peptides, and the full-length phospho-ERK protein using STEP mutants with different structural features. STEP was found to be a highly efficient ERK tyrosine phosphatase that required both its N-terminal regulatory region and key residues in its active site. Specifically, both KIM and KIS of STEP were required for ERK interaction. In addition to the N-terminal KIS region, S245, hydrophobic residues L249/L251, and basic residues R242/R243 located in the KIM region were important in controlling STEP activity toward phospho-ERK. Further kinetic experiments revealed subtle structural differences between STEP and HePTP that affected the interactions of their KIMs with ERK. Moreover, STEP recognised specific positions of a phospho-ERK peptide sequence through its active site, and the contact of STEP F311 with phospho-ERK V205 and T207 were crucial interactions. Taken together, our results not only provide the information for interactions between ERK and STEP, but will also help in the development of specific strategies to target STEP-ERK recognition, which could serve as a potential therapy for neurological disorders.
Keywords: ERK, phosphorylation, phosphatase, synaptic plasticity, Striatal enriched tyrosine phosphatases (STEP), neurological disorders
Introduction
Reversible tyrosine phosphorylation is one of the most important post-translational modifications steering cellular functions, including cell growth, immune responses, glucose metabolism, and neuronal activities (Hunter 2009, Yu et al. 2007, Chen et al. 2010). Specifically, protein tyrosine phosphorylation in the nervous system is precisely regulated both spatially and temporally by two groups of enzymes, protein tyrosine kinases and protein tyrosine phosphatases, to maintain diverse neuronal activities. Although numerous studies have identified pertinent roles for kinases in synaptic activity and cognition, the actions of tyrosine phosphatases in these processes have recently become appreciated (Hendriks et al. 2009, Fitzpatrick & Lombroso 2011). In particular, striatal-enriched protein tyrosine phosphatase (STEP) has been identified as a brain-specific tyrosine phosphatase and is implicated in several neuronal degenerative diseases in which increased STEP levels or phosphatase activities are observed (Baum et al. 2010).
STEP belongs to the protein tyrosine phosphatase (PTP) superfamily of which members have the signature CX5R motif in their active site and utilise a negatively charged cysteine for nucleophilic attack during hydrolytic reactions (Tonks 2006). Immunohistochemistry results have revealed that STEP is expressed specifically in the central nervous system (Fitzpatrick & Lombroso 2011). At least four STEP transcriptional isoforms have been identified and characterised; STEP46 and STEP61 are the two major isoforms with phosphatase activities (Sharma et al. 1995). The expression of both STEP46 and STEP61 is enriched in medium spiny neurons of the striatum, but their cellular localisations are different: STEP46 is mainly localised to the cytosol, whereas STEP61 has an additional 172 residues at its N-terminus that localise it to post-synaptic densities and endoplasmic reticulum (Baum et al. 2010).
As a member of the PTP superfamily, STEP participates in neuronal activities by regulating the phosphorylation states of key components of synaptic plasticity, including subunits of NMDAR and AMPAR and such kinases as Fyn, p38, and Pyks (Zhang et al. 2008, Xu et al. 2012, Baum et al. 2010). In particular, STEP negatively regulates the activation of ERK, which is the central hub of the phosphorylation networks that respond to extracellular stimulation. In neuronal cells, ERK activation plays important roles in spine stabilisation and transmitting action potentials. Accordingly, increased STEP activity accompanied by impaired ERK function has been implicated in neuronal degenerative diseases. Furthermore, STEP-knockout mice display increased ERK activation (Venkitaramani et al. 2009) and improved hippocampal learning and memory (Venkitaramani et al. 2011). All these results indicate that specifically inhibiting STEP activity toward phospho-ERK has therapeutic potential in neuronal degenerative diseases.
A negative regulation of STEP activity can be achieved by developing specific STEP inhibitors that target the phosphatase active site or by disrupting the interactions of STEP with its substrates. However, the underlying catalytic mechanisms of STEP towards its substrates remain unknown. In this study, we aimed to determine the molecular mechanism of STEP in the dephosphorylation of phospho-ERK, the key substrate of STEP for neuronal activity modulation, using combined molecular and enzymologic approaches. Our results reveal the contributions of key elements in mediating specific ERK-STEP recognition and identify peptide sequence selectivity in the STEP active site, findings that will help in discovering new STEP substrates and developing specific strategies to inhibit phospho-ERK dephosphorylation by STEP, potentially curing some neuronal diseases.
Material and Methods
Materials
Para-nitrophenyl phosphate (pNPP) was obtained from Bio Basic Inc. The Tyr(P)-containing peptides were synthesised and HPLC-purified by China Peptides Co. The Ni2+-NTA resin and HiTrap Q FF column used in protein purification were purchased from Bio Basic Inc. and GE Healthcare, respectively. The phospho-specific anti-ERK1/2-pT202/pY204 antibody was obtained from Cell Signaling, the anti-flag M2 antibody was purchased from Sigma, the antibody the β-Actin Antibody (C4) and the phospho-tyrosine pY-350 antibody was obtained from Santa Cruz Biotechnology. The fully sequenced human PTPN5 cDNA was purchased from Thermo Scientific. The expression plasmid for the STEP catalytic domain (STEP-CD) was a generous gift from Dr. Knapp at target discovery institute, U.K., and the plasmids expressing ERK2 and MEK1 used in the preparation of phospho-ERK were generous gifts from Dr. Lefkowitz at Duke University, U.S.A. The nerve growth factor (NGF) was purchased from Sino Biological Inc.
Cell Culture and Immunoblotting
PC12 cells were cultured as previously described (Morooka & Nishida 1998). The cells were transfected with STEP or mutants using Lipofectamine2000 (invitrogen) for 48 hours. After 8 hours starvation, cells were treated with 50ng/ml NGF for 0min, 2min, 5min, 15min, 30min, 60min and 120min at 37°C and then lysed. The protein concentration of extracts was measured by the BCA Protein Quantitation Kit (Beyotime). Equal amounts of cell lysates were denatured in 2×SDS loading buffer and boiled for 10min. Protein samples were then subjected to western blot.
Phosphatase assay using pNPP and phospho-tyrosine-containing peptides
The kinetic parameters for pNPP and Tyr(P)-containing peptides were determined as described previously (Liu et al. 2012a, Yu et al. 2011, Pan et al. 2013) All experiments were performed at 37°C in a buffer containing 50 mM succinic (pH 6.0), 1 mM EDTA, 2 mM DTT, and an ionic strength of 0.15 M adjusted with NaCl. STEP-catalysed pNPP hydrolysis was terminated by adding 120 μl 1 M NaOH, and the enzyme activity was monitored by measuring the absorbance at 405 nm. When Tyr(P)-containing peptides were used as the substrate, the reaction was stopped by adding BIOMOL GREEN™ (ENZO), and the released phosphate was determined by measuring the absorbance at 620 nm. The kinetic parameters were obtained by fitting the data to the Michaelis-Menten equation [Eq. 1]. The Tyr(P)-containing peptide hydrolysis was also continuously monitored at 305 nm by measuring the increase in tyrosine fluorescence with excitation at 280 nm as described (Vetter et al. 2000).
| [Eq. 1] |
Enzyme-coupled continuous spectrophotometric assay for phospho-ERK dephosphorylation
The kinetic parameters for the dephosphorylation of phospho-ERK2 proteins were determined using an MESG-coupled continuous spectrophotometric assay as described previously (Huang et al. 2004, Zheng et al. 2012, Zhang et al. 2011). MESG was synthesised and purified as described (Webb 1992), and the purity of MESG was quantified by HPLC and mass spectrometry. All assays were performed at 25°C in an MESG-coupled system containing 50 mM MOPS (pH 7.0), 100 mM NaCl, 0.1 mM EDTA, 100 μM MESG, and 0.1 mg/ml PNPase; the reactions were monitored at OD360. The initial rates were determined and fitted to the Michaelis-Menten equation to obtain Km and Kcat. In the case of a substrate concentration << Km, the Kcat/Km ratio was acquired by fitting the data to the following equation:
| [Eq. 2] |
Data analysis and software
The data were analysed using ImageJ and GraphPad software, and all data are presented as the mean ± standard error. The statistical comparisons were performed using ANOVA. The model of the STEP constructs was drawn using DOG (Domain Graph) 2.0 (Ren et al. 2009). The model of the STEP structure was generated from the previously resolved structure of STEP (PDB 2CJZ) (Eswaran et al. 2006) by Coot (Emsley et al. 2010) and the PyMOL Molecular Graphics System (Version 1.5.0.4 Schrödinger, LLC).
Because of page limitations, all other materials and methods are described in the supplemental material.
Results
STEP is a tyrosine-specific phospho-ERK2 phosphatase in vitro.
ERK activation is a pivotal step in many types of long-term memory and psychostimulant drug actions. Full activation of ERK requires double phosphorylation of both Thr202 and Tyr204 in its activation loop, sites that are dephosphorylated by several different phosphatases within specific cellular contexts(Patterson et al. 2009, Paul et al. 2003, Piserchio et al. 2012a) (Li et al. 2013). Both in corticostriatal culture and in vivo, STEP regulates neuronal activities mainly by targeting temporal ERK activation-loop phosphorylation (Paul et al. 2003, Valjent et al. 2005, Venkitaramani et al. 2009). Although cellular studies have detected the interaction of ERK with STEP (Munoz et al. 2003), direct quantitative measurement of phospho-ERK dephosphorylation by STEP in vitro with purified proteins has not been reported.
To begin to understand the molecular mechanism of phospho-ERK dephosphorylation by STEP, we prepared double-phosphorylated ERK and several protein phosphatases at high purity to compare the activities of different phosphatases toward phospho-ERK (Fig 1A and 1B). Unlike STEP, the Ser/Thr phosphatase PPM1A selectively dephosphorylates pT202 of ERK both in vivo and in vitro (Zhou et al. 2002, Li et al. 2013); in contrast, two other tyrosine phosphatases, BDP-1 and PTP-MEG2, have not been directly linked to phospho-ERK dephosphorylation. Using these phosphatases as controls, we investigated whether STEP is an efficient and tyrosine-specific ERK phosphatase in vitro.
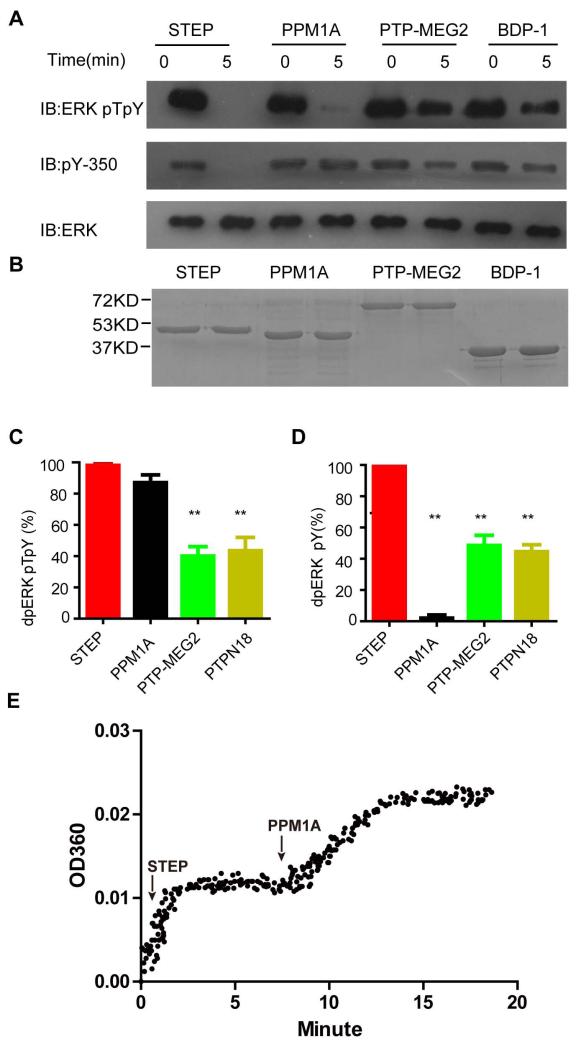
Figure 1. STEP is a tyrosine-specific ERK2 phosphatase in vitro.
A: Dephosphorylation of double-phosphorylated ERK2-pT202pY204 by several phosphatases, including STEP, PPM1A, PTP-MEG2, and BDP1. The reaction was terminated after 5 minutes. Dually phosphorylated ERK2-pT202pY204 (0.5 μM) was incubated with 20 nM purified STEP, PPM1A, PTP-MEG2, or BDP1. The phospho-tyrosine antibody pY-350 was used to monitor the dephosphorylation of ERK2 at pY204, and the phospho-specific ERK1/2-pT202/pY204 antibody was used to monitor the dephosphorylation of ERK2 at either pT202 or pY204.
B: SDS-PAGE of purified protein phosphatases
C-D: Statistical analysis of ERK dephosphorylation at pT202/pY204 (C) or pY204 (D) by different phosphatases within 5 min. All experiments were conducted at least in triplicate. **P<0.01.
E: The progressive curve of ERK2-pT202pY204 dephosphorylation by STEP and PPM1A. 1.2μM ERK2-pT202pY204 was firstly treated by 100nM STEP. Once the reaction reached a plateau, 100nM PPM1A was added.
We first examined ERK dephosphorylation by different phosphatases using a specific antibody that recognises ERK activation-loop phosphorylation (pT202EpY204). Compared to PTP-MEG2 and BDP1, both STEP and PPM1A displayed efficient catalytic activity toward dual-phosphorylated ERK with equimolar phosphatase inputs (Fig 1). To examine whether STEP specifically dephosphorylated pY204 rather than pT202, we next monitored dephosphorylation on residue pY204 using the specific phospho-tyrosine antibody pY350. Although STEP removed most of the phospho-tyrosine on double-phosphorylated ERK, PPM1A showed little effect on pY204 (Fig 1A and D). This result confirmed that STEP hydrolysed pY204, but did not exclude the possibility that STEP dephosphorylated pT202. Thus, we next monitored the time course of ERK2-pT202pY204 dephosphorylation by sequentially adding STEP and PPM1A. Once reaction reached plateau, STEP treatment only lead to one equivalent of inorganic phosphate release, compared to input ERK protein. Subsequent inputting PPM1A resulted in another equivalent of inorganic phosphate release (Fig 1E). The PPM1A was a Ser/Thr specific phosphatse. Therefore, PPM1A treated curve reflected dephosphorylation of pT202, and STEP treated curve corresponded to dephosphorylation of pY204.
Taken together, these results demonstrate that STEP is an efficient ERK phosphatase that selectively recognises pY204 in vitro, whereas PPM1A is an ERK pT202-specific phosphatase.
Kinetic parameters of dephosphorylation of phospho-ERK by STEP
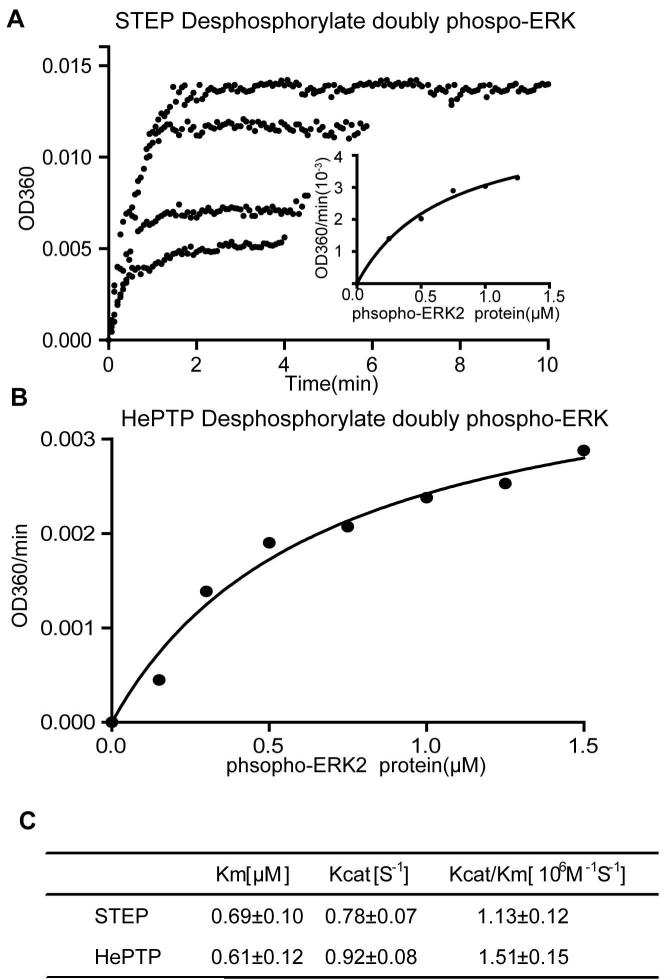
The above results demonstrated that STEP efficiently dephosphorylates double-phosphorylated ERK on pY204 in vitro. However, the kinetic constant of the enzyme is difficult to determine by western blotting. Therefore, to measure the kcat and Km of STEP in ERK dephosphorylation accurately, we utilised a previously established continuous spectrophotometric enzyme-coupled assay to characterise the reaction (Zheng et al. 2012, Zhou et al. 2002). Fig 2A displays the progressive curve of STEP-catalysed ERK dephosphorylation at several different phospho-ERK concentrations by monitoring the increase of absorbance at OD360. All of the initial rates of ERK dephosphorylation by STEP were taken together and fitted to the Michaelis-Menten equation to acquire kcat and Km. The results revealed that ERK-pT202pY204 was a highly efficient substrate of purified STEP in vitro, with a kcat of 0.78 s−1 and Km of 690 nM at pH 7.0 and 25°C (Fig 2A and 2C). For comparison, we also measured the dephosphorylation of ERK at pT202pY204 by HePTP, a previously characterised ERK phosphatase (Fig 2B) (Zhou et al. 2002). The measured kinetic constants for HePTP were similar to those previously published (Fig 2C). In conclusion, STEP is a highly efficient ERK phosphatase in vitro and is comparable to another known ERK phosphatase, HePTP.
Figure 2. Kinetic studies of double-phosphorylated ERK2-pT202/pY204 protein dephosphorylated by STEP.
A: The progressive curve of ERK2-pT202/pY204 dephosphorylation by STEP at different substrate concentrations. The inorganic phosphate released from ERK2-pT202/pY204 dephosphorylated by STEP was measured using a enzyme-coupled assay. The initial rate at different substrate concentrations was analysed by the Michaelis-Menten equation (small figure).
B: HePTP was used as a positive control. The substrate-dependent initial rate of HePTP catalysis toward the phospho-ERK protein was analysed by the Michaelis-Menten equation.
C: The summary of Km and kcat fitted from Fig A and B.
The STEP N-terminal KIM and KIS regions are required for phospho-ERK dephosphorylation
The substrate specificities of PTPs are governed by combinations of active site selectivity and regulatory domains or motifs(Alonso et al. 2004). STEP contains a unique 16-amino acid kinase interaction motif (KIM) at its N-terminal region that has been shown to be required for its interaction with ERK by GST pull-down assays in cells (Munoz et al. 2003, Pulido et al. 1998, Zuniga et al. 1999). KIM is linked to the STEP catalytic domain by the kinase-specificity sequence (KIS), which is involved in differential recognition of MAP kinases and is affected by reducing reagents (Munoz et al. 2003). To further elucidate the contribution of these N-terminal regulatory regions to phospho-ERK dephosphorylation by STEP, we made a series of deletion or truncation mutants in the STEP N-terminus and examined their activity toward pNPP, the double phospho-peptide containing pT202pY204 derived from the ERK activation loop, and dually phosphorylated ERK proteins (Fig 3).
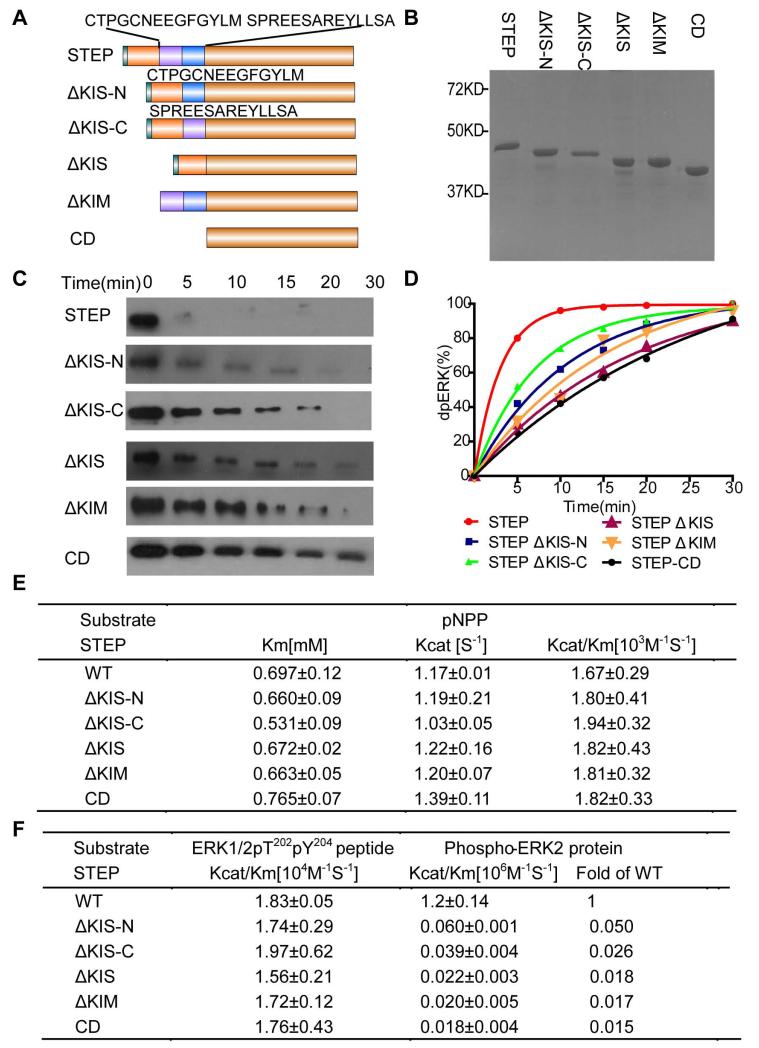
Figure 3. Dephosphorylation of ppERK-pT202/pY204 by a series of N-terminal truncation or deletion mutants of STEP.
A: Domain organisation of the STEP constructs with different truncation or deletion mutations at the N-terminus. KIM: kinase interaction motif; KIS: kinase-specificity sequence; CD: catalytic domain.
B: SDS-PAGE of purified STEP N-terminal truncation or deletion mutants.
C: The time course of ERK2-pT202/pY204 dephosphorylation, as monitored by immunoblotting with a specific phospho-ERK antibody. ERK2-pT202/pY204 (1 μM) was incubated with the STEP derivatives (10 nM) for 0-30 min.
D: The data in panel C were quantified and analysed. The curve represents three separate experiments.
E and F: Kinetic parameters of the wild-type and truncation or deletion mutants of STEP toward three different substrates: pNPP (E), phospho-peptide derived from the ERK2-pT202/pY204 activation loop, and full-length ERK2-pT202/pY204 (F).
The five N-terminal truncation/deletion derivatives of STEP included STEP-CD (deletion of both KIM and KIS), STEP- †KIM (deletion of KIM), STEP-†KIS (deletion of the 28-amino acid KIS), STEP-†KIS-N (deletion of the N-terminal 14 amino acids of KIS), and STEP-†KIS-C (deletion of the C-terminal 14 amino acids of KIS) (Fig 3A). All the STEP truncations and deletions had a good yield in E. coli and were purified to homogeneity (Fig 3B). After purification, we first examined the intrinsic phosphatase activity of these derivatives by measuring the kinetic constants for pNPP and found that the truncations had little effect on the kcat and Km for pNPP, which agreed with the distance of these N-terminal sequences from the active site (Fig 3E).
We next monitored the time course of ERK dephosphorylation by the different derivatives using western blotting (Fig 3C and D). Although little phosphorylated ERK could be detected after 5 minutes in the presence of full-length STEP, ERK phosphorylation was still detected at 15 minutes in the presence of STEP-CD, STEP-†KIM, STEP-†KIS, or STEP-†KIS-C. STEP-†KIS-N also exhibited a slower rate in dephosphorylating ERK compared to wild-type STEP. To accurately determine the effects of each of the N-terminal truncations, we measured the kcat/Km of ERK dephosphorylation by a continuous spectrophotometric enzyme-coupled assay. In comparison to wild-type STEP, all truncations decreased the kcat/Km ratio by 50–60-fold, with the exception of STEP-†KIS-N, which decreased the ratio by only 20-fold (Fig 3F). To determine whether the truncations decreased the activity toward phospho-ERK via recognition of the ERK activation loop sequence, we measured the STEP truncation activity toward the ERK pT202pY204 phospho-peptide. All truncations had kcat/Km ratios for this phospho-ERK peptide that were comparable to the wild-type phosphatase, suggesting that these truncations do not affect STEP activity through a loss of phospho-peptide sequence recognition. Therefore, KIM, the N-terminal portion of KIS, and the C-terminal part of KIS are required for ERK dephosphorylation by STEP. These motifs contribute to dephosphorylation through protein-protein interactions rather than by affecting the intrinsic activity of STEP or its recognition of the ERK phospho-peptide sequence.
Residues of the STEP KIM region responsible for efficient phospho-ERK dephosphorylation
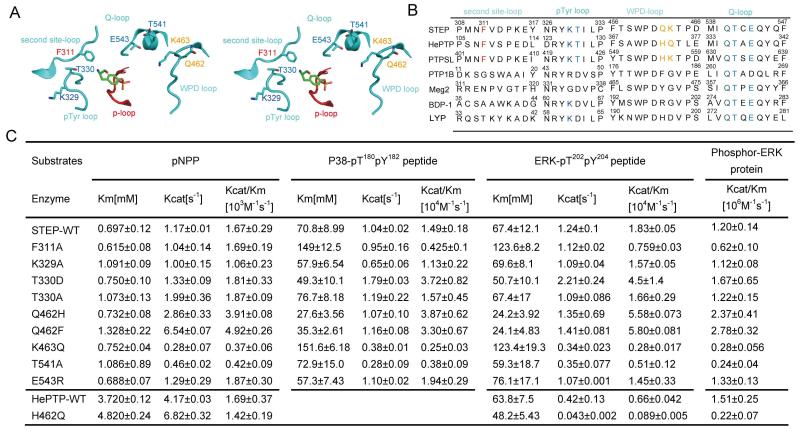
In addition to STEP, at least two known ERK tyrosine phosphatases (HePTP and PTP-SL) and most dual-specificity MAP kinase phosphatases have a KIM that mediates their interactions with ERK(Francis et al. 2011a) (Zhou et al. 2002). Biochemical and structural experiments have revealed that two conserved basic residues followed by the hydrophobic ΦA-X-ΦB motif mediate ERK-phosphatase interactions through STEP binding to the CD site and a hydrophobic groove located on the ERK surface, respectively (Fig 4A) (Liu et al. 2006, Piserchio et al. 2012b, Huang et al. 2004, Zuniga et al. 1999). Based on our previous crystallographic work on the ERK-MKP3 interaction, we also generated a structural model of ERK in complex with STEP-KIM to facilitate our mutagenesis design (Fig 4C, methods in supplemental materials).
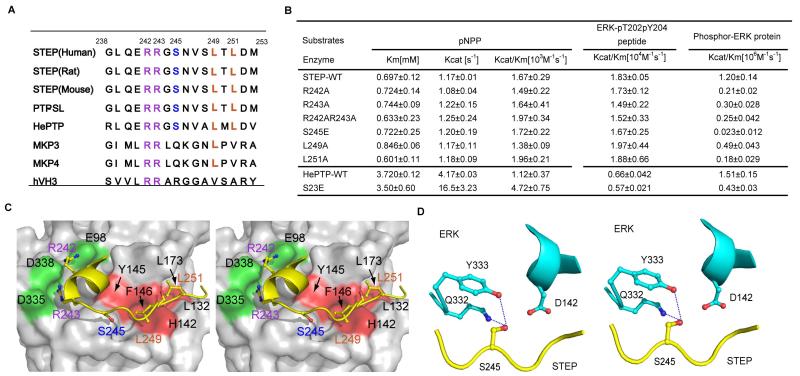
Figure 4. STEP KIM residues necessary for efficient phospho-ERK dephosphorylation.
A: Sequence alignment of KIMs from different phosphatases, including STEP from different species (human, rat, and mouse), PTP-SL, HePTP, MKP3, MKP4, and hVH3. Important interacting residues are highlighted with colour: the conserved hydrophobic residues are yellowish brown, the conserved polar residue S is blue, and the conserved basic residues are purple.
B: Kinetic parameters of wild-type and STEP /HePTP KIM mutants toward three different substrates, pNpp, phospho-ERK peptide, and full-length phospho-ERK protein.
C: The structural model of STEP-KIM binding to ERK, as derived from the previously solved structure of ERK complexed with the MKP3 KIM (PDB 2FYS). The hydrophobic patch of ERK2 (consisting of L132, L173, H142, F146 and Y145) interacting with L249 and L251 of STEP is marked in red. The acidic patch of D335 and D338 (termed CD) and the nearby residues, which are interacting with R242 and R243, are marked in purple. The peptide is coloured by the atom type (carbon, yellow; oxygen, red; nitrogen, blue).
D. Stereo-view of STEP S245 and its surrounding ERK residues. The H-bond is depicted as blue dashed lines. STEP is coloured in yellow, and ERK is coloured in cyan.
To gain insight into how KIM mediates the dephosphorylation of ERK by STEP, we first mutated the conserved basic residue R242 or R243 and the hydrophobic residue L249 or L251 and monitored the effects of these mutants on STEP catalysis. Similar to the STEP-†KIM deletion, these mutations did not affect STEP activity toward pNPP or the phospho-peptide derived from the ERK activation loop (Fig 4B). However, the mutation of either R242A or R243A decreased the kcat/Km ratio of the reaction toward the phospho-ERK protein by 4- or 6-fold, respectively (Fig 4B). These results suggest that these mutations mainly impaired the binding of STEP to ERK.
We next examined the effects of mutations in the conserved hydrophobic ΦA-X-ΦB motif of STEP. Our structural model predicted that STEP L249 sits in a pocket defined by H142, Y145 and F146, of ERK, whereas STEP L251 is located in the hydrophobic pocket defined by ERK L132 and L173 (Fig 4C). Mutation of L249A or L251A decreased the kcat/Km for phospho-ERK by 2.5-fold or 7-fold, respectively (Fig 4B). Thus, we conclude that both conserved hydrophobic residues in the ΦA-X-ΦB motif and the arginine located in KIM are important for efficient ERK dephosphorylation by STEP.
S245, located in the STEP KIM, is an important regulatory site in the dephosphorylation of phospho-ERK by STEP
It is worth noting that STEP activity is downregulated by the phosphorylation of Ser245 in KIM, which is mediated by the activation of D1 dopamine receptor stimulated by psychostimulant drugs (Valjent et al. 2005, Paul et al. 2000). Conversely, NMDA receptor activation leads to STEP dephosphorylation at Ser245 by calcineurin, activating STEP (Paul et al. 2003, Poddar et al. 2010). Therefore, S245 is an important regulatory site of STEP.
To determine whether phosphorylation of S245 directly regulates STEP activity toward phospho-ERK, we generated an S245E STEP phosphorylation mimic mutation. This mutation did not affect the intrinsic phosphatase activity of STEP or its activity toward phospho-ERK peptide; however, it decreased the kcat/Km ratio for the phospho-ERK protein 50-fold (Fig 4B). The effect of the S245E mutation was more pronounced than any single point mutation tested in KIM and was comparable to the effect of the KIM deletions (Fig 3C). In a previous study, the corresponding S245 phosphorylation mimic mutant of HePTP (S23D) exhibited little difference in ERK dephosphorylation compared to the wild-type HePTP (Huang et al. 2004). Because the HePTP S23D mutation is not directly comparable to the STEP S245E mutation, due to the shorter side chain of Asp compared to Glu, we also constructed the HePTP S23E mutant. The HePTP S23E mutation decreased the activity of STEP toward phospho-ERK three-fold, which was much less than the effect of the STEP S245E mutation (Fig 4B).
The drastic change in ERK dephosphorylation by the STEP S245E mutant could be explained by our structural model in which the STEP S245 side chain makes a hydrogen bond with the side chain of ERK Y333 or Q332 and is close to the negatively charged residue D142 (Fig 4D). The S245E mutant or the phosphorylation of S245 may disrupt important hydrogen bonds and generate electrostatic repulsion for D142, hindering the entire STEP KIM region from binding to ERK.
The amino acid sequence surrounding the central phospho-Tyr is recognised by STEP
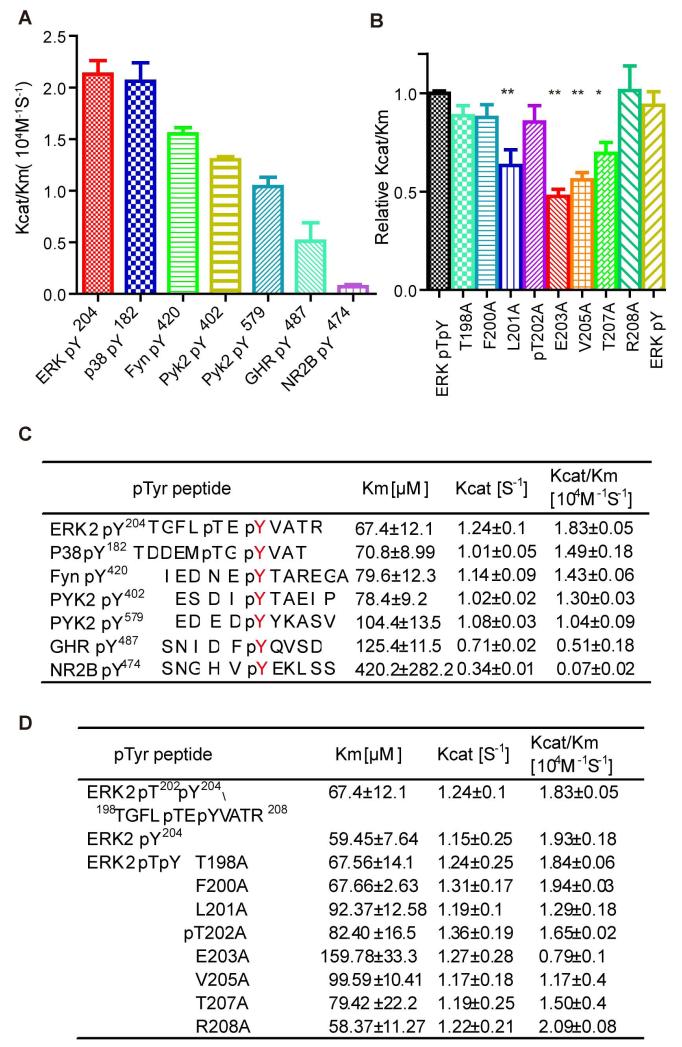
The active site configuration also contributes substantially to the substrate specificity of PTPs. In many cases, the active site of classic PTPs accommodates phospho-tyrosine (pY) and also harbours key residues to recognise the amino acids surrounding pY(Salmeen et al. 2000, Barr et al. 2009, Yu et al. 2011). Several tyrosine phosphatases display a kcat/Km for their target phospho-peptide that is orders of magnitude higher than kcat/Km for pY alone. In particular, Lyp, a phosphatase that plays important roles in the immune response, has selectivity for the amino acid sequence surrounding the central phospho-tyrosine, as determined through the examination of the Lyp activity toward an “inverse alanine-scanning” combinatory library (Yu et al. 2011). Therefore, we next probed the substrate specificity of STEP using a series synthesised phospho-peptides.
In addition to ERK, the NMDA channel subunit NR2B, growth hormone receptor (GHR), and several kinases, including PYK, FYN, and p38, are regulated by STEP and are potential STEP substrates (Baum et al. 2010). We generated the corresponding phospho-peptides derived from these proteins and measured the kinetic parameters for STEP catalysis of their dephosphorylation (Fig 5A and C). The peptide derived from phospho-ERK was the best STEP substrate, followed by the peptides derived from p38 and PYK; conversely, the peptide derived from NR2B was a relatively poor substrate. The kcat/Km of STEP toward the NR2B phospho-peptide was no better than toward pNPP, indicating that other regions of NR2B in addition to the phosphorylation site may contribute to STEP recognition.
Figure 5. Kinetics of the STEP catalysed dephosphorylation of phospho-peptides derived from known STEP substrates and an Ala-scanning peptide library of pp-ERK2-pT202pY204.
A: Statistical analysis and bar graph of the kcat/Km ratios of the STEP-catalysed dephosphorylation of phospho-peptides derived from known STEP substrates.
B: Statistical analysis and bar graph of STEP dephosphorylating the Ala-scanning phospho-peptide library derived from the pp-ERK2-pT202pY204 peptide. (**represents P<0.01 compared to the non-mutated ERKpT202/pY204 phospho-peptide. The data are the average of at least three independent measurements.)
C and D: kcat, Km, and kcat/Km of STEP dephosphorylation of various phospho-peptides.
In addition to NR2B and GHR, all other phospho-peptides tested had a kcat/Km over 104 s−1 M−1, approximately 10-fold better than pNPP. All these sequences had a common acidic or polar residue at the pY-2 position or a small residue at the pY+1 or pY+2 position. To study the contribution of each individual side chain on either side of the central pY, we examined an alanine-scanning ERK-pY204 peptide library in which each amino acid surrounding the central pY was substituted with alanine (Fig 5B and D). The largest effects of alanine scanning were observed at pY-1 (E203) and pY+1 (V205); each mutation decreased kcat/Km by 2-fold. Mutation of pY-3 (L201) or pY+3 (T207) also decreased kcat/Km by 1.6-fold. Therefore, the positions pY±1 and pY±3 contribute the most to peptide substrate recognition by STEP (Fig 5B and D).
Determinants of phospho-ERK recognition in the STEP active site
As described above, STEP exhibited substrate specificity at the pY-3, pY-1, pY+1, and pY+3 positions. STEP belongs to the classical PTP subfamily, all members of which have a conserved active site of 9 Å in depth and 6 Å in width (Tonks 2013, Wang et al. 2003). The active site of classical PTPs is defined by several surrounding loops, including a WPD loop, a Q loop, a pY-binding loop, and a second-site loop (Fig 6A), which play key roles in defining the specific amino acid sequence surrounding the central phospho-tyrosine for substrates (Salmeen et al. 2000, Barr et al. 2009, Yu et al. 2011). Therefore, we compared the sequences of these loops in several classic tyrosine phosphatases and selected mutations at key positions (Fig 6B) to inspect the contribution of residues inside the STEP active site to STEP substrate selectivity.
Figure 6. Determinants localized in the STEP active site for phospho-ERK dephosphorylation.
A: Stereo-view of STEP residues in the active site. The important loops defining the catalytic centre of STEP are red (P loop) and cyan (WPD loop, pTyr loop, Q-loop and second site-loop). Mutated residues are coloured by the atom type (carbon, cyan; oxygen, red; nitrogen, blue).
B. Sequence alignment of several protein tyrosine phosphatases: STEP, HePTP, PTP-SL, PTP1B, PTP-MEG2, BDP-1 and LYP. C. Kinetic parameters of STEP/HePTP and its active-site mutants toward pNPP, the phospho-p38 peptide, the phospho-ERK peptide and full-length phospho-ERK protein.
In contrast to the dual-specificity phosphatase subfamily, all classic PTPs have a deep binding pocket that is designed to accommodate pY and is defined by a unique pY-binding loop on one side. Several key residues in the pY-binding loop, such as Y46, R47, and D48 of PTP1B and Y60, K61, and D62 of LYP, have been well characterised in terms of peptide substrate or inhibitor recognition (Sun et al. 2003, Yu et al. 2011, Sarmiento et al. 1998, Salmeen et al. 2000). We mutated K329 of STEP to an alanine and measured the activity of the mutant (Fig 6B and Supplemental Fig S1). Although the K329A mutation decreased the activity of STEP toward pNPP and the phospho-peptide derived from ERK weakly, it did not affect the catalytic ability of STEP to dephosphorylate the full-length ERK protein (Fig 6C and Supplemental Fig S1). We next examined T330 of STEP, which is typically an aspartic acid in classic PTPs but is a threonine in ERK tyrosine phosphatases (Fig 6B). Previous studies have shown that the conserved aspartic acid in the pY-binding loop of PTP1B and LYP is a determinant of the phospho-peptide orientation through forming specific H-bonds with the peptide backbone amide; mutation of this aspartic acid to alanine significantly reduces the activity of these tyrosine phosphatases toward phospho-peptide-substrates (Sarmiento et al. 1998). Accordingly, the T330D mutation did not affect STEP activity toward pNPP but did increase its activity toward both ERK and a p38-derived phospho-peptide 2–3-fold. This observation was consistent with previous findings for HePTP (Huang et al. 2004). In contrast, the mutation T330A did not affect STEP activity towards either pNPP or phospho-peptide. All of the above results indicate that the presence of the aspartic acid, more than the other residues examined, is important in STEP binding to phospho-peptide substrates. However, when we tested the T330D and T330A mutants for phospho-ERK activity, a slight 1.3-fold increase of kcat/Km for T330D was observed (Fig 6C). This result suggests that phospho-ERK dephosphorylation by STEP does not require an aspartic acid at the 330 position of STEP. Similar results were also obtained for another ERK phosphatase, HePTP (Huang et al. 2004). Without the conserved Asp to define the mainchain conformation of the peptide, the complexes ERK:STEP or ERK:HePTP may stabilize the conformation of the activation segment of ERK through other, uncharacterized molecular mechanisms.
Residues in the WPD-loop are near the active site and are potential determinants of substrates recognition. Two residues following the WPD motif are different among many classical PTPs. In STEP, these two residues are Q462-K463, whereas the corresponding residues in HePTP and PTP-SL are H237-Q238 and H555-K556 respectively (Fig 6B). STEP Q462H or Q462F mutations, which mimic the counterpart residues in HePTP, PTP-SL or PTP1B, significantly lower the Km for the phospho-ERK-peptide and increase the activity toward the phospho-ERK protein. Consistent with these observations, the HePTP H237Q mutation significantly impairs its activity toward the phospho-ERK protein (Fig 6C and Supplemental Fig S1). However, the STEP K463Q mutation, which mimic the corresponding Q238 residue in HePTP, decrease hte STEP activity for either phospho-ERK peptide or phospho-ERK protein more than 4-fold (Fig 6C and Supplemental Fig S1). Taken together, these results demonstrate that the residues Q462 and K463 in the WPD loop of STEP are important for ERK-STEP interaction. Although the combined contribution of Q462-K463 in STEP toward phospho-ERK may not differ significantly compared to H237-Q238 in HePTP, the conformational variance of these residues in the active site may facilitate the development of specific STEP inhibitors.
The Q-loop harbours a conserved glutamine that coordinates a water molecule for phospho-enzyme hydrolysis (Zhang 2003). In the crystal structure of STEP complexed with phospho-tyrosine, the side chains of T541 and E543 in the Q-loop faced to the active site (Fig 6A). Therefore, we evaluated the mutations of these two residues for their effects on phospho-ERK recognition. The mutation of the conserved E543 to basic, charged arginine had no effect on the activity of STEP, whereas the mutation T541A decreased STEP activity 2-fold toward all substrates (Fig 6C and Supplemental Fig S1). The effect of T541A may have been due to a conformational change of the catalytic Q540 residue.
Lastly, based on the complex structure model, we mutated F311 in the second-site loop(Barr et al. 2009) (Fig 6A). Interestingly, F311A did not affect the STEP intrinsic activity toward pNPP but decreased activity toward both the ERK phospho-peptide and full-length protein by 2-fold (Fig 6C and supplemental Fig S1). It is also worth noting that F311 is conserved in all 3 known ERK tyrosine phosphatases, though its corresponding residues in other PTPs, such as PTP1B, PTP-MEG2, BDP1, and LYP, exhibit significant variety. Therefore, F311 is likely one determinant of STEP active site recognition of peptide substrates and phospho-ERK proteins.
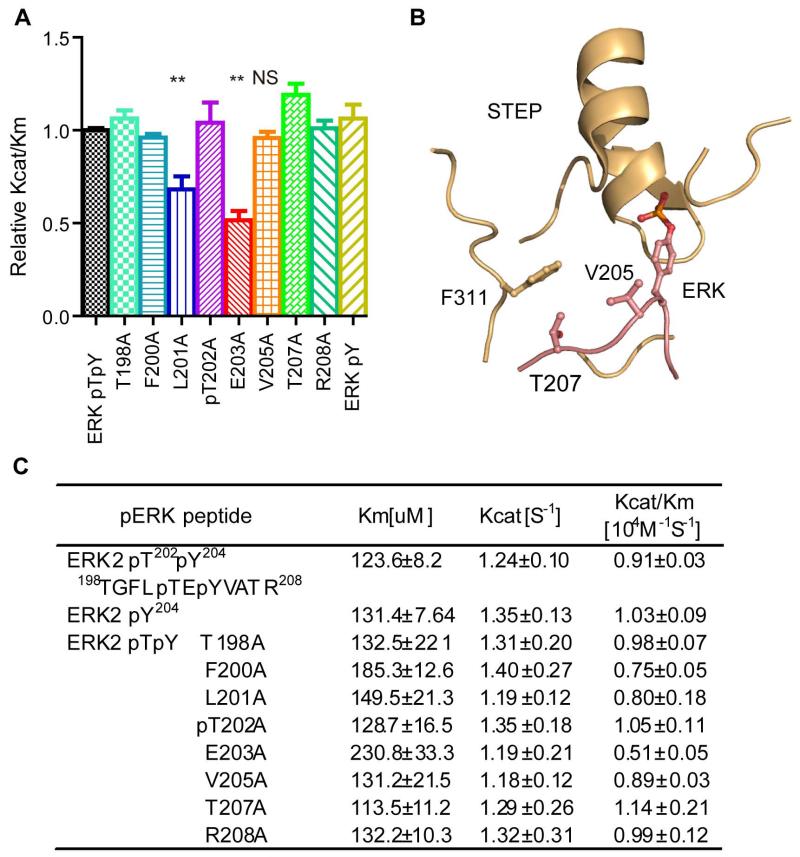
To further delineate the molecular mechanism by which F311 enables STEP to recognise phospho-ERK, we inspected the activity of F311A toward the alanine-scanning library of the ERK-pY204 peptide (Fig 7A and C). Although the L201A and E203A mutations in the ERK peptide decreased STEP F311A activity, the V205A and T207A mutations in ERK had no effect on recognition by STEP F311A, in contrast to the effects of these mutations on wild-type STEP (Fig 7A, C and Fig 5B, D). In our simulated structure model, F311 is situated close to V205 and T207 of ERK, possibly creating strong Van der Waals interactions between these three residues (Fig 7B). Therefore, our results reveal that F311 governs the STEP recognition of phospho-ERK via interaction with V205 and T207 of ERK.
Figure 7. The interaction of F311 of STEP with V205 and T207 of phospho-ERK are important in the STEP-catalysed dephosphorylation of phospho-ERK2.
A: Statistical analysis and bar graph of STEP-F311A dephosphorylating the phospho-ERK peptide and its alanine mutations. (**represents P<0.01 compared to the non-mutated ERKpT202/pY204 phospho-peptide. NS represents non-significance compared to the non-mutated ERKpT202/pT204 phospho-peptide. The data are the average of at least three independent measurements.)
B: The structural model of the STEP–ERK interaction in the region around STEP F311. STEP is coloured in light yellow, and ERK is coloured in pink.
C: Kinetic parameters of STEP F311A for the alanine-scanning phospho-peptide library.
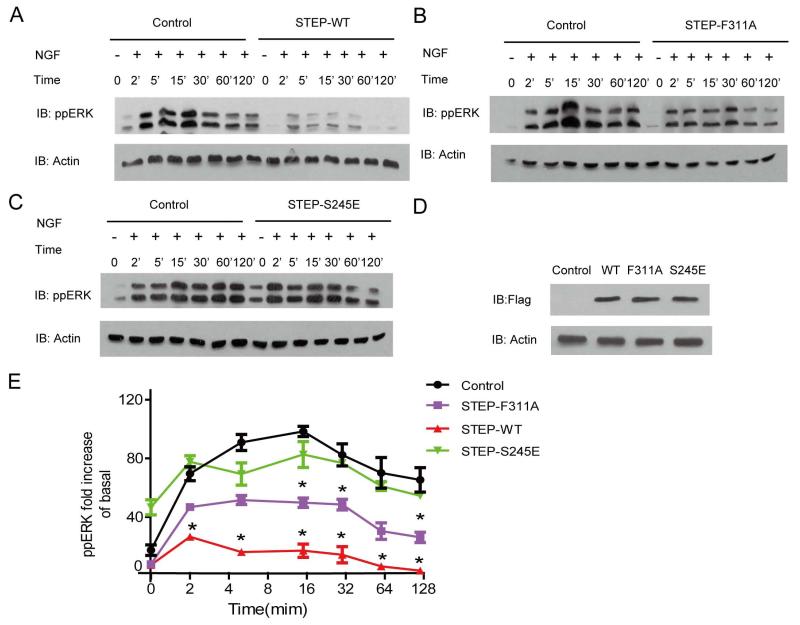
Cellular effects of STEP mutants on NGF induced ERK phosphorylation
To extend the relevance of the biochemical results of the STEP and ERK interaction into a cellular context, we examined the effects of specific STEP mutants on the dynamics of NGF induced ERK phosphorylation in PC12 cells. In control cells, NGF induced prolonged ERK activation which peaked from 5 to 15 minutes. Overexpression of wild type STEP significantly suppressed NGF induced ERK phosphorylation, and the peak ERK phosphorylation occurred at 2 minutes (Fig 8A). With an equal amount of overexpression compared to the wild type protein, the STEP F311A active site mutant reduced the effect of the wild type STEP by approximately half (Fig 8B, D and E). The phosphorylation mimic mutant S245E in the KIM region nearly abolished the effect of STEP on ERK phosphorylation (Fig 8C). The S245E mutant only showed slight effects on ERK phosphorylation from 5 to 15 minutes (Fig 8E). In the unstimulated state, the STEP S245E mutant increased ERK phosphorylation (Fig 8C and E).
Figure 8. Cellular effects of STEP mutants on NGF induced ERK phosphorylation.
A, B and C: Time course of NGF induced ERK phosphorylation with or without STEP or its mutants. PC12 cells were incubated with 50ng/mL NGF for the indicated times. Levels of phospho-ERK1/2 were monitored by phospho-ERK1/2-pT202pY204 antibody.
D: Blots showing expression levels of transfected flag-STEP-WT or mutants. E: Statistical analysis of ERK phosphorylation for the western-blot of A, B and C. * represents P<0.01 compared to the control. Graph representing ERK2 phosphorylation levels were calculated from quantitative analysis of at least three representative experiments.
Discussion
Specific inhibition of STEP activity toward phospho-ERK has great therapeutic potential, as supported by the observation of downregulated ERK activity and increased STEP activity in neuronal degenerative diseases (Baum et al. 2010, Venkitaramani et al. 2011, Venkitaramani et al. 2009). Although the crystal structure of the catalytic domain of STEP has been solved and the importance of the N-terminal region of STEP in the ERK-STEP interaction has been demonstrated by GST pull-down and co-IP experiments, no small molecules that selectively block STEP-ERK interactions have been discovered, partially due to the lack of detailed information on their binding (Munoz et al. 2003, Eswaran et al. 2006). Although a complex crystal structure of STEP bound to phospho-ERK will greatly help in designing STEP inhibitors, alternative methods, such as chemical labelling or enzymologic characterisation, could also substantially contribute to our understanding of the recognition of phospho-ERK by STEP at a quantitative level(Liu et al. 2012b, Kahsai et al. 2011, Zhang et al. 2011). For example, pioneered structural studies of HePTP complexed with inactive or active ERK, and HePTP, PTP-SL or STEP with inactive P38 have been performed with SAXS (small-angle X-ray scattering) and NMR spectrometry, which revealed the extended and dynamic complex formation that occurs during these interactions(Francis et al. 2011b, Francis et al. 2011a, Francis et al. 2013). These methods can provide dynamic structural information that crystallography can not.
In this study, we analysed the dephosphorylation of phospho-ERK by STEP using purified proteins and phospho-peptides. The kinetic constants obtained through these experiments provided detailed information on the contributions of specific residues and alternative structural elements to the dephosphorylation of phospho-ERK by STEP. In the N-terminal KIM of STEP, we quantified the contribution of the conserved hydrophobic residues L249 and L251 to phospho-ERK recognition. Interestingly, the L251A mutation decreased kcat/Km by 7-fold, similar to the 8-fold decrease of the L29A mutant of HePTP, whereas the L249A mutation of STEP only decreased kcat/Km by 2.5-fold, in contrast to the 10-fold decrease of the L27A mutant of HePTP. The 4-fold kinetic constant difference between L249A of STEP and L27A of HePTP suggest that these two phosphatase KIMs bind to ERK in different ways. Consistent with this hypothesis, we previously found that the combined mutation of the two consecutive conserved arginines to alanine in HePTP (R20 and R21) further decreased ERK dephosphorylation by 10-fold compared to either single R-to-A mutation(Huang et al. 2004). In contrast, the analogous combined mutation in STEP (R242 and R243) did not promote a further decrease, a difference that was not detected by previous GST pull-down assays (Munoz et al. 2003, Huang et al. 2004). Furthermore, although the S245E mutation greatly affected phospho-ERK dephosphorylation by STEP, the corresponding S23D or S23E mutations of HePTP had less effects (Huang et al. 2004). Taken together, these results demonstrate that there are differences in the interactions of the conserved STEP KIM with the ERK CD region among different ERK phosphatases, even though most KIM residues are conserved. Therefore, it is conceivable that specific inhibition of phospho-ERK dephosphorylation by STEP could be achieved by targeting the KIM region.
In addition to the regulatory region, our previous studies with other PTP members have demonstrated that the active site of tyrosine phosphatases contributes substantially to substrate recognition (Sun et al. 2003, Yu et al. 2011, Sarmiento et al. 2000). Although the crystal structure of the STEP active site has been solved, the determinants of STEP substrate specificity in the active site have not been determined, mainly due to the lack of biochemical characterisation (Eswaran et al. 2006). In contrast to the Y46-R47-D48 motif in the substrate recognition loop of PTP1B or Y60-K61-D62 in LYP, KIM tyrosine phosphatases including PTP-SL, HePTP and STEP possess the Y-K-T motif in the corresponding positions (Critton et al. 2008). Interestingly, the HePTP T106D mutation was shown to facilitate coordination of the bound phospho-peptide and may facilliate crystallization of the HePTP-phospho-peptide complex(Critton et al. 2008). Similarly, in the crystal structures of PTP1B in complex with the phospho-peptide or peptide-like inhibitors or LYP in complex with the phospho-peptide, the conserved D48 and D62 are required for defining the orientation of the phospho-peptide (Sarmiento et al. 2000). Mutation of D48A in PTP1B significantly impairs phospho-peptide or inhibitor interaction (Sarmiento et al. 2000, Sun et al. 2003). In agreement with this observation the STEP T330D mutant showed enhanced interaction with the phospho-ERK peptide of more than 2-fold. Combined with previous structural studies for HePTP in complex with phospho-peptides, T106 may decrease HePTP binding toward phospho-substrates (Critton et al. 2008); One can hypothesis that the phospho-segment is bound to wile type STEP without a defined conformation, and that the residues surrounding the central pY contribute less to the ERK–STEP interaction. However, when we examined STEP activity toward several phospho-peptides derived from known STEP substrates, the phosphatase displayed approximately 10-fold higher activity toward most of the phospho-peptides compared to the small artificial substrate pNPP, suggesting that residues flanking the central pY also contributed to STEP substrate recognition. To identify the specific residues located in the phospho-peptide sequence that contributed to STEP binding, we employed alanine-scanning mutations at residues surrounding the central pY and measured the STEP activity toward these phospho-peptides. Four specific positions (pY±1 and pY±3) of the phospho-ERK peptide were identified as contributing to STEP recognition. These results were comparable to recent studies of VHR, another ERK phosphatase. The study demonstrated that the positions of (pY±1 and pY-2 and pY-3) were determinants for VHR substrate specificity (Luechapanichkul et al. 2013). It was worth to note that either the mutation of pT202 to either T or to A did not significantly reduce the kcat/Km of STEP toward ERK-pY204 peptides. Therefore, the observed common acidic side chain in the pY-2 position does not contribute to STEP substrate specificity. These results also suggest that STEP does not discriminate between double- and single-phosphorylated ERK as substrates.
We then used site-directed mutagenesis to examine specific residues located in important loops surrounding the STEP active site for phospho-peptide recognition. Unlike the previously characterised PTP1B or LYP, with residues in the substrate recognition loop and Q-loop that contribute substantially to phospho-peptide or peptide mimicking inhibitor recognition (Sarmiento et al. 2000, Sun et al. 2003, Yu et al. 2011), mutations of the corresponding loops in STEP did not affect its activity toward phospho-ERK. However, a specific residue located in the second-site loop, F311, was identified as an important residue and one determinant of the STEP interaction with phospho-ERK via phospho-ERK V205 and T207. Furthermore, the mutation of two residues in the WPD loop of STEP to residues in other PTPs’ significantly affected the activity toward either the phospho-peptide or phospho-ERK protein, suggesting that the conformation varies among different PTPs in this region (Fig 6). Therefore, both the second-site loop and the WPD loop contribute to the substrate specificity of STEP, and specific inhibitors may be developed by targeting the specific residues F311, Q462 and K463 in the active site.
Finally, after we overexpressed the wild type STEP in PC12 cells, we observed that STEP has more profound effects on NGF induced ERK phosphorylation after 2 minutes. Consistent with the biochemical studies, the STEP F311A active site mutant reduced the effect of the STEP wild type by approximately half, whereas the S245E phospho-mimic mutant significantly decreased its effect on ERK phosphorylation.
Therefore, both S245 in the KIM domain and the F311 in the active site contribute to recognition of the phospho-ERK by STEP in cells. .
In summary, we demonstrated that STEP was an efficient, tyrosine-specific ERK phosphatase in vitro. STEP recognised the pY±1 and pY±3 positions of the substrate peptide sequences, with a unique peptide orientation. The interaction between F311 of STEP and V205 and T207 of phospho-ERK, the Q462 and K463 in WPD loop and the specific residues located in KIM were identified as key determinants for phospho-ERK recognition by STEP. In addition, kinetic studies revealed that structural differences in the KIM and ERK interface exist between STEP and HePTP. Therefore, both the KIM-ERK interface and the STEP active site could be targeted to specifically disrupt the STEP-ERK interaction, which has therapeutic potential for neurological disorders.
Supplementary Material
Acknowledgment
This work was supported by grants from the from the National Key Basic Research Program of China (2013CB967700 to Dr. X.Y, 2012CB910402 to Dr. JP.S and Dr. XF.A); National Natural Science Foundation of China (81171062 to Dr. Q.P; 31100580, 31271505 to Dr. JP.S; 31000362 and 31270857 to Dr. X.Y; 81100836 to Dr. T.X), the Foundation of Program for New Century Excellent Talents in University, China (NCET-09-0531 to X.Y.), Foundation for Excellent Young and Middle-Aged Scientists of Shandong Province, China (BS2011SW020 to Dr. JP.S), the Independence Innovation Foundation of Shandong University (2012TS114 to Dr. JP.S) and the grants from the National Institutes of Health (HL095556 and HL108922 to Y.H). The authors declare no conflict of interest.
Abbreviations
- STEP
Striatal-enriched tyrosine phosphatases
- ERK
Extracellular signal-regulated protein kinase
- pNPP
Para-nitrophenyl phosphate
- PTP
Protein tyrosine phosphatase
- KIM
Kinase interaction motif
- KIS
Kinase-specific sequence
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Barr AJ, Ugochukwu E, Lee WH, et al. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum ML, Kurup P, Xu J, Lombroso PJ. A STEP forward in neural function and degeneration. Communicative & integrative biology. 2010;3:419–422. doi: 10.4161/cib.3.5.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sun JP, Liu J, Yu X. [Research progress of several protein tyrosine phosphatases in diabetes] Sheng li xue bao: [Acta physiologica Sinica] 2010;62:179–189. [PubMed] [Google Scholar]
- Critton DA, Tortajada A, Stetson G, Peti W, Page R. Structural basis of substrate recognition by hematopoietic tyrosine phosphatase. Biochemistry. 2008;47:13336–13345. doi: 10.1021/bi801724n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, von Kries JP, Marsden B, et al. Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases. The Biochemical journal. 2006;395:483–491. doi: 10.1042/BJ20051931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick CJ, Lombroso PJ. The Role of Striatal-Enriched Protein Tyrosine Phosphatase (STEP) in Cognition. Frontiers in neuroanatomy. 2011;5:47. doi: 10.3389/fnana.2011.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DM, Kumar GS, Koveal D, Tortajada A, Page R, Peti W. The Differential Regulation of p38alpha by the Neuronal Kinase Interaction Motif Protein Tyrosine Phosphatases, a Detailed Molecular Study. Structure. 2013 doi: 10.1016/j.str.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DM, Rozycki B, Koveal D, Hummer G, Page R, Peti W. Structural basis of p38alpha regulation by hematopoietic tyrosine phosphatase. Nat Chem Biol. 2011a;7:916–924. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DM, Rozycki B, Tortajada A, Hummer G, Peti W, Page R. Resting and active states of the ERK2:HePTP complex. J Am Chem Soc. 2011b;133:17138–17141. doi: 10.1021/ja2075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks WJ, Dilaver G, Noordman YE, Kremer B, Fransen JA. PTPRR protein tyrosine phosphatase isoforms and locomotion of vesicles and mice. Cerebellum. 2009;8:80–88. doi: 10.1007/s12311-008-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Zhou B, Zhang ZY. Molecular determinants of substrate recognition in hematopoietic protein-tyrosine phosphatase. J Biol Chem. 2004;279:52150–52159. doi: 10.1074/jbc.M407820200. [DOI] [PubMed] [Google Scholar]
- Hunter T. Tyrosine phosphorylation: thirty years and counting. Current opinion in cell biology. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahsai AW, Xiao K, Rajagopal S, Ahn S, Shukla AK, Sun J, Oas TG, Lefkowitz RJ. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat Chem Biol. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Gong Z, Pan C, et al. PPM1A functions as an ERK phosphatase. The FEBS journal. 2013 [Google Scholar]
- Liu J, Chen M, Li R, et al. Biochemical and functional studies of lymphoid-specific tyrosine phosphatase (Lyp) variants S201F and R266W. PloS one. 2012a;7:e43631. doi: 10.1371/journal.pone.0043631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012b;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sun JP, Zhou B, Zhang ZY. Structural basis of docking interactions between ERK2 and MAP kinase phosphatase 3. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luechapanichkul R, Chen X, Taha HA, Vyas S, Guan X, Freitas MA, Hadad CM, Pei D. Specificity profiling of dual specificity phosphatase vaccinia VH1-related (VHR) reveals two distinct substrate binding modes. J Biol Chem. 2013;288:6498–6510. doi: 10.1074/jbc.M112.449611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morooka T, Nishida E. Requirement of p38 mitogen-activated protein kinase for neuronal differentiation in PC12 cells. J Biol Chem. 1998;273:24285–24288. doi: 10.1074/jbc.273.38.24285. [DOI] [PubMed] [Google Scholar]
- Munoz JJ, Tarrega C, Blanco-Aparicio C, Pulido R. Differential interaction of the tyrosine phosphatases PTP-SL, STEP and HePTP with the mitogen-activated protein kinases ERK1/2 and p38alpha is determined by a kinase specificity sequence and influenced by reducing agents. The Biochemical journal. 2003;372:193–201. doi: 10.1042/BJ20021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Liu HD, Gong Z, et al. Cadmium is a potent inhibitor of PPM phosphatases and targets the M1 binding site. Sci Rep. 2013;3:2333. doi: 10.1038/srep02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- Paul S, Nairn AC, Wang P, Lombroso PJ. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat Neurosci. 2003;6:34–42. doi: 10.1038/nn989. [DOI] [PubMed] [Google Scholar]
- Paul S, Snyder GL, Yokakura H, Picciotto MR, Nairn AC, Lombroso PJ. The Dopamine/D1 receptor mediates the phosphorylation and inactivation of the protein tyrosine phosphatase STEP via a PKA-dependent pathway. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2000;20:5630–5638. doi: 10.1523/JNEUROSCI.20-15-05630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piserchio A, Francis DM, Koveal D, Dalby KN, Page R, Peti W, Ghose R. Docking interactions of hematopoietic tyrosine phosphatase with MAP kinases ERK2 and p38alpha. Biochemistry. 2012a;51:8047–8049. doi: 10.1021/bi3012725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piserchio A, Francis DM, Koveal D, Dalby KN, Page R, Peti W, Ghose R. Docking Interactions of Hematopoietic Tyrosine Phosphatase with MAP Kinases ERK2 and p38alpha. Biochemistry. 2012b doi: 10.1021/bi3012725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar R, Deb I, Mukherjee S, Paul S. NR2B-NMDA receptor mediated modulation of the tyrosine phosphatase STEP regulates glutamate induced neuronal cell death. Journal of neurochemistry. 2010;115:1350–1362. doi: 10.1111/j.1471-4159.2010.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido R, Zuniga A, Ullrich A. PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. The EMBO journal. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell research. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6:1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y, Lawrence DS, Almo SC, Zhang ZY. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39:8171–8179. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]
- Sarmiento M, Zhao Y, Gordon SJ, Zhang ZY. Molecular basis for substrate specificity of protein-tyrosine phosphatase 1B. J Biol Chem. 1998;273:26368–26374. doi: 10.1074/jbc.273.41.26368. [DOI] [PubMed] [Google Scholar]
- Sharma E, Zhao F, Bult A, Lombroso PJ. Identification of two alternatively spliced transcripts of STEP: a subfamily of brain-enriched protein tyrosine phosphatases. Brain research. Molecular brain research. 1995;32:87–93. doi: 10.1016/0169-328x(95)00066-2. [DOI] [PubMed] [Google Scholar]
- Sun JP, Fedorov AA, Lee SY, Guo XL, Shen K, Lawrence DS, Almo SC, Zhang ZY. Crystal structure of PTP1B complexed with a potent and selective bidentate inhibitor. J Biol Chem. 2003;278:12406–12414. doi: 10.1074/jbc.M212491200. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Pascoli V, Svenningsson P, et al. Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:491–496. doi: 10.1073/pnas.0408305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaramani DV, Moura PJ, Picciotto MR, Lombroso PJ. Striatal-enriched protein tyrosine phosphatase (STEP) knockout mice have enhanced hippocampal memory. The European journal of neuroscience. 2011;33:2288–2298. doi: 10.1111/j.1460-9568.2011.07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaramani DV, Paul S, Zhang Y, et al. Knockout of striatal enriched protein tyrosine phosphatase in mice results in increased ERK1/2 phosphorylation. Synapse. 2009;63:69–81. doi: 10.1002/syn.20608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter SW, Keng YF, Lawrence DS, Zhang ZY. Assessment of protein-tyrosine phosphatase 1B substrate specificity using “inverse alanine scanning”. The Journal of biological chemistry. 2000;275:2265–2268. doi: 10.1074/jbc.275.4.2265. [DOI] [PubMed] [Google Scholar]
- Wang WQ, Sun JP, Zhang ZY. An overview of the protein tyrosine phosphatase superfamily. Curr Top Med Chem. 2003;3:739–748. doi: 10.2174/1568026033452302. [DOI] [PubMed] [Google Scholar]
- Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kurup P, Bartos JA, Patriarchi T, Hell JW, Lombroso PJ. Striatal-enriched protein-tyrosine phosphatase (STEP) regulates Pyk2 kinase activity. The Journal of biological chemistry. 2012;287:20942–20956. doi: 10.1074/jbc.M112.368654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen M, Zhang S, et al. Substrate specificity of lymphoid-specific tyrosine phosphatase (Lyp) and identification of Src kinase-associated protein of 55 kDa homolog (SKAP-HOM) as a Lyp substrate. J Biol Chem. 2011;286:30526–30534. doi: 10.1074/jbc.M111.254722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sun JP, He Y, Guo X, Liu S, Zhou B, Hudmon A, Zhang ZY. Structure, inhibitor, and regulatory mechanism of Lyp, a lymphoid-specific tyrosine phosphatase implicated in autoimmune diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19767–19772. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Venkitaramani DV, Gladding CM, Kurup P, Molnar E, Collingridge GL, Lombroso PJ. The tyrosine phosphatase STEP mediates AMPA receptor endocytosis after metabotropic glutamate receptor stimulation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:10561–10566. doi: 10.1523/JNEUROSCI.2666-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Wu JW, Wang ZX. Mitogen-activated protein kinase (MAPK) phosphatase 3-mediated cross-talk between MAPKs ERK2 and p38alpha. J Biol Chem. 2011;286:16150–16162. doi: 10.1074/jbc.M110.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY. Mechanistic studies on protein tyrosine phosphatases. Prog Nucleic Acid Res Mol Biol. 2003;73:171–220. doi: 10.1016/s0079-6603(03)01006-7. [DOI] [PubMed] [Google Scholar]
- Zheng LS, Zhang YY, Wu JW, Wu Z, Zhang ZY, Wang ZX. A continuous spectrophotometric assay for mitogen-activated protein kinase kinases. Analytical biochemistry. 2012;421:191–197. doi: 10.1016/j.ab.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Zhou B, Wang ZX, Zhao Y, Brautigan DL, Zhang ZY. The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. The Journal of biological chemistry. 2002;277:31818–31825. doi: 10.1074/jbc.M203969200. [DOI] [PubMed] [Google Scholar]
- Zuniga A, Torres J, Ubeda J, Pulido R. Interaction of mitogen-activated protein kinases with the kinase interaction motif of the tyrosine phosphatase PTP-SL provides substrate specificity and retains ERK2 in the cytoplasm. The Journal of biological chemistry. 1999;274:21900–21907. doi: 10.1074/jbc.274.31.21900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.