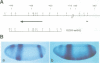
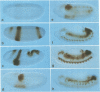
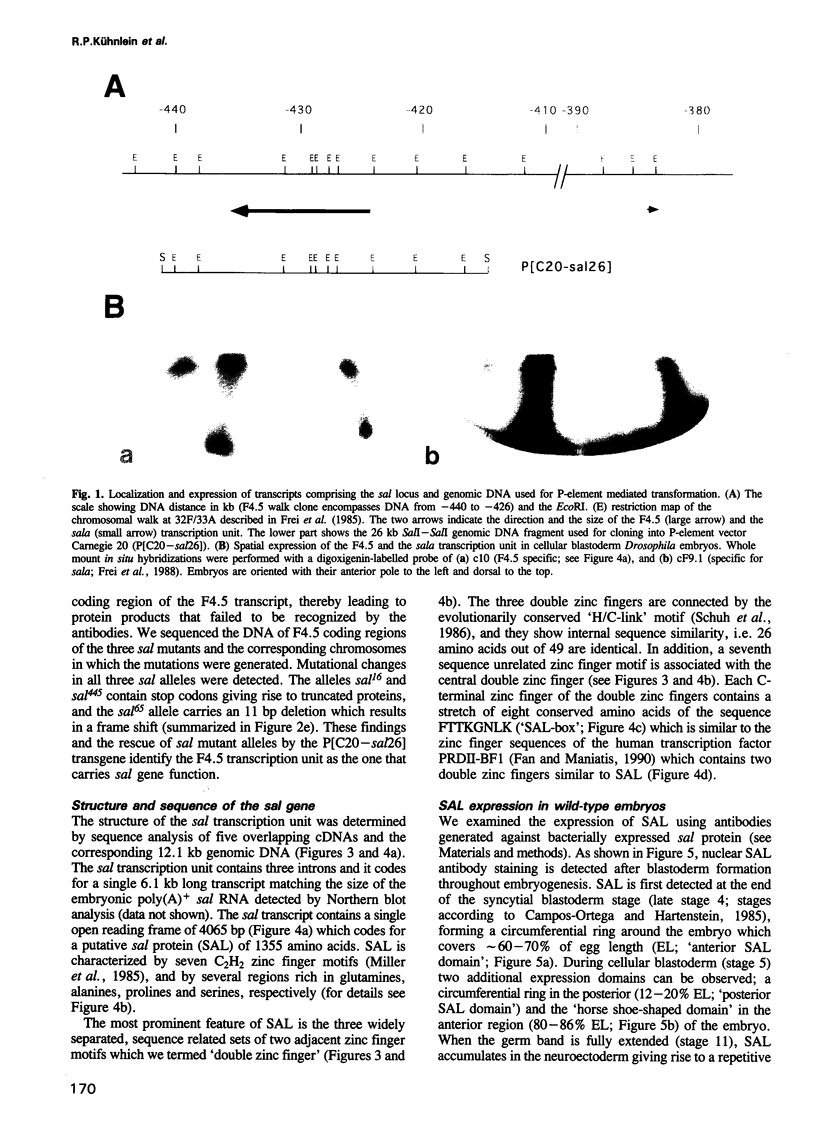
Abstract
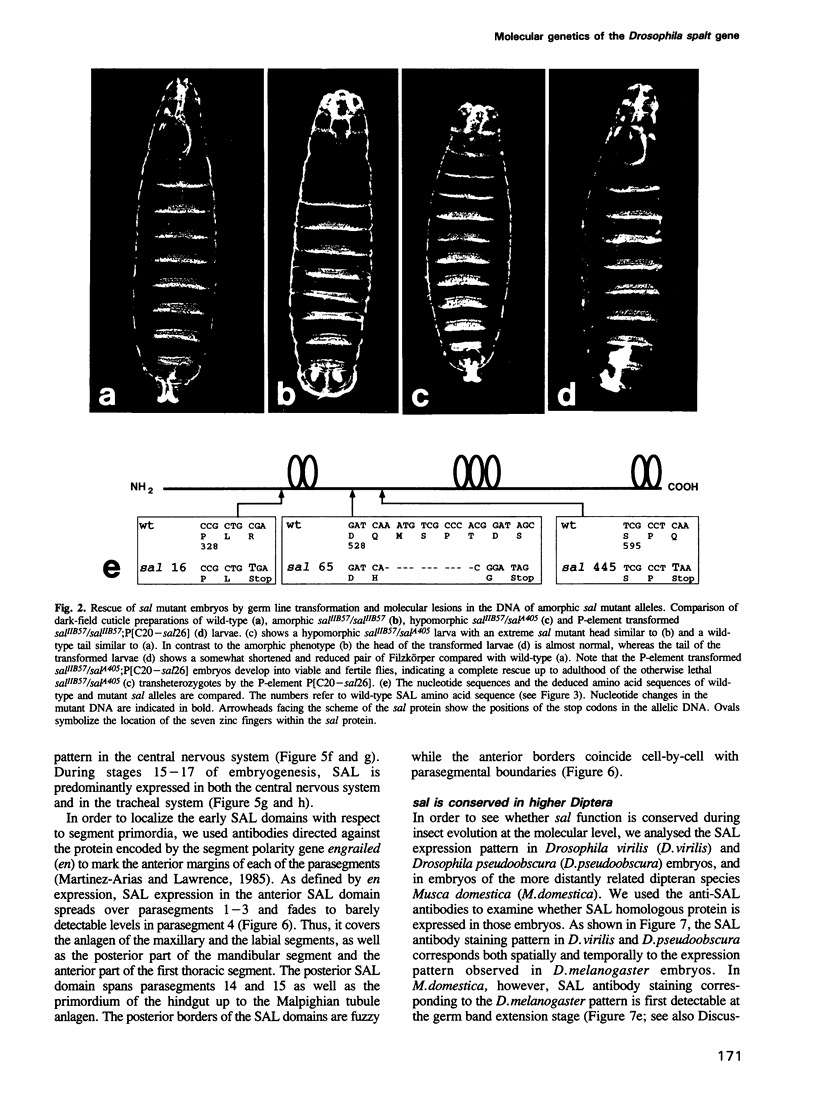
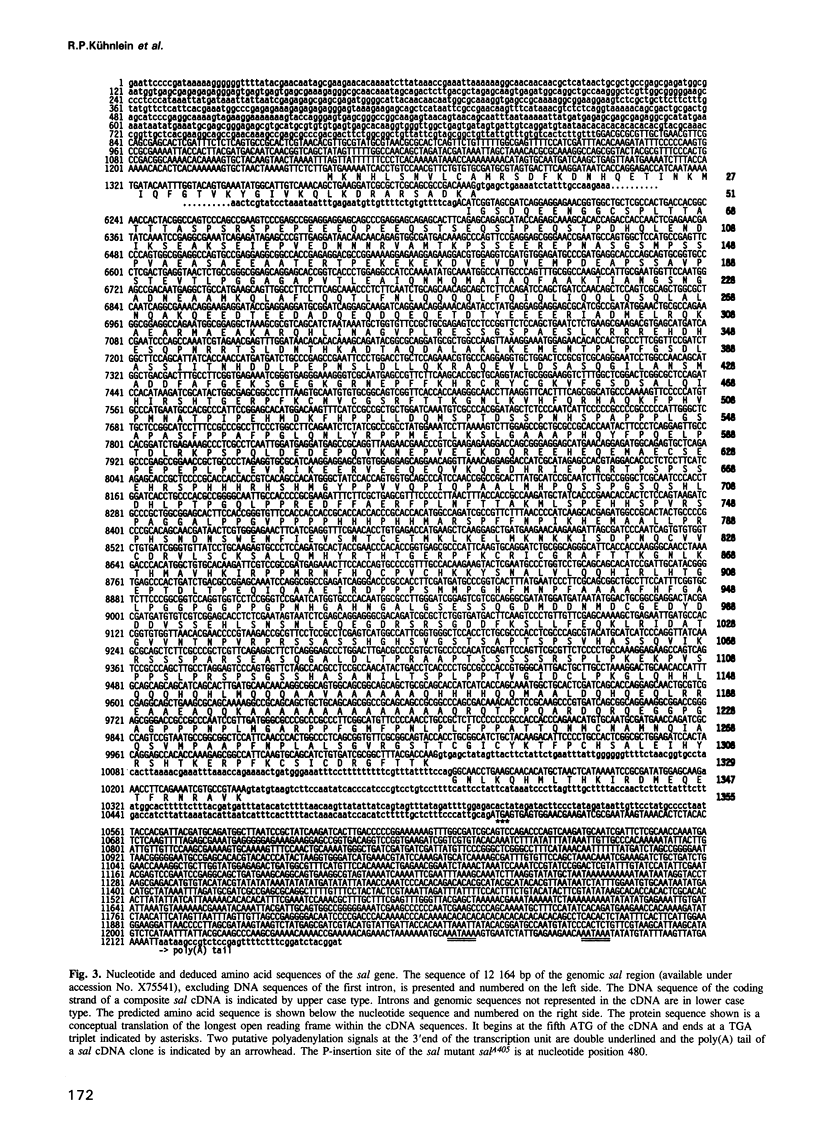
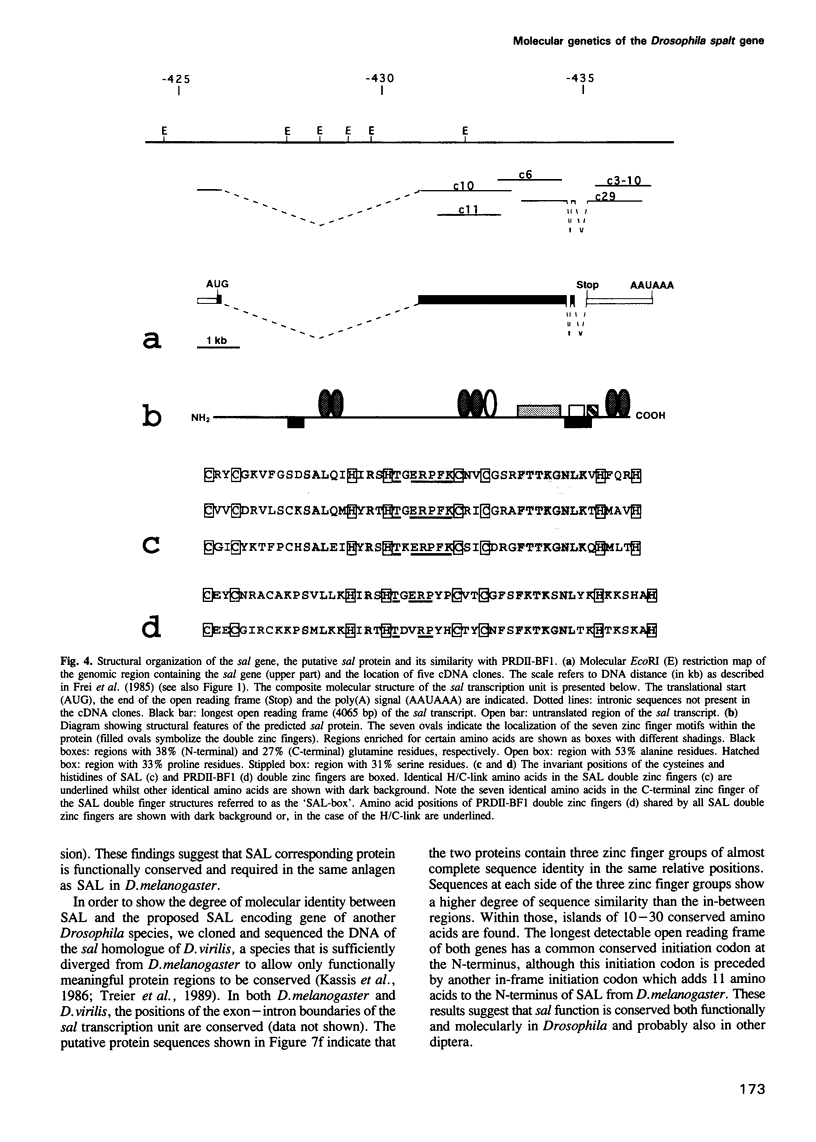
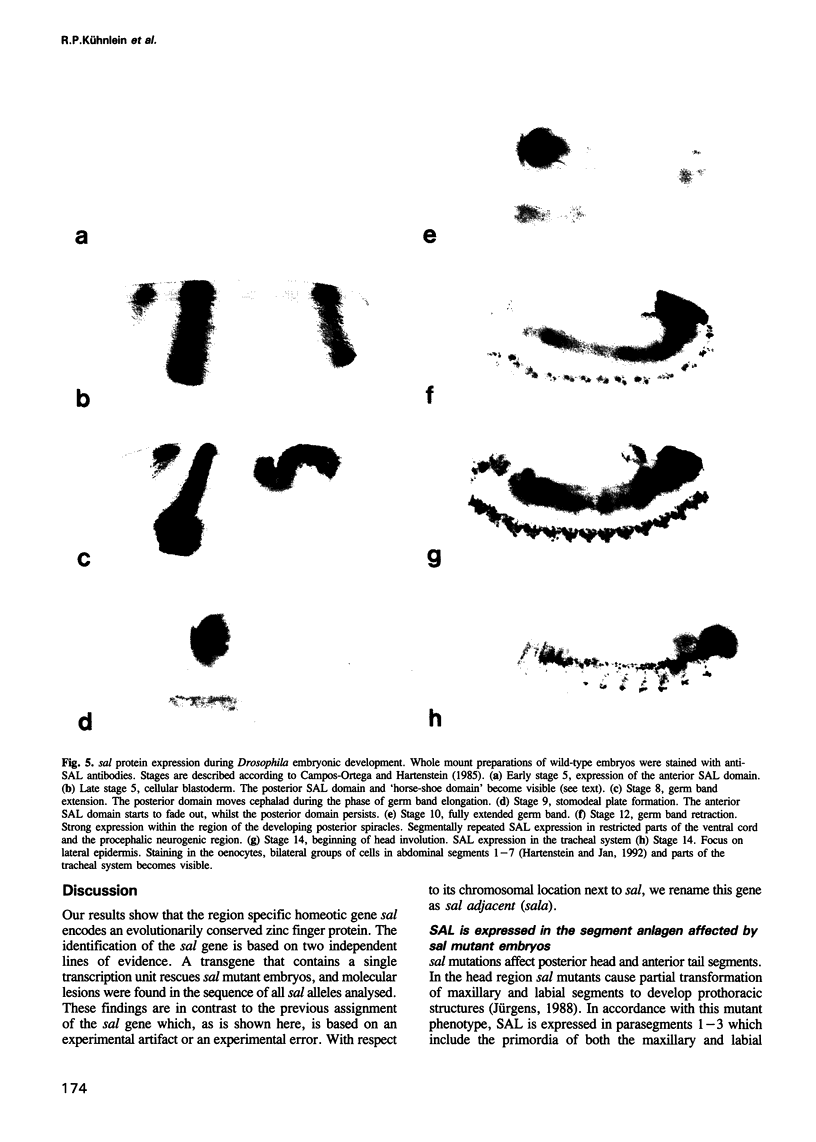
The region specific homeotic gene spalt (sal) of Drosophila melanogaster promotes the specification of terminal pattern elements as opposed to segments in the trunk. Our results show that the previously reported sal transcription unit was misidentified. Based on P-element mediated germ line transformation and DNA sequence analysis of sal mutant alleles, we identified the transcription unit that carries sal function. sal is located close to the misidentified transcription unit, and it is expressed in similar temporal and spatial patterns during embryogenesis. The sal gene encodes a zinc finger protein of novel structure composed of three widely spaced 'double zinc finger' motifs of internally conserved sequences and a single zinc finger motif of different sequence. Antibodies produced against the sal protein show that sal is first expressed at the blastoderm stage and later in restricted areas of the embryonic nervous system as well as in the developing trachea. The antibodies detect sal homologous proteins in corresponding spatial and temporal patterns in the embryos of related insect species. Sequence analysis of the sal gene of Drosophila virilis, a species which is phylogenetically separated by approximately 60 million years, suggests that the sal function is conserved during evolution, consistent with its proposed role in head formation during arthropod evolution.
Full text
PDF


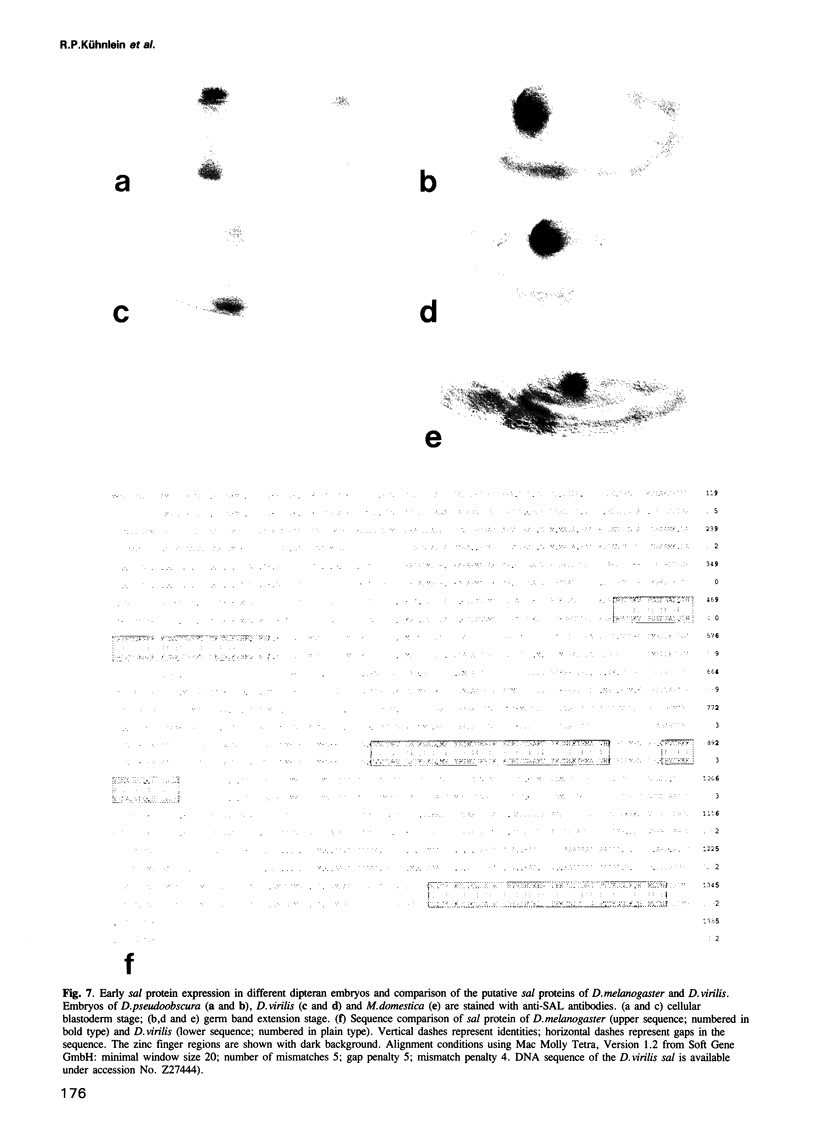



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Schier A., Gehring W. J. Homeodomain proteins and the regulation of gene expression. Curr Opin Cell Biol. 1990 Jun;2(3):485–495. doi: 10.1016/0955-0674(90)90132-x. [DOI] [PubMed] [Google Scholar]
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Bellen H. J., O'Kane C. J., Wilson C., Grossniklaus U., Pearson R. K., Gehring W. J. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989 Sep;3(9):1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- Berg J. M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988 Jan;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Hall J. C., Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 1988 Dec 1;7(12):3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Fan C. M., Maniatis T. A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev. 1990 Jan;4(1):29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- Frei E., Baumgartner S., Edström J. E., Noll M. Cloning of the extra sex combs gene of Drosophila and its identification by P-element-mediated gene transfer. EMBO J. 1985 Apr;4(4):979–987. doi: 10.1002/j.1460-2075.1985.tb03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E., Schuh R., Baumgartner S., Burri M., Noll M., Jürgens G., Seifert E., Nauber U., Jäckle H. Molecular characterization of spalt, a homeotic gene required for head and tail development in the Drosophila embryo. EMBO J. 1988 Jan;7(1):197–204. doi: 10.1002/j.1460-2075.1988.tb02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul U., Seifert E., Schuh R., Jäckle H. Analysis of Krüppel protein distribution during early Drosophila development reveals posttranscriptional regulation. Cell. 1987 Aug 14;50(4):639–647. doi: 10.1016/0092-8674(87)90037-7. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Postma J. P., Brown R. S., Argos P. A model for the tertiary structure of the 28 residue DNA-binding motif ('zinc finger') common to many eukaryotic transcriptional regulatory proteins. Protein Eng. 1988 Sep;2(3):209–218. doi: 10.1093/protein/2.3.209. [DOI] [PubMed] [Google Scholar]
- Han K., Manley J. L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993 Mar;7(3):491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 1988 Jan;7(1):189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Lewis R., Wakimoto B. Cytogenetic Analysis of Chromosome 3 in DROSOPHILA MELANOGASTER: The Homoeotic Gene Complex in Polytene Chromosome Interval 84a-B. Genetics. 1980 Jan;94(1):115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Licht J. D., Grossel M. J., Figge J., Hansen U. M. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990 Jul 5;346(6279):76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986 Dec 11;324(6097):537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Lawrence P. A. Parasegments and compartments in the Drosophila embryo. Nature. 1985 Feb 21;313(6004):639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Michael W. M., Bowtell D. D., Rubin G. M. Comparison of the sevenless genes of Drosophila virilis and Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5351–5353. doi: 10.1073/pnas.87.14.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Reuter D., Schuh R., Jäckle H. The homeotic gene spalt (sal) evolved during Drosophila speciation. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5483–5486. doi: 10.1073/pnas.86.14.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Röder L., Vola C., Kerridge S. The role of the teashirt gene in trunk segmental identity in Drosophila. Development. 1992 Aug;115(4):1017–1033. doi: 10.1242/dev.115.4.1017. [DOI] [PubMed] [Google Scholar]
- Rüther U., Müller-Hill B. Easy identification of cDNA clones. EMBO J. 1983;2(10):1791–1794. doi: 10.1002/j.1460-2075.1983.tb01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh R., Aicher W., Gaul U., Côté S., Preiss A., Maier D., Seifert E., Nauber U., Schröder C., Kemler R. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986 Dec 26;47(6):1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Sommer R., Tautz D. Segmentation gene expression in the housefly Musca domestica. Development. 1991 Oct;113(2):419–430. doi: 10.1242/dev.113.2.419. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Bernholz J. T., Wilson C., Gibson G., Schuh R., Gehring W. J. Identification of target genes of the homeotic gene Antennapedia by enhancer detection. Genes Dev. 1991 Dec;5(12B):2467–2480. doi: 10.1101/gad.5.12b.2467. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989 May 19;57(4):645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- el-Baradi T., Pieler T. Zinc finger proteins: what we know and what we would like to know. Mech Dev. 1991 Nov;35(3):155–169. doi: 10.1016/0925-4773(91)90015-x. [DOI] [PubMed] [Google Scholar]