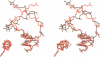Abstract
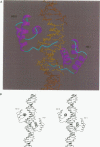
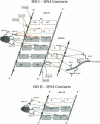
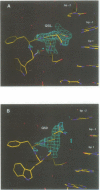
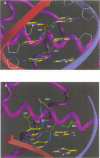
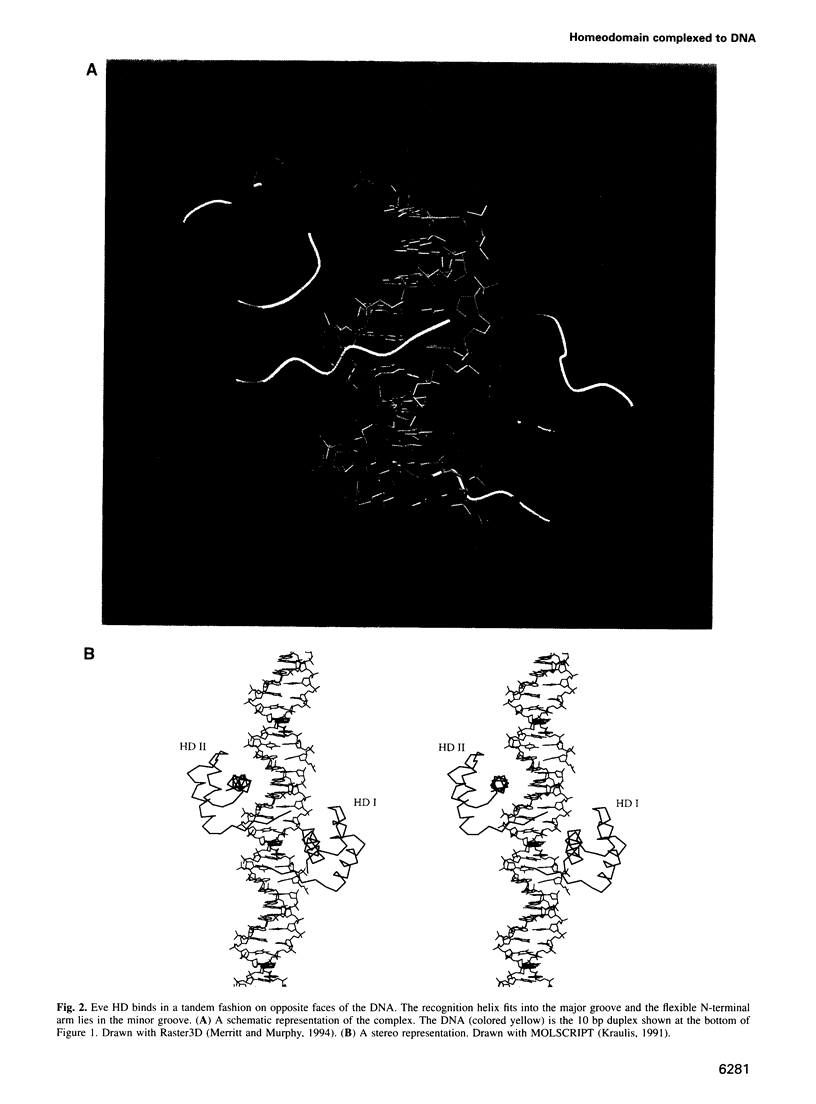
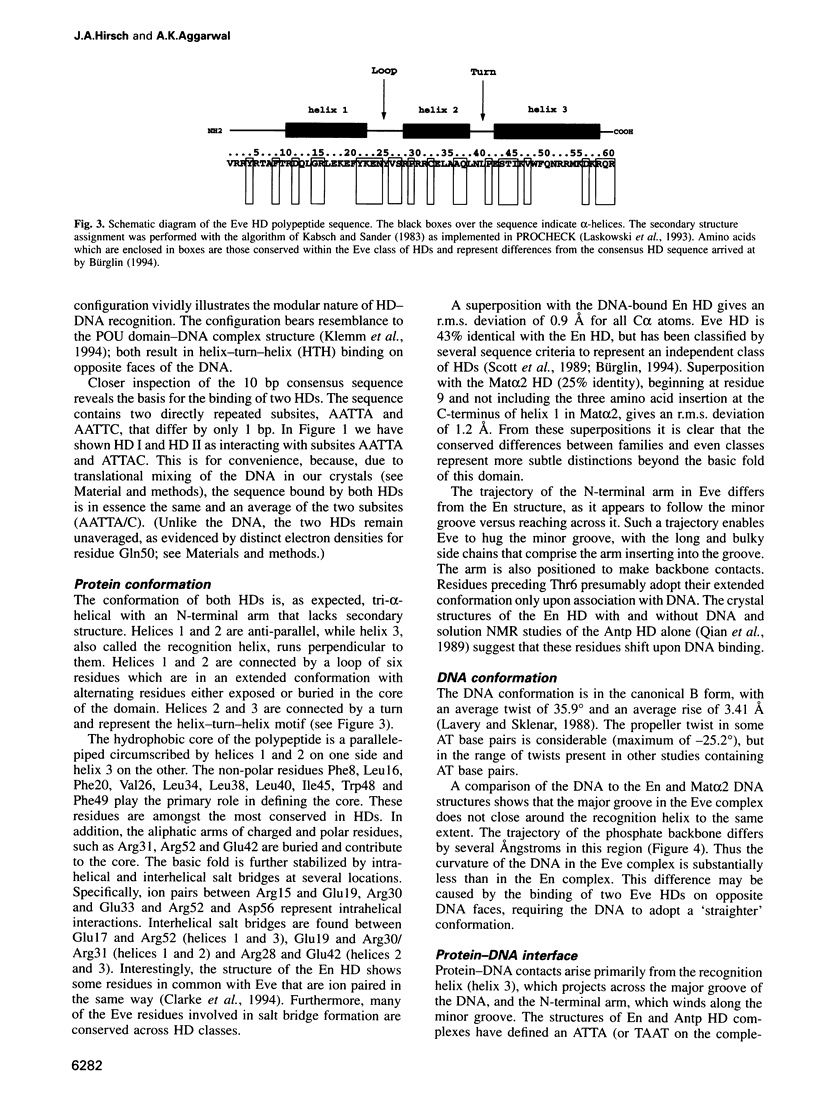
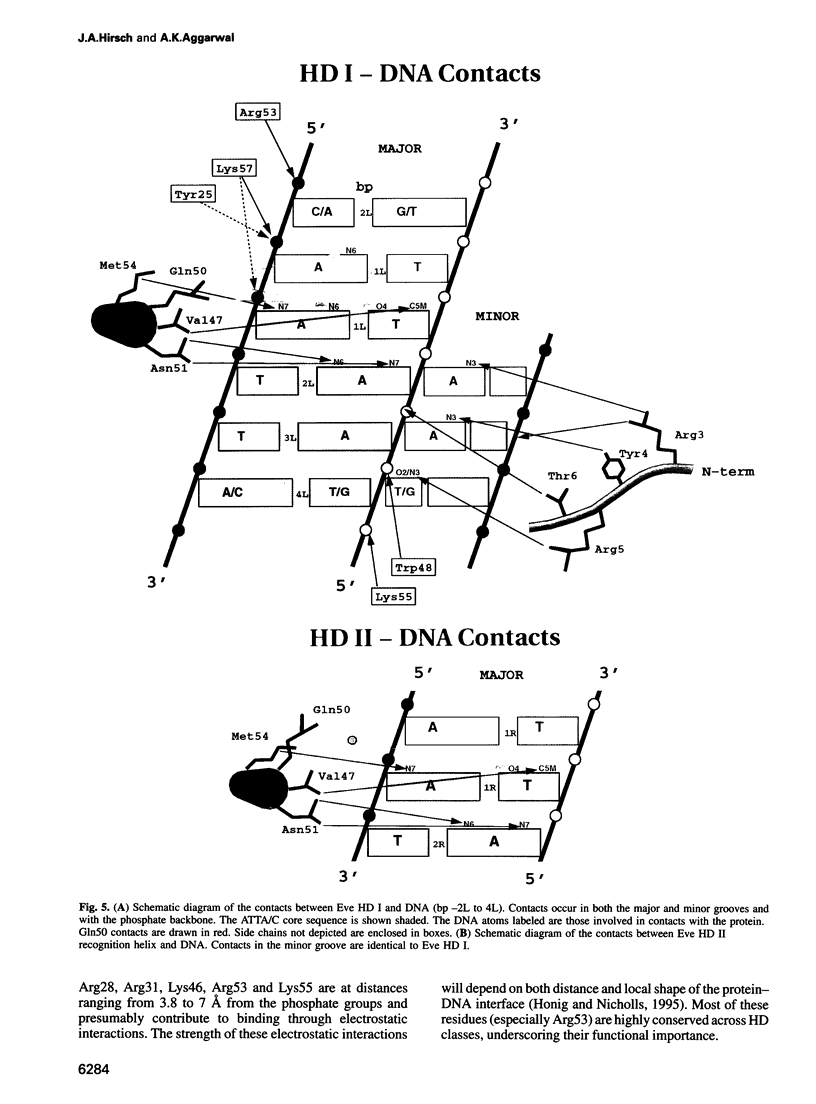
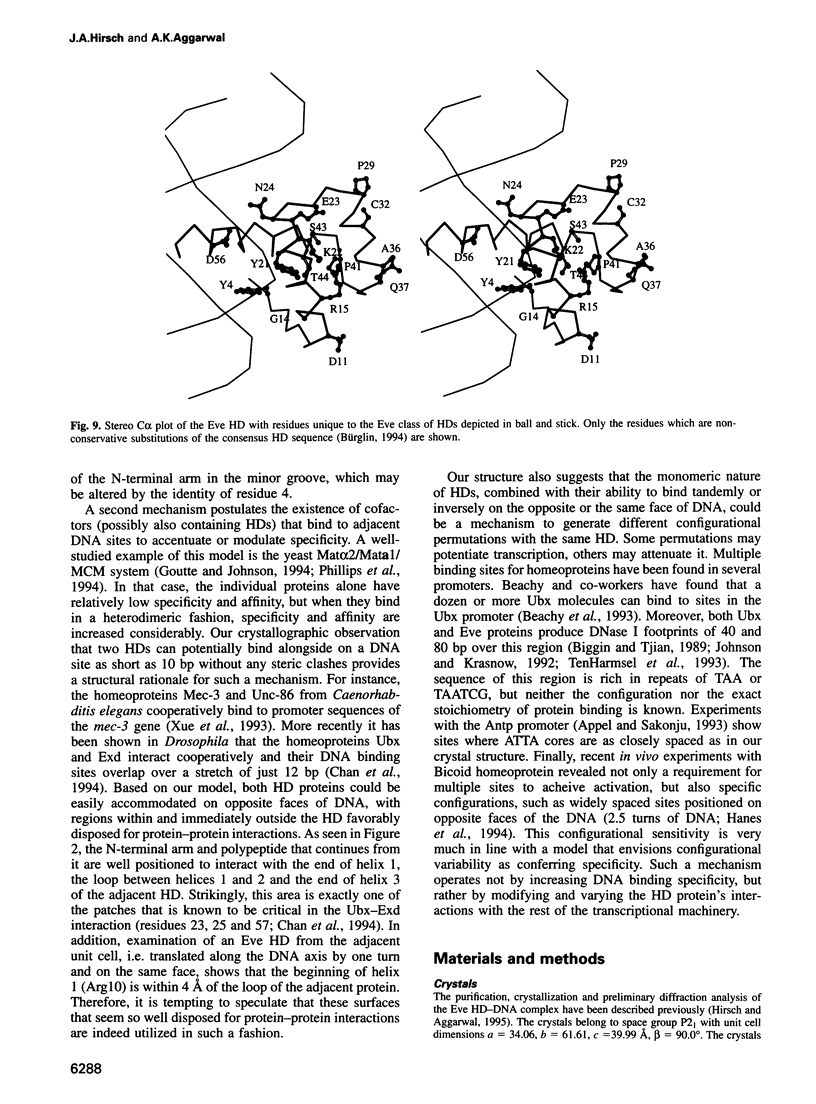
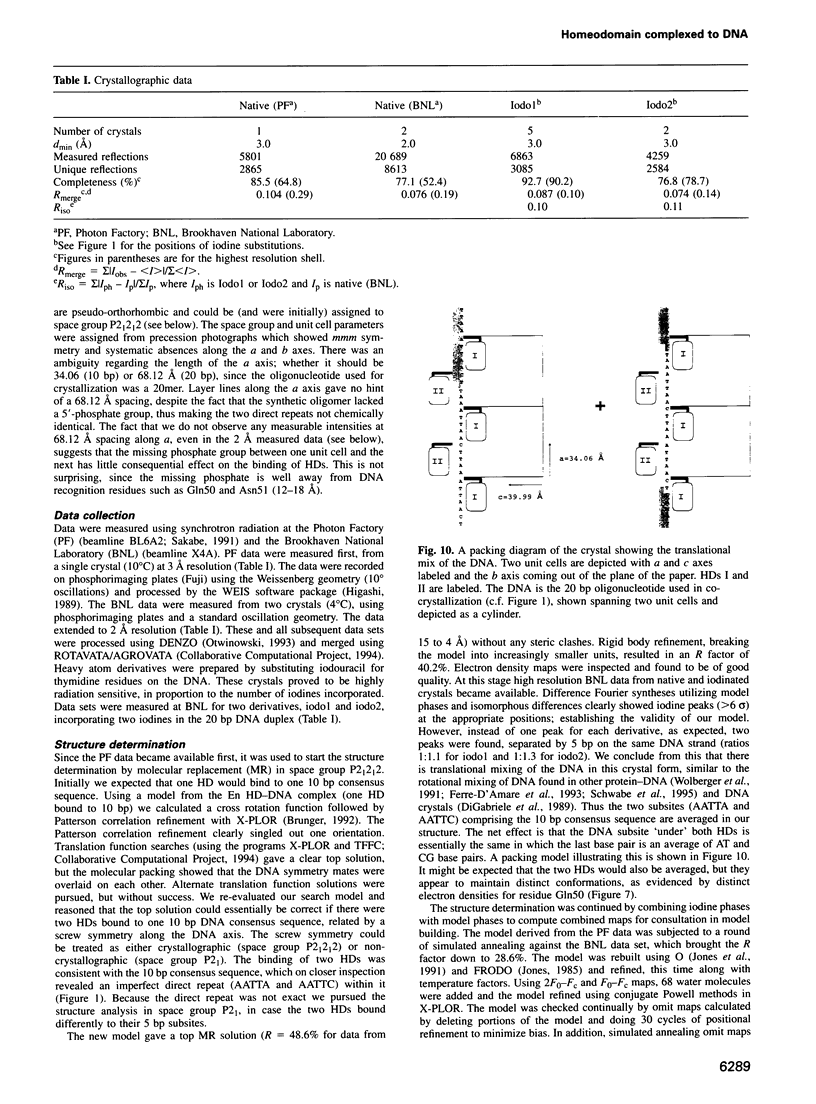
even-skipped is a homeobox gene important in controlling segment patterning in the embryonic fruit fly. Its homeobox encodes a DNA binding domain which binds with similar affinities to two DNA consensus sequences, one AT-rich, the other GC-rich. We describe a crystallographic analysis of the Even-skipped homeodomain complexed to an AT-rich oligonucleotide at 2.0 A resolution. The structure reveals a novel arrangement of two homeodomains bound to one 10 bp DNA sequence in a tandem fashion. This arrangement suggests a mechanism for the homeoproteins' regulatory specificity. In addition, the functionally important residue Gln50 is observed in multiple conformations making direct and water-mediated hydrogen bonds with the DNA bases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ades S. E., Sauer R. T. Differential DNA-binding specificity of the engrailed homeodomain: the role of residue 50. Biochemistry. 1994 Aug 9;33(31):9187–9194. doi: 10.1021/bi00197a022. [DOI] [PubMed] [Google Scholar]
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B., Sakonju S. Cell-type-specific mechanisms of transcriptional repression by the homeotic gene products UBX and ABD-A in Drosophila embryos. EMBO J. 1993 Mar;12(3):1099–1109. doi: 10.1002/j.1460-2075.1993.tb05751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian H., Gruss P. A murine even-skipped homologue, Evx 1, is expressed during early embryogenesis and neurogenesis in a biphasic manner. EMBO J. 1990 Jun;9(6):1839–1852. doi: 10.1002/j.1460-2075.1990.tb08309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy P. A., Varkey J., Young K. E., von Kessler D. P., Sun B. I., Ekker S. C. Cooperative binding of an Ultrabithorax homeodomain protein to nearby and distant DNA sites. Mol Cell Biol. 1993 Nov;13(11):6941–6956. doi: 10.1128/mcb.13.11.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989 Aug 11;58(3):433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Billeter M., Qian Y. Q., Otting G., Müller M., Gehring W., Wüthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993 Dec 20;234(4):1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- Chan S. K., Jaffe L., Capovilla M., Botas J., Mann R. S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994 Aug 26;78(4):603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Kissinger C. R., Desjarlais J., Gilliland G. L., Pabo C. O. Structural studies of the engrailed homeodomain. Protein Sci. 1994 Oct;3(10):1779–1787. doi: 10.1002/pro.5560031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Esposito M., Morelli F., Acampora D., Migliaccio E., Simeone A., Boncinelli E. EVX2, a human homeobox gene homologous to the even-skipped segmentation gene, is localized at the 5' end of HOX4 locus on chromosome 2. Genomics. 1991 May;10(1):43–50. doi: 10.1016/0888-7543(91)90482-t. [DOI] [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988 Sep 23;54(7):1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessain S., Gross C. T., Kuziora M. A., McGinnis W. Antp-type homeodomains have distinct DNA binding specificities that correlate with their different regulatory functions in embryos. EMBO J. 1992 Mar;11(3):991–1002. doi: 10.1002/j.1460-2075.1992.tb05138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Q., Smouse D., Goodman C. S. Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature. 1988 May 26;333(6171):376–378. doi: 10.1038/333376a0. [DOI] [PubMed] [Google Scholar]
- Ekker S. C., Young K. E., von Kessler D. P., Beachy P. A. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991 May;10(5):1179–1186. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker S. C., von Kessler D. P., Beachy P. A. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992 Nov;11(11):4059–4072. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiella A., D'Esposito M., Rambaldi M., Acampora D., Balsofiore S., Stornaiuolo A., Mallamaci A., Migliaccio E., Gulisano M., Simeone A. Isolation and mapping of EVX1, a human homeobox gene homologous to even-skipped, localized at the 5' end of HOX1 locus on chromosome 7. Nucleic Acids Res. 1991 Dec 11;19(23):6541–6545. doi: 10.1093/nar/19.23.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Florence B., Handrow R., Laughon A. DNA-binding specificity of the fushi tarazu homeodomain. Mol Cell Biol. 1991 Jul;11(7):3613–3623. doi: 10.1128/mcb.11.7.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Warrior R., Tugwood J., Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 1988 Dec;2(12B):1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Affolter M., Bürglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Ghosh G., van Duyne G., Ghosh S., Sigler P. B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995 Jan 26;373(6512):303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. Recognition of a DNA operator by a dimer composed of two different homeodomain proteins. EMBO J. 1994 Mar 15;13(6):1434–1442. doi: 10.1002/j.1460-2075.1994.tb06397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J. H., Spolar R. S., Record M. T., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989 Oct 20;209(4):801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Han K., Manley J. L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993 Mar;7(3):491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991 Jan 25;251(4992):426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Riddihough G., Ish-Horowicz D., Brent R. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol. 1994 May;14(5):3364–3375. doi: 10.1128/mcb.14.5.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hirsch J. A., Aggarwal A. K. Purification, crystallization, and preliminary X-ray diffraction analysis of even-skipped homeodomain complexed to DNA. Proteins. 1995 Mar;21(3):268–271. doi: 10.1002/prot.340210311. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hoey T., Warrior R., Manak J., Levine M. DNA-binding activities of the Drosophila melanogaster even-skipped protein are mediated by its homeo domain and influenced by protein context. Mol Cell Biol. 1988 Nov;8(11):4598–4607. doi: 10.1128/mcb.8.11.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995 May 26;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Jiang J., Hoey T., Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 1991 Feb;5(2):265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- Johnson F. B., Krasnow M. A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992 Nov;6(11):2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- Joly J. S., Joly C., Schulte-Merker S., Boulekbache H., Condamine H. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development. 1993 Dec;119(4):1261–1275. doi: 10.1242/dev.119.4.1261. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klemm J. D., Rould M. A., Aurora R., Herr W., Pabo C. O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994 Apr 8;77(1):21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988 Aug;6(1):63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Sigler P. B. The stereochemistry and biochemistry of the trp repressor-operator complex. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):113–126. doi: 10.1016/0167-4781(90)90047-6. [DOI] [PubMed] [Google Scholar]
- Manoukian A. S., Krause H. M. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes Dev. 1992 Sep;6(9):1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- Merritt E. A., Murphy M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Miles A., Miller D. J. Genomes of diploblastic organisms contain homeoboxes: sequence of eveC, an even-skipped homologue from the cnidarian Acropora formosa. Proc Biol Sci. 1992 May 22;248(1322):159–161. doi: 10.1098/rspb.1992.0057. [DOI] [PubMed] [Google Scholar]
- Newman M., Strzelecka T., Dorner L. F., Schildkraut I., Aggarwal A. K. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995 Aug 4;269(5224):656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. H., Ball E. E., Goodman C. S. Changing role of even-skipped during the evolution of insect pattern formation. Nature. 1992 May 28;357(6376):339–342. doi: 10.1038/357339a0. [DOI] [PubMed] [Google Scholar]
- Phillips C. L., Stark M. R., Johnson A. D., Dahlquist F. W. Heterodimerization of the yeast homeodomain transcriptional regulators alpha 2 and a1 induces an interfacial helix in alpha 2. Biochemistry. 1994 Aug 9;33(31):9294–9302. doi: 10.1021/bi00197a033. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Resendez-Perez D., Gehring W. J., Wüthrich K. The des(1-6)antennapedia homeodomain: comparison of the NMR solution structure and the DNA-binding affinity with the intact Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):4091–4095. doi: 10.1073/pnas.91.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J. W., Chapman L., Rhodes D. The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation. Structure. 1995 Feb 15;3(2):201–213. doi: 10.1016/s0969-2126(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- TenHarmsel A., Austin R. J., Savenelli N., Biggin M. D. Cooperative binding at a distance by even-skipped protein correlates with repression and suggests a mechanism of silencing. Mol Cell Biol. 1993 May;13(5):2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J., Gönczy P., Vashishtha M., Harris E., Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989 Nov 3;59(3):553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Xue D., Tu Y., Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993 Sep 3;261(5126):1324–1328. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]