Abstract
Study Objectives
To identify factors associated with variability in rifampin plasma pharmacokinetics and explore the relationship between rifampin pharmacokinetics and change in efavirenz plasma pharmacokinetics with rifampin coadministration.
Methods
In this randomized, cross-over study, 12 healthy volunteers received either efavirenz 600 mg/day or efavirenz 600 mg with rifampin 600 mg/day for 8 days. After a washout period of at least 2 weeks, subjects crossed over to the alternate 8-day regimen. Samples were obtained for pharmacokinetic assessment on day 8 of each study cycle. Drugs concentrations were determined by a validated HPLC. Pharmacokinetic parameters were calculated using noncompartmental analysis. Multivariate analysis was used to examine factors associated with rifampin pharmacokinetics. Spearman correlation analysis was used to investigate relationship between rifampin pharmacokinetics and change in efavirenz plasma pharmacokinetics with rifampin coadministration.
Measurements and Main Results
Of 11 evaluable subjects, the median interquartile range (IQR) rifampin Cmax, AUC0-24h, and weight-normalized clearance were 8.9 (7.3-13.8) μg/mL, 48.8 (29.6-67.4) μg•hr/mL, and 0.19 (0.11-0.29) L/hr/kg, respectively. SLCO1B1c.388A→G and SLCO1B1c.463C→A polymorphisms jointly had significant effect on rifampin Cmax (R2=0.75). Male sex and SLCO1B1c.463C→A polymorphism together influenced rifampin AUC0-24h (R2=0.52) and weight-normalized clearance (R2=0.65). All four subjects with rifampin Cmax < 8 μg/mL (lower end of the normal range) had c.463CA genotype. Rifampin Cmax and AUC0-24h had no significant relationship with the efavirenz AUC0-24h ratio or weight-normalized clearance ratio in the presence versus absence of rifampin (P>0.05).
Conclusions
Males with the SLCO1B1c.463CA genotype are at increased risk of lower rifampin plasma exposure. However, plasma rifampin concentrations did not correlate with the extent of induction of efavirenz clearance by rifampin during coadministration.
Keywords: Rifampin, Pharmacokinetics, pharmacogenetics, efavirenz, drug interactions
Rifampin is an essential component of short-course chemotherapy for tuberculosis (TB).1 Findings from randomized clinical studies demonstrate that the addition of rifampin to treatment regimens during the induction and the continuation phases of TB treatment reduce rates of treatment failure and relapse.2,3 In vitro studies suggest that rifampin demonstrates concentration-dependent killing of Mycobacterium tuberculosis, with the rifampin area under the concentration-time curve (AUC) to minimum inhibitory concentration (MIC) ratio identified as the best parameter that correlated best with reduction in bacterial counts.4, 5 In the hollow-fiber model of TB, maximal microbial killing was achieved by rifampin daily AUC0-24h/MIC ratio of 24.14 and rifampin prevented resistance to itself at peak concentration (Cmax) to MIC ratio of ≥ 175.4 Clinically, low plasma exposure to the rifamycins has been associated with TB treatment failure and emergence of rifamycin resistance.6-8 The organic anion transporter polypeptide 1B1 (OATP1B1) encoded by the solute carrier organic anion transporter family member 1B1 (SLCO1B1) gene mediates the hepatic uptake and elimination of a range of drugs, including rifampin.9 Data from recent studies show that SLCO1B1 c.463C→A and rs4149032 polymorphisms are associated with low rifampin plasma concentrations.10, 11 While these findings are novel, they are yet to be replicated or confirmed in other studies. Taken together, genetic testing and clinical information may be used to identify patients at risk of low rifampin plasma exposure and could enable individualized dosing designed to optimize treatment outcomes.
Concomitant administration of rifampin with efavirenz-containing antiretroviral therapy in HIV and TB co-infected patients is necessary to reduce mortality rates.12-14 Rifampin is a potent inducer of hepatic cytochrome P450 enzyme activity, and concomitant administration with efavirenz results in reduced efavirenz concentrations in some, but not all patients.15-17 The variable effect of rifampin on efavirenz plasma pharmacokinetics may be due, in part, to inter-individual variations in rifampin plasma concentrations or uptake of rifampin into hepatocytes, but this hypothesis has not been previously evaluated, to the best of our knowledge. Using stored samples and data from subjects in a previous drug-drug interaction study,16 we sought to identify factors associated with variable rifampin pharmacokinetics. In addition, we explored the relationship between variable rifampin pharmacokinetics and change in efavirenz plasma exposure and clearance.
Methods
Study Population and Study Design
Stored plasma and DNA samples from 12 healthy volunteers who participated in a drug-drug interaction study to investigate the effect of rifampin on efavirenz pharmacokinetics,16 were used in the current study. The Institutional Review Board of Miriam Hospital in Providence, Rhode Island reviewed and approved the study. All subjects signed written informed consent, as well as specimen-banking consent for future analysis. All subjects were healthy, HIV-seronegative, self-identified as Caucasian or African-American, and had normal kidney and liver function and no significant medical conditions. Subjects were randomized based on ethnicity to efavirenz 600 mg daily or efavirenz 600 mg plus rifampin 600 mg daily on days 1 – 8. After sampling for efavirenz plasma concentrations on day 8 and at least a two-week washout period, subjects crossed-over to the alternate study regimen, and the procedures were repeated. Blood samples for pharmacokinetic analysis were collected on day 8 of each study period after at least an 8-hour fast. A light standard meal was provided after the 0.5 hour sampling. Blood samples were obtained at 0, 0.5, 1, 1.5, 2, 3, 4, 6, 8, 10, 12, and 24 hours after the final dose on day 8. Heparinized blood was centrifuged at 3000 RPM (1800 g) for 10 minutes and plasma separated and stored in labeled tubes at − 80°C until rifampin assay. The total duration of storage of samples until rifampin assay was 2 to 4 years.
Sample and pharmacokinetic analysis
Rifampin plasma concentrations were determined using HPLC with UV detection as previously described.11 The standard curves for plasma rifampin concentrations covered a range of 0.5 to 50 μg/ml. The within-day precision (coefficient of variation [CV]) of validation quality control samples was 2.4% to 4.6%, and the overall validation precision was 6.3% to 7.1%. The absolute recovery of rifampin from serum was 95.5%. The observed Cmax for each subject were observed by inspection of the serum concentration- time graphs. Calculations of the AUC from time 0 to 24hr (AUC0–24hr), apparent oral clearance (CL/F) clearance, and volume of distribution (Vz/F) were performed using noncompartmental analysis (Phoenix Software; Pharsight Corporation, Mountain View, CA). Efavirenz AUC0-24h ratios (in the presence versus absence of rifampin) and body weight-normalized CL/F ratio used in the secondary analysis was previously reported.16
Enzymes and nuclear receptor genotyping
Stored DNA was genotyped for SLCO1B1c.388G→A (*1b, rs2306283), c.463C→A (*4, rs11045819), c.521T→C (*5, rs4149056), and rs4149032 (intron 2 haplotype tagging SNP; tSNP) using SNP genotyping kits (5′ nuclease assay, and TaqMan assay; Applied biosystems, Foster City, CA) on an ABI PRISM 7300 HT system (Applied biosystems, Foster City, CA) according to manufacturer instructions. Allelic frequencies were verified using published NCBI SNP database values. The genotype frequencies agreed with the predicted Hardy-Weinberg distribution.18
Statistical Analysis
Statistical analyses were performed using Software R for Windows 2.15.1 and SAS 9.3 (SAS Institute Inc, Cary, NC). Multivariate analysis with variable selection by the Smoothly Clipped Absolute Deviation (SCAD) method,19, 20 based on 2-fold cross validation was used to find the effects of patient factors (sex, race, age, body mass index, and SLCO1B1 polymorphisms) on rifampin pharmacokinetic parameters (Cmax, AUC, weight normalized CL/F and Vz/F). The reduced model was re-fitted for post-model selection inference. The SCAD approach has the oracle property, meaning that, in the asymptotic sense, they perform as well as if the analyst had known in advance which coefficients were zero and which were nonzero. Although the estimated coefficients of selected variables from post-model selection inference under current data setting may not be significant, the selected variables together have significant effect on the dependent variables, and their significance is guaranteed with a large sample size as suggested by the oracle property of the SCAD.20 The relationship between rifampin concentrations and efavirenz AUC and clearance (CL/F) ratio (in the presence versus absence of rifampin) was assessed by Spearman rank order correlation test. A P value < 0.05 was considered significant.
Results
Study population
Twelve subjects (6 Caucasian and 6 African American) completed the two arms of the study. Distribution of the selected SLCO1B1 genetic polymorphisms among the 12 volunteers is shown in Table 1. The distribution of the SNPs in the combined population was consistent with the predicted Hardy-Weinberg distribution (P < 0.05).
Table 1. Distribution of SLCO1B1 single nucleotide polymorphisms in 12 healthy volunteers.
| Characteristic | African American | Caucasian | Combined population |
|---|---|---|---|
| SLCO1B1c.388G→A (*1b) | |||
| GG (reference) | 3 (50.0%) | 1 (16.7%) | 4 (33.3%) |
| GA | 2 (33.3%) | 5 (83.3%) | 7 (58.3%) |
| AA | 1 (16.7%) | 0 (0%) | 1 (8.3%) |
| Allelic frequency % | 33.3% | 41.6% | 37.5% |
| P value* | 0.4 | ||
| SLCO1B1 rs4149032 (intron tSNP) | |||
| TT (reference) | 2 (33.3%) | 1 (16.7%) | 3 (25.0%) |
| TC | 2 (33.3%) | 3 (50%) | 5 (41.7%) |
| CC | 2 (33.3%) | 2 (33.3%) | 4 (33.3%) |
| Allelic frequency % | 50.0% | 58.3% | 54.2% |
| P value* | 0.6 | ||
| SLCO1B1 c.463C→A (*4) | |||
| CC (reference) | 4 (66.7%) | 2 (33.3%) | 6 (50.0%) |
| CA | 2 (33.3%) | 4 (66.7%) | 6 (50.0%) |
| AA | 0 (0%) | 0 (0%) | 0 (0.0%) |
| Allelic frequency % | 16.7% | 33.3% | 50.0.0% |
| P value* | 0.2 | ||
| SLCO1B1 c.521T→C (*5) | |||
| TT (reference) | 5 (83.3%) | 2 (33.3%) | 7 (58.3%) |
| TC | 0 (0.0%) | 4 (66.7%) | 4 (33.3%) |
| CC | 1 (16.7%) | 0 (0.0%) | 1 (8.3%) |
| Allelic frequency % | 8.3% | 16.7% | 25.0% |
| P value* | 0.7 |
Chi square testing of Hardy-Weinberg equilibrium
One of the 12 subjects (ID B003) had undetectable efavirenz concentrations and only trace levels of rifampin during coadministration. Because of suspected non-adherence to the study protocol, this subject was excluded from further analysis. The final study population of 11 subjects included 6 Caucasians and 5 African-Americans (6 females, 5 males). Median (IQR) baseline characteristics of the 11 subjects were age 43.0 (38.0 – 48.8) years, body weight 77.3 (60.7 – 84.6) kg, and body mass index (BMI) 25.2 (23.3 – 30.6) kg/m2. The dose of rifampin and efavirenz given to all subjects was 600 mg/day for each drug, median (IQR) 7.8 (7.1 – 9.9) mg/kg.
Rifampin pharmacokinetics and factors associated with pharmacokinetic variability
The median (IQR) rifampin Cmax, AUC0-24h, weight normalized Vz/F, and weight normalized CL/F were 8.9 (7.3 – 13.8) μg/mL, 48.8 (29.6 – 67.4) μg•hr/mL, 0.45 (0.36 – 0.63) L/kg and 0.19 (0.11 – 0.29) L/hr/kg, respectively.
In multivariate analysis, SLCO1B1 c.388A→G and SLCO1B1 c.463C→A polymorphisms jointly had a significant effect on rifampin Cmax (R2=0.75). The model fitting results from post-model selection inference suggested that the subject with the SLCO1B1 c.388AA genotype had substantially higher rifampin Cmax (32.8 μg/mL) compared to subjects with the GA or GG genotypes (9.6 and 12.4 μg/mL, respectively), and subjects with the SLCO1B1 c.463CA genotype had lower median rifampin Cmax (16.8 μg/mL) than those with the CC genotype (19.7 μg/mL). In addition, 4 of the 5 subjects with the SLCO1B1 c.463CA genotype had rifampin Cmax < 8 μg/mL (target threshold level); none of those with the CC genotype had rifampin Cmax < 8 μg/mL. Male sex and the SLCO1B1c.463C→A polymorphism together influenced rifampin AUC0-24h (R2=0.52), weight-normalized CL/F (R2=0.65), and weight-normalized Vz/F (R2=0.58) (Fig. 1). Males had lower median rifampin AUC0-24h compared to females (36.6 vs. 68.1 μg•hr/mL), and subjects with the SLCO1B1 c.463CA genotype had lower median AUC0-24h than those with the c.463Cc genotype (38.3 vs. 66.4 μg•hr/mL). Conversely, males compared to females had higher median weight-normalized CL/F (0.26 vs. 0.16 L/hr/kg) and Vz/F (0.52 vs. 0.41 L/kg), while subjects with the SLCO1B1 c.463CA genotype compared to those with the c.463Cc genotype had higher median weight-normalized CL/F (0.26 vs. 0.16 L/hr/kg) and Vz/F (0.59 vs. 0.34 L/kg).
Fig. 1.
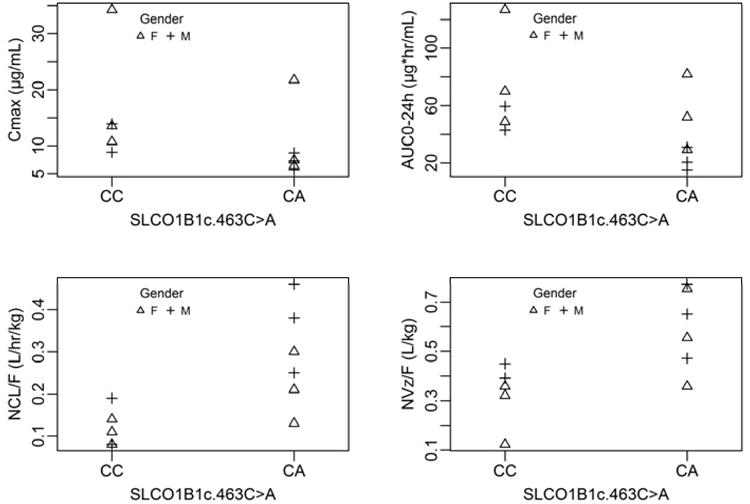
The distribution of rifampin Cmax, AUC0-24h, weight-normalized CL/F, and weight-normalized Vz/F by SLCO1B1 c.463 C→A genotype status and sex. SLCO1B1 c.463 C→A genotype status and sex were joint predictors of rifampin AUC0-24h (R2=0.52), weight-normalized CL/F (R2=0.65), and weight-normalized Vz/F (R2=0.58).
Correlation of rifampin pharmacokinetics and change in efavirenz pharmacokinetics
Individual efavirenz AUC0-24h ratios (in the presence versus absence of rifampin) ranged from 0.55 to 1.18 (median ratio 0.85). Body weight-normalized CL/F ratios ranged from 0.79 to 11.41 (median ratio 1.16). Two of the 11 subjects had CYP2B6 516GG genotype, 7 had GT genotype and 2had TT genotype.,. As previously reported, the CYP2B6 516GT genotype status had no significant relationship with AUC0-24h ratios or body weight-normalized CL/F ratios (in the presence versus absence of rifampin).16 We found no significant relationship between efavirenz AUC0-24h ratio in the presence versus absence of rifampin and rifampin Cmax (correlation coefficient (r) = 0.01, P = 0.99) or rifampin AUC0-24h (r = 0.20, P = 0.56). Similarly, we found no significant relationship between weight-normalized efavirenz clearance ratio in the presence versus absence of rifampin and rifampin Cmax (r = 0.17, P = 0.61) or AUC0-24h (r = -0.07, P = 0.84).
Discussion
This study investigated demographic and genetic factors associated with interindividual variability in rifampin pharmacokinetics in healthy volunteers who received rifampin 600 mg daily coadministered with efavirenz in a drug interaction study. In multivariate analysis, the SLCO1B1c.388A→G and SLCO1B1 c.463C→A SNPs jointly influenced Cmax, while male sex and the SLCO1B1 c.463C→A SNP were jointly associated with rifampin clearance and plasma exposure. Subjects with the SLCO1B1 c.463CA genotype had higher median weight-adjusted clearance of rifampin and lower median Cmax compared to those with CC genotype. All four subjects with rifampin Cmax < 8 μg/mL (lower end of normal range) had the SLCO1B1 c.463CA genotype. These findings substantiate the previous report that the SLCO1B1 c.463C→A polymorphism is associated with lower rifampin Cmax.11
The uptake of rifampin from blood into hepatocytes is a key step in hepatobiliary elimination of the drug. Thus, variation in uptake transporter activity might be expected to influence rifampin pharmacokinetics and treatment responsiveness. Rifampin undergoes extensive hepatic deacetylation to form the 25-deacetylated metabolite.21, 22 Uptake of rifampin from the blood into hepatocytes is mediated by the organic transporting polypeptide 1B1 (OATP1B1) encoded by the SLCO1B1 gene.23, 24 The mechanism for the increased rifampin clearance and lower plasma exposure in subjects with the SLCO1B1 c.463CA genotype could be due to gain in transporter activity associated with the SLCO1B1 c.463C→A (Pro155Thr) SNP, as previously demonstrated for lopinavir clearance.25 The high rifampin plasma concentrations observed for the subject homozygous for the SLCO1B1 c.388A→G SNP in the current study is consistent with reduction in rifampin transport activity by the expression of OATP 1B1 variant in vitro.24 The SLCO1B1 c.388A→G (Asn130Asp) SNP was associated with marked reduction in uptake of rifampin into HeLa cells.24 However, contrary to our findings, a larger study that included TB patients and healthy volunteers found no difference in rifampin exposure by SLCO1B1 c.388A→G genotype status.11 The reason(s) for the discrepant findings are not clear, but could be due to the effect of yet unknown SNPs in the different study populations or drug-drug or drug-gene interactions, since the above-mentioned study11 included patients with active TB on multiple drug therapy. Also, we had just a single subject who was homozygous for the SLCO1B1 c.388A→G variant in our study, and the results could have been due to chance. SLCO1B1 c.521T→C (Val174Ala) SNPs have been reported to decrease rifampin cellular uptake into HeLa cells by decreasing the transporting activity of OATP1B1,24 but we did not observe any differences in rifampin pharmacokinetics in vivo by this polymorphism in our small study population. However, our findings are consistent with those of a previous study that found no significant difference in rifampin exposure or clearance with the SLCO1B1 c.521T→C polymorphism.11
Rifampin is known to have variable drug-drug interactions with efavirenz, a substrate of CYP2B6. As a CYP2B6 enzyme inducer, rifampin would be expected to enhance efavirenz clearance and reduce plasma levels. However, some individuals have a paradoxical elevation of efavirenz concentration with rifampin coadministration.15, 17, 26 The paradoxical effect of rifampin coadministration on efavirenz pharmacokinetics has been associated in part with CYP2B6 561G→T polymorphisms,26-28 but it is unclear if lower rifampin plasma concentrations lead to less of an induction effect on efavirenz clearance during coadministration. We did not find a significant correlation between rifampin plasma Cmax or AUC0-24hr with the change in efavirenz AUC0-24hr ratio or clearance ratio in the presence versus absence rifampin. Rifampin binds to the nuclear receptor pregnane X receptor (PXR) in cellular cytoplasm to form ligand-nuclear receptor complexes. This initiates a series of events that ultimately upregulate the expression and activity of cytochrome P450 enzymes and the P-glycoprotein (P-gp) drug transporter. 29-31 The lack of a linear relationship between rifampin concentration and magnitude of induction may suggest that either the inducing effect of rifampin is not concentration-dependent or it is maximized at a threshold concentration or dose. In fact, the extent of induction of drug metabolizing enzymes appears to be near maximal at a dose of 300 mg daily. Larger doses do not result in further induction.29 In addition, the induction effect of rifampin may be confounded by the effect of the CYP2B6 genotype, because the slow metabolizing variants are not significantly induced by rifampin.26-28 In our sample of 11 subjects, CYP2B6 516G→T genotype status was not significantly associated with the change in the efavirenz AUC0-24hr ratio or clearance ratio in the presence versus absence rifampin.16
The main limitation of our study was the small number of subjects with known functional genetic variants, which limited our ability to draw any firm conclusions about lack of association between rifampin pharmacokinetics and some of the SLCO1B1 SNPs evaluated. In addition, we examined only four SLCO1B1 polymorphisms and were not able to account for as yet unknown genetic or non-genetic factors in our multivariate analysis. A key strength of our analysis is the use of the SCAD variable selection for the multivariate analysis. The SCAD approach has the oracle property, meaning that, in the asymptotic sense, it performs as well as if the analyst had known in advance which coefficients were zero and which were nonzero. The SCAD approach removes non-informative variables by shrinking their coefficients to zero and efficiently selects the model with important variables only, which are different from the traditional variable-selection methods (predictor by predictor) associated with inefficiency and instability. Thus the estimated coefficients of selected variables from post-model selection inference under current data setting may not be significant. However, the selected variables together have a significant effect on the dependent variables, and their significance is guaranteed with a larger sample size as suggested by the oracle property of the SCAD.
In summary, our findings confirm that the SLCO1B1 c.463C→A polymorphism is a significant determinant of low plasma rifampin Cmax. Because low rifampin Cmax may influence TB treatment success,32-34 larger clinical studies are needed to evaluate the utility of genotyping for the SLCO1B1 c.463C→A polymorphism to identify individuals at risk of low concentrations or poor treatment outcome. The lack of a significant relationship between rifampin plasma concentrations and extent of induction of efavirenz clearance suggests that higher rifampin plasma concentrations from higher dosages of rifampin may not necessarily lead to increased induction of efavirenz clearance, but this hypothesis need to be evaluated in studies evaluating higher doses of rifampin for TB treatment.
Acknowledgments
We thank the study participants, as well as the nurses and staff of at the ACTG Unit at The Miriam Hospital Clinical Research Site (U01 AI069472) and the Immunology Center Research Laboratory for all their valuable assistance in recruitment, evaluation of subjects as well as obtaining, handing and processing of the pharmacokinetic samples. This research was supported by National Institutes of Health grants (AI091448, HD071779) to Awewura Kwara. Hongmei Yang was supported in part by grant R01-HD071779. The University of Florida Infectious Disease Pharmacokinetics Laboratory analyzed the rifampin concentrations. Michael H. Court was supported by grant R01-GM061834 from the National Institute of General Medical Sciences (NIGMS).
Funding source: This research was supported by National Institutes of Health grants (AI091448, HD071779) to Dr. Kwara. Dr. Yang was supported in part by grant R01-HD071779. The University of Florida Infectious Disease Pharmacokinetics Laboratory analyzed the rifampin concentrations. Dr. Court was supported by grant R01-GM061834 from the National Institute of General Medical Sciences (NIGMS).
References
- 1.Blumberg HM, Burman WJ, Chaisson RE, et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. American journal of respiratory and critical care medicine. 2003;4:603–62. doi: 10.1164/rccm.167.4.603. [DOI] [PubMed] [Google Scholar]
- 2.Jindani A, Nunn AJ, Enarson DA. Two 8-month regimens of chemotherapy for treatment of newly diagnosed pulmonary tuberculosis: international multicentre randomised trial. Lancet. 2004;9441:1244–51. doi: 10.1016/S0140-6736(04)17141-9. [DOI] [PubMed] [Google Scholar]
- 3.Okwera A, Whalen C, Byekwaso F, et al. Randomised trial of thiacetazone and rifampicin-containing regimens for pulmonary tuberculosis in HIV-infected Ugandans. The Makerere University-Case Western University Research Collaboration. Lancet. 1994;8933:1323–8. doi: 10.1016/s0140-6736(94)90693-9. [DOI] [PubMed] [Google Scholar]
- 4.Gumbo T, Louie A, Deziel MR, et al. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrobial agents and chemotherapy. 2007;11:3781–8. doi: 10.1128/AAC.01533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayaram R, Gaonkar S, Kaur P, et al. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrobial agents and chemotherapy. 2003;7:2118–24. doi: 10.1128/AAC.47.7.2118-2124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiner M, Benator D, Burman W, et al. Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin Infect Dis. 2005;10:1481–91. doi: 10.1086/429321. [DOI] [PubMed] [Google Scholar]
- 7.Chang KC, Leung CC, Yew WW, Chan SL, Tam CM. Dosing schedules of 6-month regimens and relapse for pulmonary tuberculosis. American journal of respiratory and critical care medicine. 2006;10:1153–8. doi: 10.1164/rccm.200605-637OC. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Munsiff SS, Driver CR, Sackoff J. Relapse and acquired rifampin resistance in HIV-infected patients with tuberculosis treated with rifampin- or rifabutin-based regimens in New York City, 1997-2000. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2005;1:83–91. doi: 10.1086/430377. [DOI] [PubMed] [Google Scholar]
- 9.Giacomini KM, Huang SM, Tweedie DJ, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;3:215–36. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chigutsa E, Visser ME, Swart EC, et al. The SLCO1B1 rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: dosing implications. Antimicrob Agents Chemother. 2011;9:4122–7. doi: 10.1128/AAC.01833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner M, Peloquin C, Burman W, et al. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrobial agents and chemotherapy. 2010;10:4192–200. doi: 10.1128/AAC.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdool Karim SS, Naidoo K, Grobler A, et al. Integration of antiretroviral therapy with tuberculosis treatment. N Engl J Med. 2011;16:1492–501. doi: 10.1056/NEJMoa1014181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanc FX, Sok T, Laureillard D, et al. Earlier versus later start of antiretroviral therapy in HIV-infected adults with tuberculosis. N Engl J Med. 2011;16:1471–81. doi: 10.1056/NEJMoa1013911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havlir DV, Kendall MA, Ive P, et al. Timing of antiretroviral therapy for HIV-1 infection and tuberculosis. N Engl J Med. 2011;16:1482–91. doi: 10.1056/NEJMoa1013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedland G, Khoo S, Jack C, Lalloo U. Administration of efavirenz (600 mg/day) with rifampicin results in highly variable levels but excellent clinical outcomes in patients treated for tuberculosis and HIV. The Journal of antimicrobial chemotherapy. 2006;6:1299–302. doi: 10.1093/jac/dkl399. [DOI] [PubMed] [Google Scholar]
- 16.Kwara A, Tashima KT, Dumond JB, et al. Modest but variable effect of rifampin on steady-state plasma pharmacokinetics of efavirenz in healthy African-American and Caucasian volunteers. Antimicrob Agents Chemother. 2011;7:3527–33. doi: 10.1128/AAC.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Cortes LF, Ruiz-Valderas R, Viciana P, et al. Pharmacokinetic interactions between efavirenz and rifampicin in HIV-infected patients with tuberculosis. Clinical pharmacokinetics. 2002;9:681–90. doi: 10.2165/00003088-200241090-00004. [DOI] [PubMed] [Google Scholar]
- 18.Schaid DJ, Batzler AJ, Jenkins GD, Hildebrandt MA. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. American journal of human genetics. 2006;6:1071–80. doi: 10.1086/510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breheny P, Huang J. Coordinate Descent Algorithms for Nonconvex Penalized Regression, with Applications to Biological Feature Selection. Ann Appl Stat. 2011;1:232–53. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of the American Statistical Association. 2001;456:1348–60. [Google Scholar]
- 21.Jamis-Dow CA, Katki AG, Collins JM, Klecker RW. Rifampin and rifabutin and their metabolism by human liver esterases. Xenobiotica. 1997;10:1015–24. doi: 10.1080/004982597239994. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima A, Fukami T, Kobayashi Y, Watanabe A, Nakajima M, Yokoi T. Human arylacetamide deacetylase is responsible for deacetylation of rifamycins: rifampicin, rifabutin, and rifapentine. Biochemical pharmacology. 2011;11:1747–56. doi: 10.1016/j.bcp.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Vavricka SR, Van Montfoort J, Ha HR, Meier PJ, Fattinger K. Interactions of rifamycin SV and rifampicin with organic anion uptake systems of human liver. Hepatology. 2002;1:164–72. doi: 10.1053/jhep.2002.34133. [DOI] [PubMed] [Google Scholar]
- 24.Tirona RG, Leake BF, Wolkoff AW, Kim RB. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. The Journal of pharmacology and experimental therapeutics. 2003;1:223–8. doi: 10.1124/jpet.102.043026. [DOI] [PubMed] [Google Scholar]
- 25.Lubomirov R, di Iulio J, Fayet A, et al. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenetics and genomics. 2010;4:217–30. doi: 10.1097/FPC.0b013e328336eee4. [DOI] [PubMed] [Google Scholar]
- 26.Kwara A, Lartey M, Sagoe KW, Court MH. Paradoxically elevated efavirenz concentrations in HIV/tuberculosis-coinfected patients with CYP2B6 516TT genotype on rifampin-containing antituberculous therapy. Aids. 2011;3:388–90. doi: 10.1097/QAD.0b013e3283427e05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngaimisi E, Mugusi S, Minzi O, et al. Effect of rifampicin and CYP2B6 genotype on long-term efavirenz autoinduction and plasma exposure in HIV patients with or without tuberculosis. Clinical pharmacology and therapeutics. 2011;3:406–13. doi: 10.1038/clpt.2011.129. [DOI] [PubMed] [Google Scholar]
- 28.McIlleron HM, Schomaker M, Ren Y, et al. Effects of rifampin-based antituberculosis therapy on plasma efavirenz concentrations in children vary by CYP2B6 genotype. Aids. 2013;12:1933–40. doi: 10.1097/qad.0b013e328360dbb4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivisto KT. Pharmacokinetic interactions with rifampicin : clinical relevance. Clinical pharmacokinetics. 2003;9:819–50. doi: 10.2165/00003088-200342090-00003. [DOI] [PubMed] [Google Scholar]
- 30.Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. The Journal of pharmacology and experimental therapeutics. 2006;3:1200–9. doi: 10.1124/jpet.105.098160. [DOI] [PubMed] [Google Scholar]
- 31.Faucette SR, Zhang TC, Moore R, et al. Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. The Journal of pharmacology and experimental therapeutics. 2007;1:72–80. doi: 10.1124/jpet.106.112136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta JB, Shantaveerapa H, Byrd RP, Jr, Morton SE, Fountain F, Roy TM. Utility of rifampin blood levels in the treatment and follow-up of active pulmonary tuberculosis in patients who were slow to respond to routine directly observed therapy. Chest. 2001;5:1520–4. doi: 10.1378/chest.120.5.1520. [DOI] [PubMed] [Google Scholar]
- 33.Mitchison DA. Role of individual drugs in the chemotherapy of tuberculosis. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2000;9:796–806. [PubMed] [Google Scholar]
- 34.Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis. Drugs. 2002;15:2169–83. doi: 10.2165/00003495-200262150-00001. [DOI] [PubMed] [Google Scholar]


